

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0207.9
(758 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
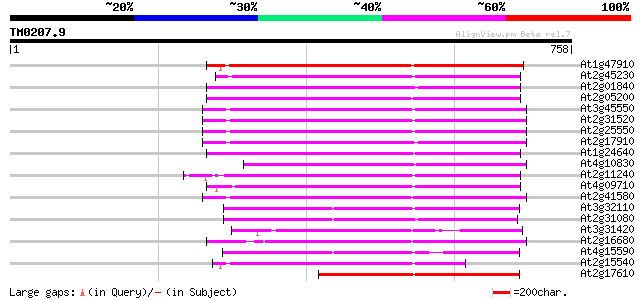
Score E
Sequences producing significant alignments: (bits) Value
At1g47910 reverse transcriptase, putative 345 5e-95
At2g45230 putative non-LTR retroelement reverse transcriptase 307 1e-83
At2g01840 putative non-LTR retroelement reverse transcriptase 307 2e-83
At2g05200 putative non-LTR retroelement reverse transcriptase 294 1e-79
At3g45550 putative protein 291 1e-78
At2g31520 putative non-LTR retroelement reverse transcriptase 290 3e-78
At2g25550 putative non-LTR retroelement reverse transcriptase 289 4e-78
At2g17910 putative non-LTR retroelement reverse transcriptase 288 6e-78
At1g24640 hypothetical protein 283 3e-76
At4g10830 putative protein 282 6e-76
At2g11240 pseudogene 281 7e-76
At4g09710 RNA-directed DNA polymerase -like protein 278 6e-75
At2g41580 putative non-LTR retroelement reverse transcriptase 274 1e-73
At3g32110 non-LTR reverse transcriptase, putative 259 5e-69
At2g31080 putative non-LTR retroelement reverse transcriptase 254 1e-67
At3g31420 hypothetical protein 252 6e-67
At2g16680 putative non-LTR retroelement reverse transcriptase 252 6e-67
At4g15590 reverse transcriptase like protein 249 5e-66
At2g15540 putative non-LTR retroelement reverse transcriptase 241 1e-63
At2g17610 putative non-LTR retroelement reverse transcriptase 227 2e-59
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 345 bits (885), Expect = 5e-95
Identities = 185/433 (42%), Positives = 267/433 (60%), Gaps = 10/433 (2%)
Query: 266 FSTLFTTSNPMGIEAATS----LVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGL 321
F LFTT+NP + A L+ +R+N +L + +VR LF +HP KA G
Sbjct: 177 FQNLFTTANPQVFDEALGEVQVLITDRIND----LLTADATECEVRAALFMIHPEKAPGP 232
Query: 322 DGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPIS 381
DG+ LF+QK W I+ D+ S LQ V + T + LIPK ++ T RPIS
Sbjct: 233 DGMTALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPIS 292
Query: 382 LCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCN 441
LCNV +K+I+ L +LK VLP++IS QSAFV GRL++DN L+A E FH ++ +
Sbjct: 293 LCNVGYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKD 352
Query: 442 GIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFD 501
MA+K DMSKAYD+VEW F+ + L KMGF W+S+IM +TTV ++V++NG P+
Sbjct: 353 KFMAIKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKGLII 412
Query: 502 PGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNV 561
P RGLRQGDPLSPYLF++C EV A I++ + ++GIK+A +P +SHLLFADD++
Sbjct: 413 PERGLRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVA--TPSPAVSHLLFADDSL 470
Query: 562 IFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDN 621
F +A +E+ + IL +Y+ V Q INF KS + + V ++K++LG+ +
Sbjct: 471 FFCKANKEQCGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSIKADIKLILGIHNLGG 530
Query: 622 YDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMS 681
YLGLP +G SKT++F FV++R ++ G LS+ G+EV+IK+VA +P Y+MS
Sbjct: 531 MGSYLGLPESLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVMS 590
Query: 682 CFILPDNLCADST 694
CF LP + + T
Sbjct: 591 CFRLPKAITSKLT 603
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 307 bits (787), Expect = 1e-83
Identities = 164/413 (39%), Positives = 244/413 (58%), Gaps = 6/413 (1%)
Query: 278 IEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFYQKFWHIVG 337
+E T LV++++N LA P + E+V+ F ++P K G DG++ YQ+FW +G
Sbjct: 409 LENLTPLVSDQMNNNLLA----PITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMG 464
Query: 338 DDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANK 397
D + + + + T + LIPKI K+ T FRPISLCNV +K+I +AN+
Sbjct: 465 DQITEMVQAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMANR 524
Query: 398 LKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRV 457
LK +LP +IS Q+AFV GRL++DN L+A+E H + +A+K D+SKAYDRV
Sbjct: 525 LKKILPSLISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRV 584
Query: 458 EWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLF 517
EWPFL A+ +GF +W+ IM+ V +V +QV++NG P P RGLRQGDPLSPYLF
Sbjct: 585 EWPFLEKAMRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLF 644
Query: 518 LICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGI 577
+IC E+ ++Q + ++G+K+AR AP ISHLLFADD++ + + E + I
Sbjct: 645 VICTEMLVKMLQSAEQKNQITGLKVAR--GAPPISHLLFADDSMFYCKVNDEALGQIIRI 702
Query: 578 LAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKT 637
+ EY Q +N+ KS + + +++ +K LG++ YLGLP SK
Sbjct: 703 IEEYSLASGQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKV 762
Query: 638 QIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFILPDNLC 690
++K+R KK+ G + LS G+E+L+KAVA +P Y MSCF +P +C
Sbjct: 763 ATLSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTIC 815
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 307 bits (786), Expect = 2e-83
Identities = 162/425 (38%), Positives = 252/425 (59%), Gaps = 2/425 (0%)
Query: 266 FSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLH 325
F+ LFTT+ E S +A ++ + L + ++VR+ +F + +A G DG
Sbjct: 755 FADLFTTTQTSDWEEIISGIAPKVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGFT 814
Query: 326 TLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNV 385
FY W ++G+DV + V I T + LIPKI H + +RPISLC
Sbjct: 815 AAFYHHLWDLIGNDVCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCTA 874
Query: 386 PFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMA 445
+KII+ L +LK L D+IS Q+AFVPG+ ++DN LVA+E H +K + +G +A
Sbjct: 875 SYKIISKILIKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVA 934
Query: 446 LKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRG 505
+K D+SKAYDRVEW FL + ++GF WV +IM VT+V ++V++NG+P P RG
Sbjct: 935 VKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRG 994
Query: 506 LRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFAR 565
+RQGDPLSPYLFL C EV S ++++ + G+KI ++ A ISHLLFADD++ F R
Sbjct: 995 IRQGDPLSPYLFLFCAEVLSNMLRKAEVNKQIHGMKITKDCLA--ISHLLFADDSLFFCR 1052
Query: 566 ATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRY 625
A+ + + I +Y++ Q IN+ KS + + + +P++ L LLG+ + +Y
Sbjct: 1053 ASNQNIEQLALIFKKYEEASGQKINYAKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGGKY 1112
Query: 626 LGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFIL 685
LGLP +G+ K ++F ++ + ++ +G LS AG+E++IKA+A +P Y M+CF+L
Sbjct: 1113 LGLPEQLGRRKVELFEYIVTKVKERTEGWAYNYLSPAGKEIVIKAIAMALPVYSMNCFLL 1172
Query: 686 PDNLC 690
P +C
Sbjct: 1173 PTLIC 1177
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 294 bits (753), Expect = 1e-79
Identities = 153/425 (36%), Positives = 243/425 (57%), Gaps = 2/425 (0%)
Query: 266 FSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLH 325
F +FT+ + + +++G L + E+V++ +F ++ +KA G DG
Sbjct: 288 FQQIFTSESDGDFSVVDEAIEPMVSQGDNDFLTRIPNDEEVKDAVFSINASKAPGPDGFT 347
Query: 326 TLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNV 385
FY +WHI+ DV + P + T + LIPK +RPI+LCN+
Sbjct: 348 AGFYHSYWHIISTDVGREIRLFFTSKNFPRRMNETHIRLIPKDLGPRKVADYRPIALCNI 407
Query: 386 PFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMA 445
+KI+ + +++++LP +IS QSAFVPGR+++DN L+ +E HF++ + MA
Sbjct: 408 FYKIVAKIMTKRMQLILPKLISENQSAFVPGRVISDNVLITHEVLHFLRTSSAKKHCSMA 467
Query: 446 LKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRG 505
+K DMSKAYDRVEW FL+ L++ GF W+ ++++ VT+V + ++NG PQ P RG
Sbjct: 468 VKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVLECVTSVSYSFLINGTPQGKVVPTRG 527
Query: 506 LRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFAR 565
LRQGDPLSP LF++C EV S L R L G+++ +++ P ++HLLFADD + F++
Sbjct: 528 LRQGDPLSPCLFILCTEVLSGLCTRAQRLRQLPGVRV--SINGPRVNHLLFADDTMFFSK 585
Query: 566 ATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRY 625
+ E + + IL+ Y K Q INF KS +++S P ++K +L ++ +Y
Sbjct: 586 SDPESCNKLSEILSRYGKASGQSINFHKSSVTFSSKTPRSVKGQVKRILKIRKEGGTGKY 645
Query: 626 LGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFIL 685
LGLP G+ K IF + ++ +K LS+AG++V++KAV +P Y MSCF L
Sbjct: 646 LGLPEHFGRRKRDIFGAIIDKIRQKSHSWASRFLSQAGKQVMLKAVLASMPLYSMSCFKL 705
Query: 686 PDNLC 690
P LC
Sbjct: 706 PSALC 710
>At3g45550 putative protein
Length = 851
Score = 291 bits (744), Expect = 1e-78
Identities = 158/438 (36%), Positives = 244/438 (55%), Gaps = 6/438 (1%)
Query: 261 FLMDIFSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASG 320
F DIF+T +P+ S V N +N L F ++ E + Q+ KA G
Sbjct: 13 FFTDIFTTNGIQVSPIDFADFPSSVTNIINSE----LTQDFRDSEIFEAICQIGDDKAPG 68
Query: 321 LDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPI 380
DGL FY++ W IVG+DV + ++ T + +IPKI+ + +RPI
Sbjct: 69 PDGLTARFYKQCWDIVGNDVIKEVKLFFESSHMKTSVNHTNICMIPKIQNPQTLSDYRPI 128
Query: 381 SLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGC 440
+LCNV +K+I+ + N+LK L I+S Q+AF+PGR++ DN ++A+E H +K +
Sbjct: 129 ALCNVLYKVISKCMVNRLKAHLNSIVSDSQAAFIPGRIINDNVMIAHEIMHSLKVRKRVS 188
Query: 441 NGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPF 500
MA+K D+SKAYDRVEW FL + + GF W+ +IM V +V + V++NG+P
Sbjct: 189 KTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCDKWIGWIMAAVKSVHYSVLINGSPHGYI 248
Query: 501 DPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDN 560
P RG+RQGDPLSPYLF++CG++ S LI+ K + + G++I AP I+HL FADD+
Sbjct: 249 SPTRGIRQGDPLSPYLFILCGDILSHLIKVKASSGDIRGVRIGN--GAPAITHLQFADDS 306
Query: 561 VIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMD 620
+ F +A +K + Y+ Q IN KS++++ V LK LL +
Sbjct: 307 LFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSLITFGSRVYGSTQTRLKTLLNIPNQG 366
Query: 621 NYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIM 680
+YLGLP G+ K ++F ++ +R ++ LS AG+E+L+K+VA +P Y M
Sbjct: 367 GGGKYLGLPEQFGRKKKEMFNYIIDRVKERTASWSAKFLSPAGKEILLKSVALAMPVYAM 426
Query: 681 SCFILPDNLCADSTGVAM 698
SCF LP + ++ + M
Sbjct: 427 SCFKLPQGIVSEIESLLM 444
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 290 bits (741), Expect = 3e-78
Identities = 160/438 (36%), Positives = 243/438 (54%), Gaps = 6/438 (1%)
Query: 261 FLMDIFSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASG 320
F +IFST +P+ S V N +N L FS ++ + + Q+ KA G
Sbjct: 548 FFTNIFSTNGIKVSPIDFADFKSTVTNTVNLD----LTKEFSDTEIYDAICQIGDDKAPG 603
Query: 321 LDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPI 380
DGL FY+ W IVG DV + +I T + +IPKI + +RPI
Sbjct: 604 PDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPI 663
Query: 381 SLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGC 440
+LCNV +K+I+ L N+LK L I+S Q+AF+PGR++ DN ++A+E H +K +
Sbjct: 664 ALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVS 723
Query: 441 NGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPF 500
MA+K D+SKAYDRVEW FL + + GF W+ +IM V +V + V++NG+P
Sbjct: 724 KTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYI 783
Query: 501 DPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDN 560
P RG+RQGDPLSPYLF++CG++ S LI + + L G++I AP I+HL FADD+
Sbjct: 784 TPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGN--GAPAITHLQFADDS 841
Query: 561 VIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMD 620
+ F +A +K + Y+ Q IN KSM+++ V ++LK +L +
Sbjct: 842 LFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSKLKQILEIPNQG 901
Query: 621 NYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIM 680
+YLGLP G+ K ++F ++ +R K+ LS AG+E+++K+VA +P Y M
Sbjct: 902 GGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAM 961
Query: 681 SCFILPDNLCADSTGVAM 698
SCF LP + ++ + M
Sbjct: 962 SCFKLPKGIVSEIESLLM 979
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 289 bits (740), Expect = 4e-78
Identities = 160/438 (36%), Positives = 242/438 (54%), Gaps = 6/438 (1%)
Query: 261 FLMDIFSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASG 320
F +IFST +P+ S V N +N L FS ++ + + Q+ KA G
Sbjct: 774 FFTNIFSTNGIKVSPIDFADFKSTVTNTVNLD----LTKEFSDTEIYDAICQIGDDKAPG 829
Query: 321 LDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPI 380
DGL FY+ W IVG DV + +I T + +IPKI + +RPI
Sbjct: 830 PDGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPI 889
Query: 381 SLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGC 440
+LCNV +K+I+ L N+LK L I+S Q+AF+PGR++ DN ++A+E H +K +
Sbjct: 890 ALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVS 949
Query: 441 NGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPF 500
MA+K D+SKAYDRVEW FL + + GF W+ +IM V +V + V++NG+P
Sbjct: 950 KTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYI 1009
Query: 501 DPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDN 560
P RG+RQGDPLSPYLF++CG++ S LI + + L G++I AP I+HL FADD+
Sbjct: 1010 TPTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGN--GAPAITHLQFADDS 1067
Query: 561 VIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMD 620
+ F +A +K + Y+ Q IN KSM+++ V + LK +L +
Sbjct: 1068 LFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSRLKQILEIPNQG 1127
Query: 621 NYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIM 680
+YLGLP G+ K ++F ++ +R K+ LS AG+E+++K+VA +P Y M
Sbjct: 1128 GGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAM 1187
Query: 681 SCFILPDNLCADSTGVAM 698
SCF LP + ++ + M
Sbjct: 1188 SCFKLPKGIVSEIESLLM 1205
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 288 bits (738), Expect = 6e-78
Identities = 163/439 (37%), Positives = 251/439 (57%), Gaps = 8/439 (1%)
Query: 261 FLMDIFSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASG 320
F ++F++ + ++ +E + V + +N L + +V +F ++ A G
Sbjct: 370 FFENLFTSTYILTHNNHLEGLQAKVTSEMNHN----LIQEVTELEVYNAVFSINKESAPG 425
Query: 321 LDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPI 380
DG LF+Q+ W +V + + V P T + LIPKI + RPI
Sbjct: 426 PDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPI 485
Query: 381 SLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGC 440
SLC+V +KII+ L +LK LP I+S QSAFVP RL++DN LVA+E H ++
Sbjct: 486 SLCSVLYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRIS 545
Query: 441 NGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPF 500
MA K DMSKAYDRVEWPFL + + +GF W+S+IM+ VT+V + V++NG P
Sbjct: 546 KEHMAFKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHI 605
Query: 501 DPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIA-RNVSAPVISHLLFADD 559
P RG+RQGDPLSP LF++C E ++ + A ++GI+ + VS ++HLLFADD
Sbjct: 606 IPTRGIRQGDPLSPALFVLCTEALIHILNKAEQAGKITGIQFQDKKVS---VNHLLFADD 662
Query: 560 NVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAM 619
++ +AT++E + L++Y ++ Q+IN +KS +++ +NV + +K G+
Sbjct: 663 TLLMCKATKQECEELMQCLSQYGQLSGQMINLNKSAITFGKNVDIQIKDWIKSRSGISLE 722
Query: 620 DNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYI 679
+YLGLP + SK +F F+KE+ +L G +LS+ G+EVL+K++A +P Y+
Sbjct: 723 GGTGKYLGLPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYV 782
Query: 680 MSCFILPDNLCADSTGVAM 698
MSCF LP NLC T V M
Sbjct: 783 MSCFKLPKNLCQKLTTVMM 801
>At1g24640 hypothetical protein
Length = 1270
Score = 283 bits (724), Expect = 3e-76
Identities = 163/425 (38%), Positives = 239/425 (55%), Gaps = 2/425 (0%)
Query: 266 FSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLH 325
F LF +SNP G S + R+++ L S ++++E +F + P A G DG+
Sbjct: 356 FGNLFKSSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMS 415
Query: 326 TLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNV 385
LF+Q +W VG+ V + P T L LIPK + RPISLC+V
Sbjct: 416 ALFFQHYWSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSV 475
Query: 386 PFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMA 445
+KII+ +A +L+ LP+I+S QSAFV RL+TDN LVA+E H +K + MA
Sbjct: 476 LYKIISKIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMA 535
Query: 446 LKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRG 505
+K DMSKAYDRVEW +LRS L +GF + WV++IM V++V + V++N P RG
Sbjct: 536 VKSDMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRG 595
Query: 506 LRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFAR 565
LRQGDPLSP+LF++C E + L+ + L GI+ + N P++ HLLFADD++ +
Sbjct: 596 LRQGDPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFSEN--GPMVHHLLFADDSLFLCK 653
Query: 566 ATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRY 625
A+RE++ ++ IL Y Q IN +KS +++ V ++ LG+ Y
Sbjct: 654 ASREQSLVLQKILKVYGNATGQTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTY 713
Query: 626 LGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFIL 685
LGLP SK + ++K+R +KL LS+ G+EVL+K+VA +P + MSCF L
Sbjct: 714 LGLPECFSGSKVDMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKL 773
Query: 686 PDNLC 690
P C
Sbjct: 774 PITTC 778
>At4g10830 putative protein
Length = 1294
Score = 282 bits (721), Expect = 6e-76
Identities = 148/382 (38%), Positives = 220/382 (56%), Gaps = 2/382 (0%)
Query: 317 KASGLDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATH 376
KA G DGL FY+ W IVG DV + +I T + +IPKI +
Sbjct: 806 KAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSD 865
Query: 377 FRPISLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKK 436
+RPI+LCNV +KII+ L +LK L I+S Q+AF+PGRLV DN ++A+E H +K +
Sbjct: 866 YRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTR 925
Query: 437 IYGCNGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNP 496
MA+K D+SKAYDRVEW FL + + GF W+ +IM V +V + V++NG P
Sbjct: 926 KRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIP 985
Query: 497 QVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLF 556
P RG+RQGDPLSPYLF++C ++ + LI+ ++ + GI+I V P ++HL F
Sbjct: 986 HGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIGNGV--PGVTHLQF 1043
Query: 557 ADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGV 616
ADD++ F ++ +K + Y+ Q IN KSM+++ V N LK +LG+
Sbjct: 1044 ADDSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMITFGSRVHGTTQNRLKNILGI 1103
Query: 617 KAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIP 676
++ +YLGLP G+ K +F ++ ER K+ LS AG+E+++K+VA +P
Sbjct: 1104 QSHGGGGKYLGLPEQFGRKKRDMFNYIIERVKKRTSSWSAKYLSPAGKEIMLKSVAMSMP 1163
Query: 677 AYIMSCFILPDNLCADSTGVAM 698
Y MSCF LP N+ ++ + M
Sbjct: 1164 VYAMSCFKLPLNIVSEIEALLM 1185
>At2g11240 pseudogene
Length = 1044
Score = 281 bits (720), Expect = 7e-76
Identities = 163/459 (35%), Positives = 250/459 (53%), Gaps = 15/459 (3%)
Query: 235 GRNVI*LMSFGMMRAEDGWRMRTYPGFL---MDIFSTLFTTSNPMGIEAATSLVANRLNK 291
GR VI F ++ EDG G L + F LF+ + G AAT + +
Sbjct: 195 GRTVI--NKFSVIEKEDGVPEYEEAGILNVISEYFQKLFSANE--GARAAT------IKE 244
Query: 292 GHLAILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSVLQGR 351
++ + E+++ F +H KA G DG F+Q W VG ++ S
Sbjct: 245 AIKPFISPEQNPEEIKSACFSIHADKAPGPDGFSASFFQSNWMTVGPNIVLEIQSFFSSS 304
Query: 352 VSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIISGPQS 411
TI T + LIPKI+ +RPI+LC V +KII+ L+ +L+ +L +IIS QS
Sbjct: 305 TLQPTINKTHITLIPKIQSLKRMVDYRPIALCTVFYKIISKLLSRRLQPILQEIISENQS 364
Query: 412 AFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEKMGF 471
AFVP R DN L+ +E H++K MA+K +MSKAYDR+EW F++ +++MGF
Sbjct: 365 AFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAVKTNMSKAYDRIEWDFIKLVMQEMGF 424
Query: 472 PVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRK 531
W+S+I+ +TTV + +LNG+ Q P RGLRQGDPLSP+LF+IC EV S L ++
Sbjct: 425 HQTWISWILQCITTVSYSFLLNGSAQGAVTPERGLRQGDPLSPFLFIICSEVLSGLCRKA 484
Query: 532 IGASHLSGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINF 591
L G+++++ P ++HLLFADD + F R+ + IL +Y++ Q+IN
Sbjct: 485 QLDGSLLGLRVSK--GNPRVNHLLFADDTIFFCRSDLKSCKTFLCILKKYEEASGQMINK 542
Query: 592 DKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKL 651
KS +++SR P E + +LG++ + +YLGLP + G+ K +F + +R ++
Sbjct: 543 SKSAITFSRKTPDHIKTEAQQILGIQLVGGLGKYLGLPKMFGRKKRDLFNQIVDRIRQRS 602
Query: 652 KGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFILPDNLC 690
LS AG+ ++K+V +P Y MSCF L +LC
Sbjct: 603 LSWSSRFLSTAGKTTMLKSVLASMPTYTMSCFKLLVSLC 641
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 278 bits (712), Expect = 6e-75
Identities = 156/429 (36%), Positives = 237/429 (54%), Gaps = 10/429 (2%)
Query: 266 FSTLFTTSNPMGI----EAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGL 321
F +FTTSN + EA + ++++ N+ + I + +++E LF + KA G
Sbjct: 334 FQNIFTTSNNSDLQVVQEALSPIISSHCNEELIKISSLL----EIKEALFSISADKAPGP 389
Query: 322 DGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPIS 381
DG F+ +W I+ DV+ S + T + LIPKI + +RPI+
Sbjct: 390 DGFSASFFHAYWDIIEADVSRDIRSFFVDSCLSPRLNETHVTLIPKISAPRKVSDYRPIA 449
Query: 382 LCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCN 441
LCNV +KI+ L +L+ L ++IS QSAFVPGR + DN L+ +E HF++
Sbjct: 450 LCNVQYKIVAKILTRRLQPWLSELISLHQSAFVPGRAIADNVLITHEILHFLRVSGAKKY 509
Query: 442 GIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFD 501
MA+K DMSKAYDR++W FL+ L ++GF W+ ++M V TV + ++NG+PQ
Sbjct: 510 CSMAIKTDMSKAYDRIKWNFLQEVLMRLGFHDKWIRWVMQCVCTVSYSFLINGSPQGSVV 569
Query: 502 PGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNV 561
P RGLRQGDPLSPYLF++C EV S L ++ + GI++AR +P ++HLLFADD +
Sbjct: 570 PSRGLRQGDPLSPYLFILCTEVLSGLCRKAQEKGVMVGIRVAR--GSPQVNHLLFADDTM 627
Query: 562 IFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDN 621
F + + IL +Y+ Q IN KS +++S P +K+ L +
Sbjct: 628 FFCKTNPTCCGALSNILKKYELASGQSINLAKSAITFSSKTPQDIKRRVKLSLRIDNEGG 687
Query: 622 YDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMS 681
+YLGLP G+ K IF + +R ++ LS AG+++L+KAV +P+Y M
Sbjct: 688 IGKYLGLPEHFGRRKRDIFSSIVDRIRQRSHSWSIRFLSSAGKQILLKAVLSSMPSYAMM 747
Query: 682 CFILPDNLC 690
CF LP +LC
Sbjct: 748 CFKLPASLC 756
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 274 bits (701), Expect = 1e-73
Identities = 155/438 (35%), Positives = 242/438 (54%), Gaps = 6/438 (1%)
Query: 261 FLMDIFSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASG 320
F D+FS+ + +S +E V +N+ L + +++ + +F ++ A G
Sbjct: 121 FFEDLFSSSYPSSMDSVLEGFNKRVTEDMNQD----LTKKVNEQEIYKAVFSINAESAPG 176
Query: 321 LDGLHTLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPI 380
DG LF+Q+ W +V + + Q + P T L LIPKI K RPI
Sbjct: 177 PDGFTALFFQRQWPLVKNQIISDIELFFQTGILPEDWNHTHLCLIPKITKPARMADIRPI 236
Query: 381 SLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGC 440
SLC+V +KII+ L+ +LK LP I+S QSAFV RLV+DN ++A+E H ++
Sbjct: 237 SLCSVMYKIISKILSARLKKYLPVIVSPTQSAFVAERLVSDNIILAHEIVHNLRTNEKIS 296
Query: 441 NGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPF 500
M K DMSKAYDRVEWPFL+ L +GF W++++M V++V + V++NG P
Sbjct: 297 KDFMVFKTDMSKAYDRVEWPFLKGILLALGFNSTWINWMMACVSSVSYSVLINGQPFGHI 356
Query: 501 DPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDN 560
P RGLRQGDPLSP+LF++C E ++ + +SGI+ N + P ++HLLFADD
Sbjct: 357 TPHRGLRQGDPLSPFLFVLCTEALIHILNQAEKIGKISGIQF--NGTGPSVNHLLFADDT 414
Query: 561 VIFARATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMD 620
++ +A++ E + + L++Y + Q+IN +KS +++ V + G++
Sbjct: 415 LLICKASQLECAEIMHCLSQYGHISGQMINSEKSAITFGAKVNEETKQWIMNRSGIQTEG 474
Query: 621 NYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIM 680
+YLGLP SK +F F+KE+ +L G +LS+ G+++L+K++A P Y M
Sbjct: 475 GTGKYLGLPECFQGSKQVLFGFIKEKLQSRLSGWYAKTLSQGGKDILLKSIAMAFPVYAM 534
Query: 681 SCFILPDNLCADSTGVAM 698
+CF L LC T V M
Sbjct: 535 TCFRLSKTLCTKLTSVMM 552
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 259 bits (661), Expect = 5e-69
Identities = 154/400 (38%), Positives = 218/400 (54%), Gaps = 3/400 (0%)
Query: 289 LNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSVL 348
L++ + L PFS +V + M KA G DG +FYQ+ W +VG+ V F +
Sbjct: 956 LSEADFSSLTKPFSPLEVEGAIRSMGKYKAPGPDGFQPVFYQQGWEVVGESVTKFVMDFF 1015
Query: 349 QGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIISG 408
P L+VLI K+ K T FRPISLCNV FK IT + +LK V+ +I
Sbjct: 1016 SSGSFPQETNDVLVVLIAKVLKPEKITQFRPISLCNVLFKTITKVMVGRLKGVINKLIGP 1075
Query: 409 PQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEK 468
Q++F+PGRL TDN +V E H M++K G G M LKLD+ KAYDR+ W L L+
Sbjct: 1076 AQTSFIPGRLSTDNIVVVQEVVHSMRRK-KGVKGWMLLKLDLEKAYDRIRWDLLEDTLKA 1134
Query: 469 MGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALI 528
G P WV +IM V +++ NG F P RGLRQGDPLSPYLF++C E LI
Sbjct: 1135 AGLPGTWVQWIMKCVEGPSMRLLWNGEKTDAFKPLRGLRQGDPLSPYLFVLCIERLCHLI 1194
Query: 529 QRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQV 588
+ I A IKI++ S P +SH+ FADD ++FA A+ ++ ++G+L ++ Q
Sbjct: 1195 ESSIAAKKWKPIKISQ--SGPRLSHICFADDLILFAEASIDQIRVLRGVLEKFCGASGQK 1252
Query: 589 INFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDW 648
++ +KS + +S+NV + G+KA + +YLG+P + + + F V +R
Sbjct: 1253 VSLEKSKIYFSKNVLRDLGKRISEESGMKATKDLGKYLGVPILQKRINKETFGEVIKRFS 1312
Query: 649 KKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFILPDN 688
+L G K LS AGR L KAV I + MS LP +
Sbjct: 1313 SRLAGWKGRMLSFAGRLTLTKAVLTSILVHTMSTIKLPQS 1352
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 254 bits (649), Expect = 1e-67
Identities = 152/398 (38%), Positives = 211/398 (52%), Gaps = 3/398 (0%)
Query: 289 LNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSVL 348
+++ A L F+ +V + M KA G DG +FYQ+ W VG V F L
Sbjct: 280 ISEAEKAALLQAFTKAEVVSAVKSMGRFKAPGPDGYQPVFYQQCWETVGPSVTRFVLEFF 339
Query: 349 QGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIISG 408
+ V P + LLVLI K+ K FRP+SLCNV FKIIT + +LK V+ +I
Sbjct: 340 ETGVLPASTNDALLVLIAKVAKPERIQQFRPVSLCNVLFKIITKMMVTRLKNVISKLIGP 399
Query: 409 PQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEK 468
Q++F+PGRL DN ++ E H M++K G G M LKLD+ KAYDRV W FL+ LE
Sbjct: 400 AQASFIPGRLSIDNIVLVQEAVHSMRRK-KGRKGWMLLKLDLEKAYDRVRWDFLQETLEA 458
Query: 469 MGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALI 528
G W S IM VT V+ NG F P RGLRQGDPLSPYLF++C E LI
Sbjct: 459 AGLSEGWTSRIMAGVTDPSMSVLWNGERTDSFVPARGLRQGDPLSPYLFVLCLERLCHLI 518
Query: 529 QRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQV 588
+ +G I ++ S +SH+ FADD ++FA A+ + ++ +L + + Q
Sbjct: 519 EASVGKREWKPIAVSCGGSK--LSHVCFADDLILFAEASVAQIRIIRRVLERFCEASGQK 576
Query: 589 INFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDW 648
++ +KS + +S NV + G+ +YLG+P + + + F V ER
Sbjct: 577 VSLEKSKIFFSHNVSREMEQLISEESGIGCTKELGKYLGMPILQKRMNKETFGEVLERVS 636
Query: 649 KKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFILP 686
+L G K SLS AGR L KAV IP ++MS +LP
Sbjct: 637 ARLAGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLP 674
>At3g31420 hypothetical protein
Length = 1491
Score = 252 bits (643), Expect = 6e-67
Identities = 147/398 (36%), Positives = 217/398 (53%), Gaps = 39/398 (9%)
Query: 300 PFSGEKVREPLFQMHPTKASGLDGLHTLFYQKFW-----HIVGDDVAHFCLSVLQGRVSP 354
P + ++ E +F + P++ DG FYQ+FW ++ + F S L R +
Sbjct: 612 PVTELEIEESVFSVAPSRTPDPDGFTADFYQQFWPDIKQKVIDEVTRFFERSELDERHNH 671
Query: 355 GTIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIISGPQSAFV 414
T L LIPK++ FRPI+LCNV +KII+ L N+LK L I+ Q+AFV
Sbjct: 672 -----TNLCLIPKVETPTTIAKFRPIALCNVSYKIISKILVNRLKKHLGGAITENQAAFV 726
Query: 415 PGRLVTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVN 474
PGRL+T+NA++A+E ++ +K + N MALK D++KAYDR+EW FL + +MGF
Sbjct: 727 PGRLITNNAIIAHEVYYALKARKRQANSYMALKTDITKAYDRLEWDFLEETMRQMGFNTK 786
Query: 475 WVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGA 534
W+ IM VT V F V++NG+P P RG+R GDPLSPYLF++C EV S +I++
Sbjct: 787 WIERIMICVTMVRFSVLINGSPHGTIKPERGIRHGDPLSPYLFILCAEVLSHMIKQAEIN 846
Query: 535 SHLSGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKS 594
L GI++ + P ISHLLFADD++ F A + + +K E D
Sbjct: 847 KKLKGIRL--STQGPFISHLLFADDSIFFTLANQRSCTAIK----EPDTK---------- 890
Query: 595 MLSYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGR 654
++ LLG+ +YLGLP K K ++F ++ E+ K +G
Sbjct: 891 -------------RRMRHLLGIHNEGGEGKYLGLPEQFNKKKKELFNYIIEKVKDKTQGW 937
Query: 655 KEPSLSRAGREVLIKAVAQEIPAYIMSCFILPDNLCAD 692
+ LS+ G+EVL+KAVA +P Y M+ F L +C +
Sbjct: 938 SKKFLSQGGKEVLLKAVALAMPVYSMNIFKLTKEVCEE 975
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 252 bits (643), Expect = 6e-67
Identities = 152/433 (35%), Positives = 229/433 (52%), Gaps = 13/433 (3%)
Query: 266 FSTLFTTSNPMGIEAATSLVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLH 325
F LF +S P ++A R+++ L + ++ +F ++ A
Sbjct: 356 FENLFMSSYPANSQSALDGFKTRVSEEMNQELTQAVTELEIHSAVFSINVESAP------ 409
Query: 326 TLFYQKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNV 385
+K G D L + V P T L LIPK + RPISLC+V
Sbjct: 410 ----EKLECCQGSDYIEI-LGFFETGVLPQEWNHTHLYLIPKFTNPQRMSDIRPISLCSV 464
Query: 386 PFKIIT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMA 445
+KII+ L+ KLK LP I+S QSAF RL++DN L+A+E H ++ M
Sbjct: 465 LYKIISKILSFKLKKHLPSIVSPSQSAFFAERLISDNILIAHEIVHSLRTNDKISKEFMV 524
Query: 446 LKLDMSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRG 505
K DMSKAYDRVEW FL+ L +GF W+S+IM VT+V + V++NG P RG
Sbjct: 525 FKTDMSKAYDRVEWSFLQEILVALGFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERG 584
Query: 506 LRQGDPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFAR 565
+RQGDP+SP+LF++C E ++Q+ + +SGI+ N S P ++HLLF DD + R
Sbjct: 585 IRQGDPISPFLFVLCTEALIHILQQAENSKKVSGIQF--NGSGPSVNHLLFVDDTQLVCR 642
Query: 566 ATREEASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRY 625
AT+ + + L++Y + Q+IN +KS +++ V +K G+ +Y
Sbjct: 643 ATKSDCEQMMLCLSQYGHISGQLINVEKSSITFGVKVDEDTKRWIKNRSGIHLEGGTGKY 702
Query: 626 LGLPTIIGKSKTQIFRFVKERDWKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFIL 685
LGLP + SK +F ++KE+ L G + +LS+ G+E+L+K++A +P YIM+CF L
Sbjct: 703 LGLPENLSGSKQDLFGYIKEKLQSHLSGWYDKTLSQGGKEILLKSIALALPVYIMTCFRL 762
Query: 686 PDNLCADSTGVAM 698
P LC T V M
Sbjct: 763 PKGLCTKLTSVMM 775
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 249 bits (635), Expect = 5e-66
Identities = 150/402 (37%), Positives = 209/402 (51%), Gaps = 20/402 (4%)
Query: 288 RLNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFYQKFWHIVGDDVAHFCLSV 347
RL + LN PF+ ++V + M KA G DG +FYQ+ W VG+ V+ F +
Sbjct: 231 RLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGESVSKFVMEF 290
Query: 348 LQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKIIT*TLANKLKIVLPDIIS 407
+ V P + LLVL+ K+ K T FRP+SLCNV FKIIT + +LK V+ +I
Sbjct: 291 FESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRLKNVISKLIG 350
Query: 408 GPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALE 467
Q++F+PGRL DN +V E H M++K G G M LKLD+ KAYDR+ W FL LE
Sbjct: 351 PAQASFIPGRLSFDNIVVVQEAVHSMRRK-KGRKGWMLLKLDLEKAYDRIRWDFLAETLE 409
Query: 468 KMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSAL 527
G W+ IM+ V ++ NG F P RGLRQGDP+SPYLF++C E
Sbjct: 410 AAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGLRQGDPISPYLFVLCIERLCHQ 469
Query: 528 IQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQ 587
I+ +G I I++ P +SH+ FADD ++FA A+ Q
Sbjct: 470 IETAVGRGDWKSISISQ--GGPKVSHVCFADDLILFAEAS-----------------VAQ 510
Query: 588 VINFDKSMLSYSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERD 647
++ +KS + +S NV + G+ + +YLG+P + + F V ER
Sbjct: 511 KVSLEKSKIFFSNNVSRDLEGLITAETGIGSTRELGKYLGMPVLQKRINKDTFGEVLERV 570
Query: 648 WKKLKGRKEPSLSRAGREVLIKAVAQEIPAYIMSCFILPDNL 689
+L G K SLS AGR L KAV IP + MS +LP +L
Sbjct: 571 SSRLSGWKSRSLSLAGRITLTKAVLMSIPIHTMSSILLPASL 612
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 241 bits (615), Expect = 1e-63
Identities = 133/347 (38%), Positives = 203/347 (58%), Gaps = 10/347 (2%)
Query: 274 NPMGIEAATS----LVANRLNKGHLAILNTPFSGEKVREPLFQMHPTKASGLDGLHTLFY 329
NP ++A L+ +NK L S E+V+ LF ++P KA G DG+ FY
Sbjct: 348 NPQVFDSALQEVPVLITEEMNKS----LTKVISPEEVKRALFSLNPDKAPGPDGMTAFFY 403
Query: 330 QKFWHIVGDDVAHFCLSVLQGRVSPGTIIPTLLVLIPKIKKSVHATHFRPISLCNVPFKI 389
Q +W + G D+ + + T + LIPK ++ FRPISLCNV +K+
Sbjct: 404 QHYWDLTGPDLIKLVQNFHSTGFFDERLNETNICLIPKTERPRKMAEFRPISLCNVSYKV 463
Query: 390 IT*TLANKLKIVLPDIISGPQSAFVPGRLVTDNALVAYECFHFMKKKIYGCNGIMALKLD 449
I+ L+++LK +LP++IS QSAFV RL+TDN L+A E FH ++ MA+K D
Sbjct: 464 ISKVLSSRLKRLLPELISETQSAFVAERLITDNILIAQENFHALRTNPACKKKYMAIKTD 523
Query: 450 MSKAYDRVEWPFLRSALEKMGFPVNWVSFIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQG 509
MSKAYDRVEW FLR+ + KMGF WV +I+ +++V ++++LNG+P+ P RG+RQG
Sbjct: 524 MSKAYDRVEWSFLRALMLKMGFAQKWVDWIIFCISSVSYKILLNGSPKGFIKPSRGIRQG 583
Query: 510 DPLSPYLFLICGEVFSALIQRKIGASHLSGIKIARNVSAPVISHLLFADDNVIFARATRE 569
DP+SP+LF++C E A ++ + G++I+R ++P SHLLFADD++ F +A
Sbjct: 584 DPISPFLFILCTEALVAKLKDAEWHGRIQGLQISR--ASPSTSHLLFADDSLFFCKADPL 641
Query: 570 EASCVKGILAEYDKVYVQVINFDKSMLSYSRNVPSLYFNELKMLLGV 616
+ + IL Y + Q +N DKS + + V + N +K+ LG+
Sbjct: 642 QGKEIIDILRLYGEASGQQLNPDKSSVMFGHEVDNSIRNTIKVSLGI 688
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 227 bits (578), Expect = 2e-59
Identities = 113/272 (41%), Positives = 168/272 (61%), Gaps = 3/272 (1%)
Query: 418 LVTDNALVAYECFHFMKKKIYGCNGIMALKLDMSKAYDRVEWPFLRSALEKMGFPVNWVS 477
++TDN L+A+E H + K +A KLD++KA+D++EW F+ + +++MGF W +
Sbjct: 1 MITDNILIAHELIHSLHTKKL-VQPFVATKLDITKAFDKIEWGFIEAIMKQMGFSEKWCN 59
Query: 478 FIMDYVTTVLFQVMLNGNPQVPFDPGRGLRQGDPLSPYLFLICGEVFSALIQRKIGASHL 537
+IM +TT + +++NG P P RG+RQGDP+SPYL+L+C E SALIQ I A L
Sbjct: 60 WIMTCITTTTYSILINGQPVRRIIPKRGIRQGDPISPYLYLLCTEGLSALIQASIKAKQL 119
Query: 538 SGIKIARNVSAPVISHLLFADDNVIFARATREEASCVKGILAEYDKVYVQVINFDKSMLS 597
G K +RN P ISHLLFA D+++F +AT EE + +L Y+K Q +NF KS +
Sbjct: 120 HGFKASRN--GPAISHLLFAHDSLVFCKATLEECMTLVNVLKLYEKASGQAVNFQKSAIL 177
Query: 598 YSRNVPSLYFNELKMLLGVKAMDNYDRYLGLPTIIGKSKTQIFRFVKERDWKKLKGRKEP 657
+ + + +L LLG+ + + RYLGLP +G++KT F F+ + +K+
Sbjct: 178 FGKGLDFRTSEQLSQLLGIYKTEGFGRYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNK 237
Query: 658 SLSRAGREVLIKAVAQEIPAYIMSCFILPDNL 689
LS AG+EVLIK++ IP Y MSCF+LP L
Sbjct: 238 LLSPAGKEVLIKSIVTAIPTYSMSCFLLPMRL 269
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.334 0.146 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,863,716
Number of Sequences: 26719
Number of extensions: 649322
Number of successful extensions: 2119
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1911
Number of HSP's gapped (non-prelim): 98
length of query: 758
length of database: 11,318,596
effective HSP length: 107
effective length of query: 651
effective length of database: 8,459,663
effective search space: 5507240613
effective search space used: 5507240613
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0207.9


