

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0211.20
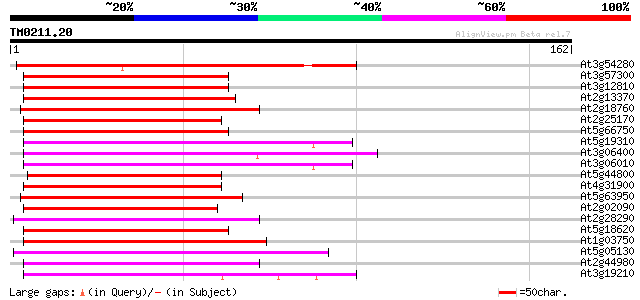
(162 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g54280 TATA box binding protein (TBP) associated factor (TAF)... 111 2e-25
At3g57300 helicase-like protein 69 1e-12
At3g12810 Snf2-related CBP activator protein, putative 62 2e-10
At2g13370 pseudogene 62 2e-10
At2g18760 putative SNF2/RAD54 family DNA repair and recombinatio... 59 1e-09
At2g25170 putative chromodomain-helicase-DNA-binding protein 58 3e-09
At5g66750 SWI2/SNF2-like protein (gb|AAD28303.1) 57 4e-09
At5g19310 homeotic gene regulator - like protein 57 6e-09
At3g06400 putative ATPase (ISW2-like) 56 1e-08
At3g06010 putative transcriptional regulator 55 2e-08
At5g44800 helicase-like protein 55 2e-08
At4g31900 putative protein 55 2e-08
At5g63950 unknown protein 54 3e-08
At2g02090 putative helicase 54 3e-08
At2g28290 putative SNF2 subfamily transcription regulator 54 4e-08
At5g18620 chromatin remodelling complex ATPase chain ISWI -like ... 51 3e-07
At1g03750 hypothetical protein 51 3e-07
At5g05130 helicase-like transcription factor-like protein 50 7e-07
At2g44980 putative transcription activator 49 9e-07
At3g19210 DNA repair protein, putative 49 1e-06
>At3g54280 TATA box binding protein (TBP) associated factor (TAF)
-like protein
Length = 2049
Score = 111 bits (277), Expect = 2e-25
Identities = 61/109 (55%), Positives = 77/109 (69%), Gaps = 13/109 (11%)
Query: 3 HVGGFGMNLTSADTLVFVEHDRNSMRDHQ-----------AMDKARRLGQEKVVNVHRLI 51
HVGG G+NLTSADTLVF+EHD N MRDHQ AMD+A RLGQ++VVNVHRLI
Sbjct: 1886 HVGGLGLNLTSADTLVFMEHDWNPMRDHQFANIELNKLWQAMDRAHRLGQKRVVNVHRLI 1945
Query: 52 MRDTLEEQVMSKERFEVSVANAANNLRLLSVKSSVDHSDGVTELVGSGD 100
MR TLEE+VMS ++F+VSVAN N S+K+ ++D + +L S +
Sbjct: 1946 MRGTLEEKVMSLQKFKVSVANTVINAENASMKTM--NTDQLLDLFASAE 1992
>At3g57300 helicase-like protein
Length = 1507
Score = 68.9 bits (167), Expect = 1e-12
Identities = 32/59 (54%), Positives = 45/59 (76%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSK 63
GG G+NLT+ADT++F E D N D QAMD+A RLGQ K V V+RLI ++T+EE+++ +
Sbjct: 1284 GGLGINLTAADTVIFYESDWNPTLDLQAMDRAHRLGQTKDVTVYRLICKETVEEKILHR 1342
>At3g12810 Snf2-related CBP activator protein, putative
Length = 1048
Score = 61.6 bits (148), Expect = 2e-10
Identities = 26/59 (44%), Positives = 39/59 (66%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSK 63
GG G+NL ADT++F + D N D QA D+ R+GQ + V+++RLI T+EE ++ K
Sbjct: 738 GGVGINLVGADTVIFYDSDWNPAMDQQAQDRCHRIGQTREVHIYRLISESTIEENILKK 796
>At2g13370 pseudogene
Length = 1738
Score = 61.6 bits (148), Expect = 2e-10
Identities = 25/61 (40%), Positives = 43/61 (69%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT+V + D N D QAM +A R+GQ++VVN++R + ++EE+++ +
Sbjct: 1032 GGLGINLATADTVVIFDSDWNPQNDLQAMSRAHRIGQQEVVNIYRFVTSKSVEEEILERA 1091
Query: 65 R 65
+
Sbjct: 1092 K 1092
>At2g18760 putative SNF2/RAD54 family DNA repair and recombination
protein
Length = 1187
Score = 58.9 bits (141), Expect = 1e-09
Identities = 29/69 (42%), Positives = 45/69 (65%)
Query: 4 VGGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSK 63
VGG G NLT A+ ++ + D N D QA ++A R+GQ+K V V+RLI R T+EE+V +
Sbjct: 804 VGGLGTNLTGANRVIIFDPDWNPSNDMQARERAWRIGQKKDVTVYRLITRGTIEEKVYHR 863
Query: 64 ERFEVSVAN 72
+ ++ + N
Sbjct: 864 QIYKHFLTN 872
>At2g25170 putative chromodomain-helicase-DNA-binding protein
Length = 1359
Score = 57.8 bits (138), Expect = 3e-09
Identities = 28/57 (49%), Positives = 39/57 (68%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVM 61
GG G+NL +ADT++ + D N D QAM +A RLGQ V ++RLI R T+EE++M
Sbjct: 674 GGLGINLATADTVIIYDSDWNPHADLQAMARAHRLGQTNKVMIYRLINRGTIEERMM 730
>At5g66750 SWI2/SNF2-like protein (gb|AAD28303.1)
Length = 764
Score = 57.0 bits (136), Expect = 4e-09
Identities = 26/59 (44%), Positives = 39/59 (66%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSK 63
GG G+NLT+ADT + + D N D QAMD+ R+GQ K V+V+RL ++E +V+ +
Sbjct: 603 GGLGINLTAADTCILYDSDWNPQMDLQAMDRCHRIGQTKPVHVYRLSTAQSIETRVLKR 661
>At5g19310 homeotic gene regulator - like protein
Length = 1041
Score = 56.6 bits (135), Expect = 6e-09
Identities = 29/96 (30%), Positives = 53/96 (55%), Gaps = 1/96 (1%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT++ + D N D QA D+A R+GQ+K V V L+ ++EE ++ +
Sbjct: 751 GGLGLNLQTADTIIIFDSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVSIGSIEEVILERA 810
Query: 65 RFEVSVANAANNLRLLSVKSSV-DHSDGVTELVGSG 99
+ ++ + L + S+ D + + E++ G
Sbjct: 811 KQKMGIDAKVIQAGLFNTTSTAQDRREMLEEIMSKG 846
>At3g06400 putative ATPase (ISW2-like)
Length = 1057
Score = 55.8 bits (133), Expect = 1e-08
Identities = 32/103 (31%), Positives = 55/103 (53%), Gaps = 1/103 (0%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +AD ++ + D N D QA D+A R+GQ+K V V R +EE+V+ +
Sbjct: 569 GGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVFRFCTESAIEEKVIERA 628
Query: 65 RFEVSV-ANAANNLRLLSVKSSVDHSDGVTELVGSGDWWIYET 106
++++ A RL KS + D + ++V G ++ +
Sbjct: 629 YKKLALDALVIQQGRLAEQKSKSVNKDELLQMVRYGAEMVFSS 671
>At3g06010 putative transcriptional regulator
Length = 1132
Score = 55.1 bits (131), Expect = 2e-08
Identities = 29/96 (30%), Positives = 53/96 (55%), Gaps = 1/96 (1%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT++ + D N D QA D+A R+GQ+K V V L+ ++EE ++ +
Sbjct: 822 GGLGLNLQTADTVIIFDSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVSVGSVEEVILERA 881
Query: 65 RFEVSVANAANNLRLLSVKSSV-DHSDGVTELVGSG 99
+ ++ + L + S+ D + + E++ G
Sbjct: 882 KQKMGIDAKVIQAGLFNTTSTAQDRREMLEEIMRKG 917
>At5g44800 helicase-like protein
Length = 2228
Score = 54.7 bits (130), Expect = 2e-08
Identities = 24/56 (42%), Positives = 41/56 (72%)
Query: 6 GFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVM 61
G G+NL +ADT++ + D N D QAM++A R+GQ K + V+RL++R ++EE+++
Sbjct: 1071 GLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSKRLLVYRLVVRASVEERIL 1126
>At4g31900 putative protein
Length = 1067
Score = 54.7 bits (130), Expect = 2e-08
Identities = 26/57 (45%), Positives = 38/57 (66%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVM 61
GG G+NL +ADT++ + D N D QAM + RLGQ V ++RLI + T+EE++M
Sbjct: 493 GGIGINLATADTVIIYDSDWNPHADLQAMARVHRLGQTNKVMIYRLIHKGTVEERMM 549
>At5g63950 unknown protein
Length = 1090
Score = 54.3 bits (129), Expect = 3e-08
Identities = 26/64 (40%), Positives = 43/64 (66%)
Query: 4 VGGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSK 63
VGG G+ LT AD ++ V+ N D+Q++D+A R+GQ K V V+RL+ T+EE++ K
Sbjct: 809 VGGLGLTLTKADRVIVVDPAWNPSTDNQSVDRAYRIGQTKDVIVYRLMTSATVEEKIYRK 868
Query: 64 ERFE 67
+ ++
Sbjct: 869 QVYK 872
>At2g02090 putative helicase
Length = 763
Score = 54.3 bits (129), Expect = 3e-08
Identities = 24/56 (42%), Positives = 36/56 (63%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQV 60
GG G+NLT ADT++ + D N D QA D+ R+GQ K V + RL+ + T++E +
Sbjct: 666 GGQGLNLTGADTVIIHDMDFNPQIDRQAEDRCHRIGQTKPVTIFRLVTKSTVDENI 721
>At2g28290 putative SNF2 subfamily transcription regulator
Length = 1331
Score = 53.9 bits (128), Expect = 4e-08
Identities = 29/71 (40%), Positives = 43/71 (59%)
Query: 2 IHVGGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVM 61
I GG G+NL +ADT++ + D N D QA +A R+GQ+K V V R +++EEQV
Sbjct: 1178 IRAGGVGVNLQAADTVILFDTDWNPQVDLQAQARAHRIGQKKDVLVLRFETVNSVEEQVR 1237
Query: 62 SKERFEVSVAN 72
+ ++ VAN
Sbjct: 1238 ASAEHKLGVAN 1248
>At5g18620 chromatin remodelling complex ATPase chain ISWI -like
protein
Length = 1069
Score = 50.8 bits (120), Expect = 3e-07
Identities = 23/59 (38%), Positives = 36/59 (60%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSK 63
GG G+NL +AD ++ + D N D QA D+A R+GQ+K V V R + +E +V+ +
Sbjct: 574 GGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVFRFCTENAIEAKVIER 632
>At1g03750 hypothetical protein
Length = 874
Score = 50.8 bits (120), Expect = 3e-07
Identities = 26/70 (37%), Positives = 44/70 (62%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL SA+ +V + + N D QA D++ R GQ++ V V RL+ +LEE V +++
Sbjct: 618 GGLGLNLVSANRVVIFDPNWNPSHDLQAQDRSFRYGQKRHVVVFRLLSAGSLEELVYTRQ 677
Query: 65 RFEVSVANAA 74
++ ++N A
Sbjct: 678 VYKQQLSNIA 687
>At5g05130 helicase-like transcription factor-like protein
Length = 862
Score = 49.7 bits (117), Expect = 7e-07
Identities = 28/91 (30%), Positives = 52/91 (56%)
Query: 2 IHVGGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVM 61
+ G G+NLT+A + + N + QAMD+ R+GQ++ V + R+I R+++EE+V+
Sbjct: 767 LKASGTGINLTAASRVYLFDPWWNPAVEEQAMDRIHRIGQKQEVKMIRMIARNSIEERVL 826
Query: 62 SKERFEVSVANAANNLRLLSVKSSVDHSDGV 92
++ + ++AN A R + V+ D V
Sbjct: 827 ELQQKKKNLANEAFKRRQKKDEREVNVEDVV 857
>At2g44980 putative transcription activator
Length = 861
Score = 49.3 bits (116), Expect = 9e-07
Identities = 23/68 (33%), Positives = 41/68 (59%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT++F E D N D QA+ +A R+GQ V L+ ++EE ++ +
Sbjct: 457 GGVGLNLVAADTVIFYEQDWNPQVDKQALQRAHRIGQISHVLSINLVTEHSVEEVILRRA 516
Query: 65 RFEVSVAN 72
++ +++
Sbjct: 517 ERKLQLSH 524
>At3g19210 DNA repair protein, putative
Length = 688
Score = 48.9 bits (115), Expect = 1e-06
Identities = 36/108 (33%), Positives = 58/108 (53%), Gaps = 12/108 (11%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQV---- 60
GG G+NL A+ LV + D N D QA + R GQ+K V V+R + T+EE+V
Sbjct: 510 GGCGLNLIGANRLVLFDPDWNPANDKQAAARVWRDGQKKRVYVYRFLSTGTIEEKVYQRQ 569
Query: 61 MSKERFEVSVANAANN------LRLLSVKSSVD--HSDGVTELVGSGD 100
MSKE + + + + L + SS++ HS+G+++ + SG+
Sbjct: 570 MSKEGLQKVIQHEQTDNSTRQVLAGMLYASSINDTHSNGLSQSLFSGN 617
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.139 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,239,236
Number of Sequences: 26719
Number of extensions: 115263
Number of successful extensions: 459
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 409
Number of HSP's gapped (non-prelim): 52
length of query: 162
length of database: 11,318,596
effective HSP length: 91
effective length of query: 71
effective length of database: 8,887,167
effective search space: 630988857
effective search space used: 630988857
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0211.20


