

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0211.14
(280 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
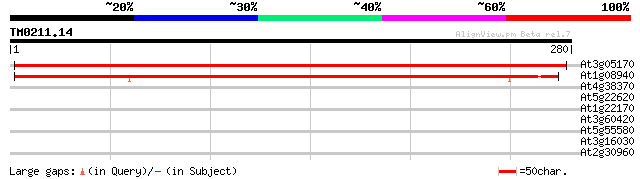
Score E
Sequences producing significant alignments: (bits) Value
At3g05170 unknown protein 422 e-118
At1g08940 unknown protein 390 e-109
At4g38370 unknown protein 36 0.026
At5g22620 phosphoglycerate mutase like protein 34 0.100
At1g22170 phosphoglycerate mutase like protein 34 0.100
At3g60420 unknown protein 33 0.13
At5g55580 Unknown protein (At5g55580) 29 3.2
At3g16030 putative receptor kinase 28 4.2
At2g30960 hypothetical protein 28 7.2
>At3g05170 unknown protein
Length = 316
Score = 422 bits (1084), Expect = e-118
Identities = 194/276 (70%), Positives = 232/276 (83%)
Query: 3 VLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPD 62
+LPKRIIL+RHGES+GNLDT+AYTTTPDH IQLT G+ QA+ AG RL +++++ SP+
Sbjct: 7 LLPKRIILVRHGESEGNLDTAAYTTTPDHKIQLTDSGLLQAQEAGARLHALISSNPSSPE 66
Query: 63 WRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRE 122
WRV FYVSPY RTRSTLRE+GRSFS++RVIGVREE RIREQDFGNFQV+ERM+ K+ RE
Sbjct: 67 WRVYFYVSPYDRTRSTLREIGRSFSRRRVIGVREECRIREQDFGNFQVKERMRATKKVRE 126
Query: 123 RFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIF 182
RFGRFFYRFPEGESAADV+DR+S F E+LWRDID+NRLH +PS++LN VIVSHGLTSR+F
Sbjct: 127 RFGRFFYRFPEGESAADVFDRVSSFLESLWRDIDMNRLHINPSHELNFVIVSHGLTSRVF 186
Query: 183 LMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPEMVADQKW 242
LMKWFKW+VEQFE LNN GN E RVME G GG+YSLA+HHTEEE+ WGLSPEM+ADQKW
Sbjct: 187 LMKWFKWSVEQFEGLNNPGNSEIRVMELGQGGDYSLAIHHTEEELATWGLSPEMIADQKW 246
Query: 243 RAGASRGAWNDQCSWYLDGFFDRLAEESDDEEASSS 278
RA A +G W + C WY FFD +A+ + E ++
Sbjct: 247 RANAHKGEWKEDCKWYFGDFFDHMADSDKECETEAT 282
>At1g08940 unknown protein
Length = 281
Score = 390 bits (1003), Expect = e-109
Identities = 186/276 (67%), Positives = 224/276 (80%), Gaps = 5/276 (1%)
Query: 3 VLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADG---C 59
+LPKRIILMRHGES GN+D AY TTPDH I LT +G +QAR AG ++R +++ C
Sbjct: 7 MLPKRIILMRHGESAGNIDAGAYATTPDHKIPLTEEGRAQAREAGKKMRALISTQSGGAC 66
Query: 60 SPDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQ 119
+WRV FYVSPY RTR+TLREVG+ FS+KRVIGVREE RIREQDFGNFQVEERM+++K+
Sbjct: 67 GENWRVYFYVSPYERTRTTLREVGKGFSRKRVIGVREECRIREQDFGNFQVEERMRVVKE 126
Query: 120 TRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTS 179
TRERFGRFFYRFPEGESAADVYDR+S F E++WRD+D+NR DPS++LNLVIVSHGLTS
Sbjct: 127 TRERFGRFFYRFPEGESAADVYDRVSSFLESMWRDVDMNRHQVDPSSELNLVIVSHGLTS 186
Query: 180 RIFLMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPEMVAD 239
R+FL KWFKWTV +FE LNNFGNCEFRVME G+ GEY+ A+HH+EEEML WG+S +M+ D
Sbjct: 187 RVFLTKWFKWTVAEFERLNNFGNCEFRVMELGASGEYTFAIHHSEEEMLDWGMSKDMIDD 246
Query: 240 QKWRAGASR-GAWNDQCSWYLDGFFDRLAEESDDEE 274
QK R R ND CS +L+ +FD L + +DDEE
Sbjct: 247 QKDRVDGCRVTTSNDSCSLHLNEYFD-LLDVTDDEE 281
>At4g38370 unknown protein
Length = 225
Score = 35.8 bits (81), Expect = 0.026
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 4/81 (4%)
Query: 7 RIILMRHGESQGN---LDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDW 63
R ++RHG+S N L S+ QL P G++QAR AG + + D
Sbjct: 11 RYWVLRHGKSIPNERGLVVSSMENGVLPEYQLAPDGVAQARLAGESFLQQLKESNIELD- 69
Query: 64 RVNFYVSPYARTRSTLREVGR 84
+V SP++RT T R V +
Sbjct: 70 KVRICYSPFSRTTHTARVVAK 90
>At5g22620 phosphoglycerate mutase like protein
Length = 482
Score = 33.9 bits (76), Expect = 0.100
Identities = 36/128 (28%), Positives = 60/128 (46%), Gaps = 19/128 (14%)
Query: 6 KRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWRV 65
KR++L+RHG+S N + + D S+ LT +G SQA + R M D D
Sbjct: 48 KRVVLVRHGQSTWN-EEGRIQGSSDFSV-LTKKGESQA-----EISRQMLID----DSFD 96
Query: 66 NFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERFG 125
+ SP R++ T + S + + + +RE D +FQ + K+ +E+FG
Sbjct: 97 VCFTSPLKRSKKTAEIIWGSRESEMIF----DYDLREIDLYSFQ----GLLKKEGKEKFG 148
Query: 126 RFFYRFPE 133
F ++ E
Sbjct: 149 EAFKQWQE 156
>At1g22170 phosphoglycerate mutase like protein
Length = 334
Score = 33.9 bits (76), Expect = 0.100
Identities = 46/212 (21%), Positives = 87/212 (40%), Gaps = 41/212 (19%)
Query: 8 IILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRV-------------- 53
+IL+RHGES N + + +T D + LT +G+ +A AG R+ +
Sbjct: 80 LILIRHGESLWN-EKNLFTGCVD--VPLTEKGVEEAIEAGKRISNIPVDVIFTSSLIRAQ 136
Query: 54 ---MAADGCSPDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQV 110
M A +V + + T +V +K + I V ++ E+ +G Q
Sbjct: 137 MTAMLAMIQHRRKKVPIILHDESEQAKTWSQVFSDETKNQSIPVIPAWQLNERMYGELQG 196
Query: 111 EERMKIIKQTRERFGR---------FFYRFPEGESAADVYDRISGFFEALWRDIDLNRLH 161
+ ++T ER+G+ + P+GES +R +F+ + +
Sbjct: 197 LNK----QETAERYGKEQVHEWRRSYDIPPPKGESLEMCAERAVAYFQ--------DNIE 244
Query: 162 HDPSNDLNLVIVSHGLTSRIFLMKWFKWTVEQ 193
+ N++I +HG + R +M K T ++
Sbjct: 245 PKLAAGKNVMIAAHGNSLRSIIMYLDKLTCQE 276
>At3g60420 unknown protein
Length = 270
Score = 33.5 bits (75), Expect = 0.13
Identities = 38/153 (24%), Positives = 61/153 (39%), Gaps = 24/153 (15%)
Query: 6 KRIILMRHGESQGNLDTS-AYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
+ +ILMRHG+ N + T L G +A R G R+R + P R
Sbjct: 12 QNVILMRHGDRLDNFEPLWTSTAARPWDPPLAQDGKDRAFRTGQRIRSQLGV----PIHR 67
Query: 65 VNFYVSPYARTRSTLREVGR----------SFSKKRVIGVREESRIREQDFGNFQVEERM 114
V +VSP+ R T EV + S K V+ + +FG ++ +
Sbjct: 68 V--FVSPFLRCIQTASEVVAALSAVDFDPIAMSSKDVLSIDNTKIKVAIEFGLSEIPHPI 125
Query: 115 KIIKQTRERFGRFFYR-------FPEGESAADV 140
I + + G+F ++ FPEG ++V
Sbjct: 126 FIKSEVAPKDGKFDFKISDLEAMFPEGTVDSNV 158
>At5g55580 Unknown protein (At5g55580)
Length = 496
Score = 28.9 bits (63), Expect = 3.2
Identities = 15/49 (30%), Positives = 25/49 (50%)
Query: 223 TEEEMLQWGLSPEMVADQKWRAGASRGAWNDQCSWYLDGFFDRLAEESD 271
T+E+ L G+SP +++K + G Y++ + RLAEE D
Sbjct: 131 TDEDDLDLGISPNATSEKKKESWRLDGRGKMSSRKYVEKLYPRLAEEID 179
>At3g16030 putative receptor kinase
Length = 850
Score = 28.5 bits (62), Expect = 4.2
Identities = 29/128 (22%), Positives = 49/128 (37%), Gaps = 23/128 (17%)
Query: 62 DWRVNFYVSP-------YARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERM 114
DW++ F + Y S L+ + R ++ + E+ + DFG ++
Sbjct: 622 DWKLRFRIMEGIIQGLLYLHKYSRLKVIHRDIKAGNIL-LDEDMNPKISDFGMARIFGAQ 680
Query: 115 KIIKQTRERFGRFFYRFPEG------ESAADVYDRISGFFEALWRDIDLNR----LHHDP 164
+ T+ G F Y PE + +DV+ F L +I R HHD
Sbjct: 681 ESKANTKRVAGTFGYMSPEYFREGLFSAKSDVFS-----FGVLMLEIICGRKNNSFHHDS 735
Query: 165 SNDLNLVI 172
LNL++
Sbjct: 736 EGPLNLIV 743
>At2g30960 hypothetical protein
Length = 260
Score = 27.7 bits (60), Expect = 7.2
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query: 88 KKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERFGR 126
++R + EE +I E+D + E+RM + R+ FGR
Sbjct: 47 RRRKLQAEEEKKIEEEDLKKAEEEKRMN--RSNRKHFGR 83
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,704,212
Number of Sequences: 26719
Number of extensions: 285663
Number of successful extensions: 647
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 642
Number of HSP's gapped (non-prelim): 9
length of query: 280
length of database: 11,318,596
effective HSP length: 98
effective length of query: 182
effective length of database: 8,700,134
effective search space: 1583424388
effective search space used: 1583424388
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0211.14


