

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0212.10
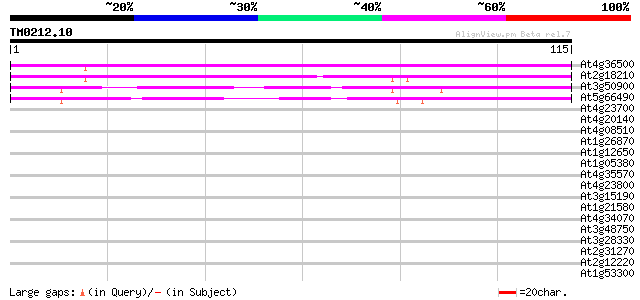
(115 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g36500 unknown protein 93 2e-20
At2g18210 unknown protein 84 1e-17
At3g50900 unknown protein 83 2e-17
At5g66490 unknown protein 76 3e-15
At4g23700 putative Na+/H+-exchanging protein 30 0.19
At4g20140 leucine rich repeat-like protein 27 1.6
At4g08510 hypothetical protein 27 2.1
At1g26870 putative NAM protein 27 2.1
At1g12650 unknown protein 27 2.1
At1g05380 unknown protein 27 2.1
At4g35570 HMG delta protein 26 2.8
At4g23800 98b like protein 26 2.8
At3g15190 30S ribosomal protein S20 26 2.8
At1g21580 hypothetical protein 26 2.8
At4g34070 putative protein 26 3.7
At3g48750 protein kinase (cdc2) 26 3.7
At3g28330 hypothetical protein 26 3.7
At2g31270 unknown protein 26 3.7
At2g12220 En/Spm-like transposon protein 26 3.7
At1g53300 unknown protein 26 3.7
>At4g36500 unknown protein
Length = 122
Score = 92.8 bits (229), Expect = 2e-20
Identities = 52/120 (43%), Positives = 69/120 (57%), Gaps = 5/120 (4%)
Query: 1 MLRSFTTRRYERLG-----KETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQ 55
MLRS +TR R G + ++ +LL +RSTS+P A S L
Sbjct: 3 MLRSLSTRTRSRRGGYERVSDDSTFSLLGAKLRRSTSVPYYAPSIKLGAGGVPTILEELP 62
Query: 56 RNPTKKANNSSKSTHPLFSFLDFRRKKKTTARPEFTRYLEYLKEGGMWDLNSNKPVIHYK 115
R +KK + K +HP+FS ++KK TT +PEF+RYLEYLKEGGMWD +N PVI+YK
Sbjct: 63 RQKSKKVKPTGKFSHPIFSLFYGKKKKSTTTKPEFSRYLEYLKEGGMWDARANAPVIYYK 122
>At2g18210 unknown protein
Length = 124
Score = 83.6 bits (205), Expect = 1e-17
Identities = 54/123 (43%), Positives = 72/123 (57%), Gaps = 9/123 (7%)
Query: 1 MLRSFTTRRYERLG-----KETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQ 55
MLR+ +TR R G + ++ +LL +RSTS+P A S +
Sbjct: 3 MLRNLSTRTRSRRGGYERVSDDSTFSLLGAKLRRSTSVPYYAPSIRLGGDFPVILEKLPR 62
Query: 56 RNPTKKANNSSKSTHPLFSFLD-FRR--KKKTTARPEFTRYLEYLKEGGMWDLNSNKPVI 112
+ PTK +SK +HP+FS D +RR KKK TA+PEF+RY EYLKE GMWDL SN PVI
Sbjct: 63 QKPTKTVV-TSKLSHPIFSLFDGYRRHNKKKATAKPEFSRYHEYLKESGMWDLRSNSPVI 121
Query: 113 HYK 115
++K
Sbjct: 122 YFK 124
>At3g50900 unknown protein
Length = 114
Score = 83.2 bits (204), Expect = 2e-17
Identities = 52/129 (40%), Positives = 71/129 (54%), Gaps = 29/129 (22%)
Query: 1 MLRSFTTRR----YERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR 56
M R+ +TR+ YE+LG E A KR +S+P+ +R +++
Sbjct: 1 MFRAMSTRKIHGGYEKLGDEEAR-------LKRVSSVPASVYGHSRNPVQ------EVKK 47
Query: 57 NPTKKANNSSKSTHPLFSFLD--FRRKKKTTAR--------PEFTRYLEYLKEGGMWDLN 106
PT K S HPLFSF D F+RKKK TA+ PEF RY+EY++EGG+WD +
Sbjct: 48 TPTAKPTGGS--VHPLFSFFDVHFQRKKKNTAKKKSLATAKPEFARYMEYVREGGVWDPS 105
Query: 107 SNKPVIHYK 115
SN PVIHY+
Sbjct: 106 SNAPVIHYR 114
>At5g66490 unknown protein
Length = 109
Score = 75.9 bits (185), Expect = 3e-15
Identities = 49/125 (39%), Positives = 70/125 (55%), Gaps = 26/125 (20%)
Query: 1 MLRSFTTRR----YERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR 56
M+R+ +TR+ Y++LG E + A L E + S+P+ + K+
Sbjct: 1 MIRALSTRKVRGSYKKLGDEEEAGAGLLE--VKPESVPTNPHGQSPKV-----------N 47
Query: 57 NPTKKANNSSKSTHPLFSFLDF---RRKKK---TTARPEFTRYLEYLKEGGMWDLNSNKP 110
P +K S+ HPLFSF + R+KKK TTA+PEF RYLEY+KEGG+WD SN P
Sbjct: 48 KPVEKTRGSA---HPLFSFFEMSLKRKKKKKSTTTAKPEFARYLEYVKEGGVWDNTSNGP 104
Query: 111 VIHYK 115
I+Y+
Sbjct: 105 AIYYR 109
>At4g23700 putative Na+/H+-exchanging protein
Length = 820
Score = 30.0 bits (66), Expect = 0.19
Identities = 20/50 (40%), Positives = 24/50 (48%)
Query: 39 SSARKMAHATFGNINLQRNPTKKANNSSKSTHPLFSFLDFRRKKKTTARP 88
SSA MAH N N K NNSS S + +F FRR + + RP
Sbjct: 498 SSAILMAHKVRRNGLPFWNKDKSENNSSSSDMVVVAFEAFRRLSRVSVRP 547
>At4g20140 leucine rich repeat-like protein
Length = 1232
Score = 26.9 bits (58), Expect = 1.6
Identities = 11/22 (50%), Positives = 16/22 (72%), Gaps = 1/22 (4%)
Query: 95 EYLKEGGMWD-LNSNKPVIHYK 115
EY+K G +WD L+ +KPV+ K
Sbjct: 1011 EYMKNGSIWDWLHEDKPVLEKK 1032
>At4g08510 hypothetical protein
Length = 551
Score = 26.6 bits (57), Expect = 2.1
Identities = 20/97 (20%), Positives = 37/97 (37%), Gaps = 6/97 (6%)
Query: 14 GKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSKSTHPLF 73
G SS L + + S + +R+ S A + + +N S+K + F
Sbjct: 19 GSGVGSSNSLSDSLRNSKNRNARSRSDADSVGSPFLDRSSSTNTRRGSSNGSTKHAYSSF 78
Query: 74 SFLDFRRKKKTTARPEFTRYLEYLKEGGMWDLNSNKP 110
+F R K + + Y++ WD +S+ P
Sbjct: 79 NFNRSNRDKDRSREKDRMSYMD------PWDNDSSMP 109
>At1g26870 putative NAM protein
Length = 425
Score = 26.6 bits (57), Expect = 2.1
Identities = 20/84 (23%), Positives = 35/84 (40%), Gaps = 2/84 (2%)
Query: 2 LRSFTTRRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKK 61
LR+ + L ET++ + + K+S + + + +H F + N+ T
Sbjct: 188 LRALSHSFVSSLPPETSTDTMSNQ--KQSNTYHFSSDKILKPSSHFQFHHENMNTPKTSN 245
Query: 62 ANNSSKSTHPLFSFLDFRRKKKTT 85
+ S T FS+LDF K T
Sbjct: 246 STTPSVPTISPFSYLDFTSYDKPT 269
>At1g12650 unknown protein
Length = 248
Score = 26.6 bits (57), Expect = 2.1
Identities = 14/30 (46%), Positives = 16/30 (52%)
Query: 64 NSSKSTHPLFSFLDFRRKKKTTARPEFTRY 93
NS K + L S+LD RRKK T F Y
Sbjct: 214 NSLKESGKLTSYLDKRRKKNATKDHRFMPY 243
>At1g05380 unknown protein
Length = 1138
Score = 26.6 bits (57), Expect = 2.1
Identities = 14/64 (21%), Positives = 31/64 (47%)
Query: 20 SALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSKSTHPLFSFLDFR 79
+ +++ F++S ++ + + S M+ + + P K+ N SKST P D R
Sbjct: 240 NVVVKPPFRKSNNVDNNSESEESDMSRKSKRKKSEYSKPKKEFNTKSKSTFPELVNPDVR 299
Query: 80 RKKK 83
+++
Sbjct: 300 EERR 303
>At4g35570 HMG delta protein
Length = 125
Score = 26.2 bits (56), Expect = 2.8
Identities = 13/31 (41%), Positives = 15/31 (47%)
Query: 59 TKKANNSSKSTHPLFSFLDFRRKKKTTARPE 89
TK N K P F FLD RK+ A P+
Sbjct: 28 TKDPNRPKKPPSPFFVFLDDFRKEFNLANPD 58
>At4g23800 98b like protein
Length = 456
Score = 26.2 bits (56), Expect = 2.8
Identities = 16/61 (26%), Positives = 27/61 (44%), Gaps = 1/61 (1%)
Query: 56 RNPTKKANNSSKSTHPLFSFLDFRRKKKTTARPEFTRYLEYLK-EGGMWDLNSNKPVIHY 114
+ KK + K HP+ +FL + +++ R E +E K G W S+K Y
Sbjct: 243 KKKNKKEKDPLKPKHPVSAFLVYANERRAALREENKSVVEVAKITGEEWKNLSDKKKAPY 302
Query: 115 K 115
+
Sbjct: 303 E 303
>At3g15190 30S ribosomal protein S20
Length = 202
Score = 26.2 bits (56), Expect = 2.8
Identities = 13/41 (31%), Positives = 22/41 (52%)
Query: 58 PTKKANNSSKSTHPLFSFLDFRRKKKTTARPEFTRYLEYLK 98
PTKKA++++K + + KK+ AR + LE L+
Sbjct: 83 PTKKADSAAKRARQAEKRRVYNKSKKSEARTRMKKVLEALE 123
>At1g21580 hypothetical protein
Length = 1696
Score = 26.2 bits (56), Expect = 2.8
Identities = 12/42 (28%), Positives = 21/42 (49%)
Query: 7 TRRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHAT 48
T R +LG + + EGF+RST + + + +H+T
Sbjct: 432 TERNGKLGTHLSDEISVSEGFRRSTRQTTASKNEKEPDSHST 473
>At4g34070 putative protein
Length = 324
Score = 25.8 bits (55), Expect = 3.7
Identities = 17/72 (23%), Positives = 30/72 (41%), Gaps = 4/72 (5%)
Query: 3 RSFTTRRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKA 62
R F E+ T +++ K S+ + + K A G + ++NP KK
Sbjct: 75 RKFQLSVTEKKDASTKLYQIIRASNKSGRSIKNNRRTETTKPA----GELRGEKNPAKKG 130
Query: 63 NNSSKSTHPLFS 74
N++S S F+
Sbjct: 131 NSNSSSVDHCFT 142
>At3g48750 protein kinase (cdc2)
Length = 294
Score = 25.8 bits (55), Expect = 3.7
Identities = 11/35 (31%), Positives = 20/35 (56%)
Query: 64 NSSKSTHPLFSFLDFRRKKKTTARPEFTRYLEYLK 98
+S K + +F +LD KK + P+F++ L +K
Sbjct: 71 HSEKRLYLVFEYLDLDLKKHMDSTPDFSKDLHMIK 105
>At3g28330 hypothetical protein
Length = 349
Score = 25.8 bits (55), Expect = 3.7
Identities = 12/27 (44%), Positives = 16/27 (58%), Gaps = 1/27 (3%)
Query: 82 KKTTARPEFTRYLEYLKEGGMWDLNSN 108
K +P FT +L Y E G+W LNS+
Sbjct: 122 KVVLLKPNFT-FLIYSSETGLWSLNSD 147
>At2g31270 unknown protein
Length = 571
Score = 25.8 bits (55), Expect = 3.7
Identities = 19/62 (30%), Positives = 28/62 (44%), Gaps = 3/62 (4%)
Query: 8 RRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSK 67
+R L +E AS + TS P + +SS +A T IN+ PTK + +K
Sbjct: 259 KRVSSLMEEMASIPASKLFSSPITSTPVKTTSS---LAKPTSSQINIAPTPTKPTSTPAK 315
Query: 68 ST 69
T
Sbjct: 316 QT 317
>At2g12220 En/Spm-like transposon protein
Length = 781
Score = 25.8 bits (55), Expect = 3.7
Identities = 24/103 (23%), Positives = 36/103 (34%), Gaps = 15/103 (14%)
Query: 14 GKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKA--NNSSKSTHP 71
G + SSA + +STS S AS S + +A + Q+ K N +
Sbjct: 642 GSASKSSAGAKSAASKSTSSKSSASGSETPLENANAPSSTNQKTLQKSQCFNQGVSKENM 701
Query: 72 LFSFLDFRRKKKTTARPEFTRYLEYLKEGGMWDLNSNKPVIHY 114
+D KK+ A G W N K +H+
Sbjct: 702 KCKLMDITGKKRVVAE-------------GRWATNDPKQKVHF 731
>At1g53300 unknown protein
Length = 699
Score = 25.8 bits (55), Expect = 3.7
Identities = 12/37 (32%), Positives = 20/37 (53%)
Query: 38 SSSARKMAHATFGNINLQRNPTKKANNSSKSTHPLFS 74
S+S R ++++ GN NL+ +SS + PL S
Sbjct: 84 SNSVRSQSNSSSGNNNLRPRSDSATTSSSSHSQPLLS 120
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,456,084
Number of Sequences: 26719
Number of extensions: 88445
Number of successful extensions: 314
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 293
Number of HSP's gapped (non-prelim): 32
length of query: 115
length of database: 11,318,596
effective HSP length: 91
effective length of query: 24
effective length of database: 8,887,167
effective search space: 213292008
effective search space used: 213292008
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0212.10


