

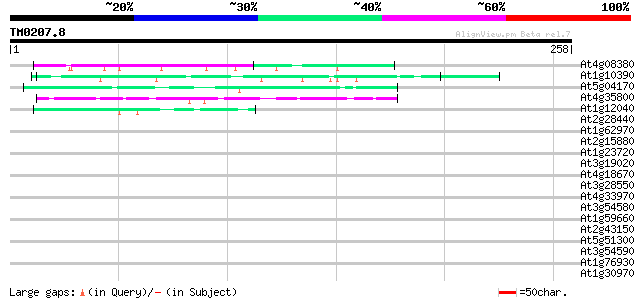
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0207.8
(258 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g08380 extensin-like protein 51 5e-07
At1g10390 unknown protein 48 5e-06
At5g04170 EF - hand Calcium binding protein - like 43 2e-04
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 42 2e-04
At1g12040 leucine-rich repeat/extensin 1 (LRX1) 41 7e-04
At2g28440 En/Spm-like transposon protein 40 0.001
At1g62970 unknown protein 39 0.002
At2g15880 unknown protein 37 0.008
At1g23720 unknown protein 37 0.014
At3g19020 hypothetical protein 35 0.030
At4g18670 extensin-like protein 35 0.052
At3g28550 unknown protein 35 0.052
At4g33970 extensin-like protein 34 0.089
At3g54580 extensin precursor -like protein 34 0.089
At1g59660 nucleoporin 98-like protein 34 0.089
At2g43150 putative extensin 33 0.12
At5g51300 unknown protein 33 0.15
At3g54590 extensin precursor -like protein 33 0.15
At1g76930 extensin 33 0.15
At1g30970 putative 11-zinc finger protein 33 0.15
>At4g08380 extensin-like protein
Length = 437
Score = 51.2 bits (121), Expect = 5e-07
Identities = 51/175 (29%), Positives = 66/175 (37%), Gaps = 18/175 (10%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDF 71
Y SPP +S+ PYA +S P+ S P+V S P Y P P Y P Y Y
Sbjct: 168 YKSPPYVYSSPPPYA--YSPPPSPYVYKSPPYVYSSPPPYAYSPPPYAYSPPPSPYVYKS 225
Query: 72 FPESDSVSKPYGYSGLAS-----EDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASED 126
P S PY YS S + Y PP + P P P +S
Sbjct: 226 PPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSP---PPSPYVYKSPPYVYSSPP 282
Query: 127 SFGYEQWLSASSKPSLGEAQPYF----PSYIPSTIPSPTSSVAAPIHWSSSSGFA 177
+ Y S P + ++ PY P Y S PSP + P +SS +A
Sbjct: 283 PYAY----SPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYA 333
Score = 46.2 bits (108), Expect = 2e-05
Identities = 53/185 (28%), Positives = 70/185 (37%), Gaps = 23/185 (12%)
Query: 12 YYSPPQPHSTLSPYA-----AAFSVNRPPFNDVSAP-FVDSDEPAYGVPPNPVHYPVHPR 65
Y SPP +S+ PYA + + PP+ S P +V S P Y P P Y P
Sbjct: 105 YKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYVYSSPPPYAYSPPPYAYSPPPS 164
Query: 66 SYGYDFFPESDSVSKPYGYSGLAS-----EDSFWYDQWPPTS-SKPSLAEPQPCFPDLNS 119
Y Y P S PY YS S + Y PP + S P A P P +
Sbjct: 165 PYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPYAYSPPPSPYVYK 224
Query: 120 MG---LASEDSFGYEQWLSASSKPSLGEAQPYF----PSYIPSTIPSPTSSVAAPIHWSS 172
+S + Y S P + ++ PY P Y S PSP + P +SS
Sbjct: 225 SPPYVYSSPPPYAY----SPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSS 280
Query: 173 SSGFA 177
+A
Sbjct: 281 PPPYA 285
Score = 43.9 bits (102), Expect = 9e-05
Identities = 52/182 (28%), Positives = 68/182 (36%), Gaps = 32/182 (17%)
Query: 12 YYSPPQPHSTLSPYAA-----AFSVNRPPFNDVSAPFVDSDEP--AYGVPPNPVHYPVHP 64
Y SPP +S+ PYA A+S P+ S P+V S P AY PP+P Y P
Sbjct: 192 YKSPPYVYSSPPPYAYSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPP 251
Query: 65 RSYGYDFFPESDSVSKPYGYSGLAS-----EDSFWYDQWPPTSSKPSLAEPQPCFPDLNS 119
Y S PY YS S + Y PP + P P P
Sbjct: 252 YVY---------SSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSP---PPSPYVYKSPP 299
Query: 120 MGLASEDSFGYEQWLSASSKPSLGEAQPYF----PSYIPSTIPSPTSSVAAPIHWSSSSG 175
+S + Y S P + ++ PY P Y S PSP + P +SS
Sbjct: 300 YVYSSPPPYAY----SPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPP 355
Query: 176 FA 177
+A
Sbjct: 356 YA 357
Score = 42.7 bits (99), Expect = 2e-04
Identities = 33/107 (30%), Positives = 44/107 (40%), Gaps = 8/107 (7%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEP--AYGVPPNPVHYPVHPRSYG- 68
Y SPP +S+ PYA +S P+ S P+V S P AY PP+P Y P Y
Sbjct: 319 YKSPPYVYSSPPPYA--YSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSS 376
Query: 69 ---YDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQP 112
Y + P + S P + + Y PP P + P P
Sbjct: 377 PPPYTYSPPPYAYSPPPPCPDVYKPPPYVYSSPPPYVYNPPPSSPPP 423
Score = 39.3 bits (90), Expect = 0.002
Identities = 50/181 (27%), Positives = 67/181 (36%), Gaps = 34/181 (18%)
Query: 10 WGYYSPPQPHSTLSP---YAA----AFSVNRPPFNDVSAPFVDSDEP--AYGVPPNPVHY 60
+ Y PP P+ SP Y++ A+S P+ S P+V S P AY PP+P Y
Sbjct: 212 YAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVY 271
Query: 61 PVHPRSYGYDFFPESDSVSKPYGYSGLAS-----EDSFWYDQWPPTSSKPSLAEPQPCFP 115
P Y S PY YS S + Y PP + P P P
Sbjct: 272 KSPPYVY---------SSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSP---PPSPYVY 319
Query: 116 DLNSMGLASEDSFGYEQWLSASSKPSLGEAQPYF----PSYIPSTIPSPTSSVAAPIHWS 171
+S + Y S P + ++ PY P Y S PSP + P +S
Sbjct: 320 KSPPYVYSSPPPYAY----SPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYS 375
Query: 172 S 172
S
Sbjct: 376 S 376
Score = 38.9 bits (89), Expect = 0.003
Identities = 52/195 (26%), Positives = 70/195 (35%), Gaps = 30/195 (15%)
Query: 10 WGYYSPPQPHSTLSP---YAA----AFSVNRPPFNDVSAPFVDSDEP--AYGVPPNPVHY 60
+ Y PP P+ SP Y++ A+S P+ S P+V S P AY PP+P Y
Sbjct: 70 YAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVY 129
Query: 61 PVHPRSYG------YDFFPESDSVSKPYGYSGLAS-----EDSFWYDQWPPTSSKPSLAE 109
P Y Y P PY YS S + Y PP + P
Sbjct: 130 KSPPYVYSSPPPYVYSSPPPYAYSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSP---P 186
Query: 110 PQPCFPDLNSMGLASEDSFGYE---QWLSASSKPSLGEAQPYF----PSYIPSTIPSPTS 162
P P +S + Y S P + ++ PY P Y S PSP
Sbjct: 187 PSPYVYKSPPYVYSSPPPYAYSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYV 246
Query: 163 SVAAPIHWSSSSGFA 177
+ P +SS +A
Sbjct: 247 YKSPPYVYSSPPPYA 261
Score = 38.5 bits (88), Expect = 0.004
Identities = 47/175 (26%), Positives = 66/175 (36%), Gaps = 32/175 (18%)
Query: 13 YSPPQP----HSTLSPYAAAFSVNRPPFNDVSAPFVDSDEP--AYGVPPNPVHYPVHPRS 66
YSPP P +S+ PY +S P+ S P+V S P AY PP+P Y P
Sbjct: 30 YSPPSPPPYVYSSPPPYT--YSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYV 87
Query: 67 YGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASED 126
Y S PY YS S + + +S P P P S
Sbjct: 88 Y---------SSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPP-----------SPY 127
Query: 127 SFGYEQWLSASSKPSLGEAQPYF----PSYIPSTIPSPTSSVAAPIHWSSSSGFA 177
+ ++ +S P + + P + P Y S PSP + P +SS +A
Sbjct: 128 VYKSPPYVYSSPPPYVYSSPPPYAYSPPPYAYSPPPSPYVYKSPPYVYSSPPPYA 182
Score = 38.1 bits (87), Expect = 0.005
Identities = 55/197 (27%), Positives = 74/197 (36%), Gaps = 38/197 (19%)
Query: 12 YYSPPQPHSTLSP---YAA----AFSVNRPPFNDVSAPFVDSDEP--AYGVPPNPVHYPV 62
Y PP P+ SP Y++ A+S P+ S P+V S P AY PP+P Y
Sbjct: 48 YSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKS 107
Query: 63 HPRSYG----YDFFPESDSV---SKPYGYSGLASEDSFWYDQWPPTS-SKPSLAEPQPCF 114
P Y Y + P S PY YS S + Y PP + S P A P
Sbjct: 108 PPYVYSSPPPYAYSPPPSPYVYKSPPYVYS---SPPPYVYSSPPPYAYSPPPYAYSPPPS 164
Query: 115 PDLNSMG---LASEDSFGYEQWLSASSKPSLGEAQPYF-----------PSYIPSTIPSP 160
P + +S + Y S P + ++ PY P Y S PSP
Sbjct: 165 PYVYKSPPYVYSSPPPYAY----SPPPSPYVYKSPPYVYSSPPPYAYSPPPYAYSPPPSP 220
Query: 161 TSSVAAPIHWSSSSGFA 177
+ P +SS +A
Sbjct: 221 YVYKSPPYVYSSPPPYA 237
Score = 36.2 bits (82), Expect = 0.018
Identities = 46/172 (26%), Positives = 62/172 (35%), Gaps = 30/172 (17%)
Query: 10 WGYYSPPQPHSTLSP---YAA----AFSVNRPPFNDVSAPFVDSDEP--AYGVPPNPVHY 60
+ Y PP P+ SP Y++ A+S P+ S P+V S P AY PP+P Y
Sbjct: 236 YAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVY 295
Query: 61 PVHPRSYGYDFFPESDSVSKPYGYSGLAS-----EDSFWYDQWPPTSSKPSLAEPQPCFP 115
P Y S PY YS S + Y PP + P P P
Sbjct: 296 KSPPYVY---------SSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSP---PPSPYVY 343
Query: 116 DLNSMGLASEDSFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAP 167
+S + Y S P + ++ PY S P SP +P
Sbjct: 344 KSPPYVYSSPPPYAY----SPPPSPYVYKSPPYVYSSPPPYTYSPPPYAYSP 391
Score = 35.8 bits (81), Expect = 0.023
Identities = 48/190 (25%), Positives = 68/190 (35%), Gaps = 49/190 (25%)
Query: 10 WGYYSPPQPHSTLSP---YAA----AFSVNRPPFNDVSAPFVDSDEP--AYGVPPN---- 56
+ Y PP P+ SP Y++ A+S P+ S P+V S P AY PP+
Sbjct: 260 YAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVY 319
Query: 57 -----------PVHYPVHPRSYGYDFFPESDSVSKPYGYSGLAS-----EDSFWYDQWPP 100
P Y P Y Y P S PY YS S + Y PP
Sbjct: 320 KSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPPYAYSPPPSPYVYKSPPYVYSSPPP 379
Query: 101 TSSKP---SLAEPQPCFPDLNSMGLASEDSFGYEQWLSASSKPSLGEAQPYFPSYIPSTI 157
+ P + + P PC D + ++ +S P + P P
Sbjct: 380 YTYSPPPYAYSPPPPC-----------PDVYKPPPYVYSSPPPYVYNPPPSSPP------ 422
Query: 158 PSPTSSVAAP 167
PSP+ S ++P
Sbjct: 423 PSPSYSYSSP 432
>At1g10390 unknown protein
Length = 1041
Score = 48.1 bits (113), Expect = 5e-06
Identities = 66/228 (28%), Positives = 89/228 (38%), Gaps = 37/228 (16%)
Query: 11 GYYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRS-YGY 69
G + PQ T SP+A+ P F S+P + PA+G +P P S +G
Sbjct: 62 GVFGAPQ---TSSPFAST-----PTFGASSSPAFGNSTPAFGA--SPASSPFGGSSGFGQ 111
Query: 70 DFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCF--PDLNSMGLASEDS 127
S S P+G S S+ +F + SS P A P F P S G S S
Sbjct: 112 KPLGFSTPQSNPFGNSTQQSQPAFGNTSF--GSSTPFGATNTPAFGAPSTPSFGATSTPS 169
Query: 128 FGYEQWLSASSKPSLGEAQ-PYF-----PSYIPSTIP----SPTSSVAAPIHWSSSSGFA 177
FG ASS P+ G P F PS+ + P SPT + + ++GF
Sbjct: 170 FG------ASSTPAFGATNTPAFGASNSPSFGATNTPAFGASPTPAFGSTGTTFGNTGFG 223
Query: 178 PLDVVPSFGDYAAENSSEFGFSGLGAGSWNQFSGFDHGKGKQVGAESS 225
S G + A N+ FG SG A + F GA S+
Sbjct: 224 ------SGGAFGASNTPAFGASGTPAFGASGTPAFGASSTPAFGASST 265
Score = 42.0 bits (97), Expect = 3e-04
Identities = 50/196 (25%), Positives = 76/196 (38%), Gaps = 27/196 (13%)
Query: 13 YSPPQPHSTLSPYAAAFSVNRPPFNDVS----APFVDSDEPAYGVPPNPVHYPVHPRSYG 68
+S PQ + P+ + ++P F + S PF ++ PA+G P P S+G
Sbjct: 116 FSTPQSN----PFGNSTQQSQPAFGNTSFGSSTPFGATNTPAFGAPSTPSFGATSTPSFG 171
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCF----PDLNSMGLAS 124
P + + P G ++ SF P A P P F + G S
Sbjct: 172 ASSTPAFGATNTP--AFGASNSPSFGATNTPAFG-----ASPTPAFGSTGTTFGNTGFGS 224
Query: 125 EDSFGYEQW--LSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSSSSGFAPLDVV 182
+FG AS P+ G + P++ S+ P+ +S SS+ F
Sbjct: 225 GGAFGASNTPAFGASGTPAFGASGT--PAFGASSTPAFGASSTPAFGASSTPAFGG-SST 281
Query: 183 PSFGDYAAENSSEFGF 198
PSFG A N+S F F
Sbjct: 282 PSFG---ASNTSSFSF 294
Score = 37.4 bits (85), Expect = 0.008
Identities = 49/196 (25%), Positives = 70/196 (35%), Gaps = 26/196 (13%)
Query: 5 GSYNDWGYYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP-FVDSDEPAYGVPPNPVHYPVH 63
GS +G + P + +P A S P F S P F ++ PA+G +P +
Sbjct: 141 GSSTPFGATNTPAFGAPSTPSFGATST--PSFGASSTPAFGATNTPAFGASNSPSFGATN 198
Query: 64 PRSYGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPP------------------TSSKP 105
++G P S +G +G S +F P SS P
Sbjct: 199 TPAFGASPTPAFGSTGTTFGNTGFGSGGAFGASNTPAFGASGTPAFGASGTPAFGASSTP 258
Query: 106 SL-AEPQPCF--PDLNSMGLASEDSFGYEQWLSAS--SKPSLGEAQPYFPSYIPSTIPSP 160
+ A P F + G +S SFG S S S P+ G++ F S + PSP
Sbjct: 259 AFGASSTPAFGASSTPAFGGSSTPSFGASNTSSFSFGSSPAFGQSTSAFGSSAFGSTPSP 318
Query: 161 TSSVAAPIHWSSSSGF 176
A SGF
Sbjct: 319 FGGAQASTPTFGGSGF 334
>At5g04170 EF - hand Calcium binding protein - like
Length = 354
Score = 42.7 bits (99), Expect = 2e-04
Identities = 46/175 (26%), Positives = 62/175 (35%), Gaps = 23/175 (13%)
Query: 7 YNDWGYYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRS 66
Y+ G P +T SPYA + ++P + SAP S +YG PP Y P
Sbjct: 28 YSSGGNNPPYGSSTTSSPYAVPYGASKPQSSSSSAPTYGSS--SYGAPPPSAPYAPSPGD 85
Query: 67 YGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSK---PSLAEPQPCFPDLNSMGLA 123
Y KPYG Y PP+ S A P+P P + G
Sbjct: 86 Y------NKPPKEKPYGGG---------YGAPPPSGSSDYGSYGAGPRPSQPSGHGGGYG 130
Query: 124 SEDSFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSSSSGFAP 178
+ G + S P + + Y P P +S +P SGFAP
Sbjct: 131 ATPPHGVSDYGSYGGAPPRPASSGHGGGY--GGYP-PQASYGSPFASLIPSGFAP 182
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 42.4 bits (98), Expect = 2e-04
Identities = 52/168 (30%), Positives = 73/168 (42%), Gaps = 23/168 (13%)
Query: 13 YSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDFF 72
YSP P + SP + ++S P ++ S P P+Y P +P + P P GY
Sbjct: 1657 YSPTSP--SYSPTSPSYSPTSPSYSPTS-PSYSPTSPSYS-PTSPAYSPTSP---GYSPT 1709
Query: 73 PESDSVSKP-YGYSGLA-SEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSFGY 130
S S + P YG + + + S Y P + PS A P P S S Y
Sbjct: 1710 SPSYSPTSPSYGPTSPSYNPQSAKYS--PSIAYSPSNARLSPASP-------YSPTSPNY 1760
Query: 131 EQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSSSSGFAP 178
S S P+ P P+Y PS SP SS A+P +S S+G++P
Sbjct: 1761 SP-TSPSYSPTSPSYSPSSPTYSPS---SPYSSGASP-DYSPSAGYSP 1803
Score = 37.0 bits (84), Expect = 0.010
Identities = 43/167 (25%), Positives = 66/167 (38%), Gaps = 27/167 (16%)
Query: 13 YSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGY--- 69
YSP P + SP + ++S P ++ S P P Y P +P + P P SYG
Sbjct: 1671 YSPTSP--SYSPTSPSYSPTSPSYSPTS-PAYSPTSPGYS-PTSPSYSPTSP-SYGPTSP 1725
Query: 70 DFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEP-QPCFPDLNSMGLASEDSF 128
+ P+S S YS P++++ S A P P P+ + + S+
Sbjct: 1726 SYNPQSAKYSPSIAYS--------------PSNARLSPASPYSPTSPNYSP----TSPSY 1767
Query: 129 GYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSSSSG 175
+ S P+ + PY P PS S P + SS+G
Sbjct: 1768 SPTSPSYSPSSPTYSPSSPYSSGASPDYSPSAGYSPTLPGYSPSSTG 1814
>At1g12040 leucine-rich repeat/extensin 1 (LRX1)
Length = 744
Score = 40.8 bits (94), Expect = 7e-04
Identities = 34/107 (31%), Positives = 41/107 (37%), Gaps = 17/107 (15%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEP---AYGVPPNP--VHYPVHPRS 66
Y PP P+S +SP A+ PP P+V S P Y PP P V+ P
Sbjct: 439 YSPPPPPYSKMSPSVRAYPPPPPPSPSPPPPYVYSSPPPPYVYSSPPPPPYVYSSPPPPP 498
Query: 67 YGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPC 113
Y Y S PY YS + + Y PP P P PC
Sbjct: 499 YVY------SSPPPPYVYS--SPPPPYVYSSPPPPPPSP----PPPC 533
Score = 33.9 bits (76), Expect = 0.089
Identities = 46/166 (27%), Positives = 58/166 (34%), Gaps = 44/166 (26%)
Query: 15 PPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVH--------YPVHPRS 66
PP P S +SP A+S PP + +S P V AY PP P YP P
Sbjct: 408 PPPPSSKMSPTVRAYSPPPPPSSKMS-PSV----RAYSPPPPPYSKMSPSVRAYPPPP-- 460
Query: 67 YGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASED 126
P S S PY YS + + Y PP S P P
Sbjct: 461 ------PPSPSPPPPYVYS--SPPPPYVYSSPPPPPYVYSSPPPPP-------------- 498
Query: 127 SFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSS 172
++ +S P + P P Y+ S+ P P S P SS
Sbjct: 499 ------YVYSSPPPPYVYSSPP-PPYVYSSPPPPPPSPPPPCPESS 537
Score = 32.7 bits (73), Expect = 0.20
Identities = 45/167 (26%), Positives = 60/167 (34%), Gaps = 27/167 (16%)
Query: 15 PPQPHSTLSP---YAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDF 71
PP P S+ P Y A + + PP + V P V P PP+PV+YP S
Sbjct: 531 PPCPESSPPPPVVYYAPVTQSPPPPSPVYYPPVTQSPP----PPSPVYYPPVTNSPP--- 583
Query: 72 FPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSFGYE 131
P S P YS +Y Q P+ PS P P S Y
Sbjct: 584 -PPSPVYYPPVTYSP-PPPSPVYYPQVTPSPPPPSPLYYPPVTPS------PPPPSPVYY 635
Query: 132 QWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSSSSGFAP 178
++ S P P Y P PSP +P+++ S + P
Sbjct: 636 PPVTPSPPPP-------SPVYYPPVTPSPPP--PSPVYYPSETQSPP 673
Score = 29.6 bits (65), Expect = 1.7
Identities = 30/115 (26%), Positives = 37/115 (32%), Gaps = 18/115 (15%)
Query: 12 YYSP--PQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRS--- 66
YY P P P Y + + PP + V P V P PP+PV+YP +S
Sbjct: 619 YYPPVTPSPPPPSPVYYPPVTPSPPPPSPVYYPPVTPSPP----PPSPVYYPSETQSPPP 674
Query: 67 -----YGYDFFPESDSVSK----PYGYSGLASEDSFWYDQWPPTSSKPSLAEPQP 112
Y P K P + Y PP S S P P
Sbjct: 675 PTEYYYSPSQSPPPTKACKEGHPPQATPSYEPPPEYSYSSSPPPPSPTSYFPPMP 729
>At2g28440 En/Spm-like transposon protein
Length = 268
Score = 40.0 bits (92), Expect = 0.001
Identities = 56/210 (26%), Positives = 66/210 (30%), Gaps = 43/210 (20%)
Query: 23 SPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDFFPESDSV---- 78
SP AA S R P D +P S P P +P P PE DS
Sbjct: 27 SPSPAAVSPGREPSTD--SPLSPSSSPEEDSPLSPSSSPEEDSPLPPSSSPEEDSPLAPS 84
Query: 79 SKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSFGYEQWLSASS 138
S P S LA S D P SS P P P + S S + L+ S
Sbjct: 85 SSPEVDSPLAPSSSPEVDSPQPPSSSPEADSPLPPSSSPEANSPQSPASSPKPESLADSP 144
Query: 139 KPSLGEAQPYFPS---------------------------YIPSTIPSPTSSVAAPIHWS 171
P QP PS Y PS PSP +A I S
Sbjct: 145 SPPPPPPQPESPSSPSYPEPAPVPAPSDDDSDDDPEPETEYFPSPAPSPELGMAQDIKAS 204
Query: 172 SSSGFAPLDVVPSFGDYAAENSSEFGFSGL 201
++G D E ++G SGL
Sbjct: 205 DAAGEELND----------ERGEDYGMSGL 224
>At1g62970 unknown protein
Length = 825
Score = 39.3 bits (90), Expect = 0.002
Identities = 50/176 (28%), Positives = 78/176 (43%), Gaps = 23/176 (13%)
Query: 14 SPPQPHSTLSPYAAAFSVNRPPFNDVSAPF-VDSDEPAYGVPPNPVHYPVHPRSYGYDFF 72
S P P S + F V++P N S PF V +PA P PV P P S +
Sbjct: 438 SKPLPVSQSLQNSNPFPVSQPSSN--SKPFPVSQPQPASN--PFPVSQP-RPNSQPFSMS 492
Query: 73 PESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSFGYEQ 132
S S ++P+ S + + PPT+SKP +++P P+ + S+
Sbjct: 493 QPS-STARPFPASQPPAASKSFPISQPPTTSKPFVSQP----PNTSKPMPVSQP------ 541
Query: 133 WLSASSKP-SLGEAQPYFPSYIPSTIPSPTSSVA-APIHWSSSSGF--APLDVVPS 184
+SKP + + P F S PS P+ +SS++ P ++S+ F P+ PS
Sbjct: 542 --PTTSKPLPVSQPPPTFQSTCPSQPPAASSSLSPLPPVFNSTQSFQSPPVSTTPS 595
Score = 32.0 bits (71), Expect = 0.34
Identities = 40/155 (25%), Positives = 58/155 (36%), Gaps = 18/155 (11%)
Query: 15 PPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDFFPE 74
PP P S P + F +++PP N P S + + P + P +
Sbjct: 379 PPFPMSQPPPTSNPFPLSQPPSNSKPFPMSQSSQNSKPFPVSQSSQKSKPL-----LVSQ 433
Query: 75 SDSVSKPYGYS-GLASEDSFWYDQWPPTSSKP-SLAEPQPCF-------PDLNSMGLASE 125
S SKP S L + + F Q P ++SKP +++PQP P NS +
Sbjct: 434 SSQRSKPLPVSQSLQNSNPFPVSQ-PSSNSKPFPVSQPQPASNPFPVSQPRPNSQPFSMS 492
Query: 126 DSFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSP 160
+ AS P+ A FP P T P
Sbjct: 493 QPSSTARPFPASQPPA---ASKSFPISQPPTTSKP 524
>At2g15880 unknown protein
Length = 727
Score = 37.4 bits (85), Expect = 0.008
Identities = 22/61 (36%), Positives = 23/61 (37%), Gaps = 1/61 (1%)
Query: 13 YSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDFF 72
YSPP P SP FS PP + P P Y PP PV P P Y
Sbjct: 614 YSPPPPPPVHSPPPPVFSPP-PPVHSPPPPVYSPPPPVYSPPPPPVKSPPPPPVYSPPLL 672
Query: 73 P 73
P
Sbjct: 673 P 673
Score = 32.3 bits (72), Expect = 0.26
Identities = 20/52 (38%), Positives = 21/52 (39%)
Query: 13 YSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHP 64
YSPP P SP + PP P V S P PP PVH P P
Sbjct: 525 YSPPPPPPVYSPPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPP 576
Score = 32.0 bits (71), Expect = 0.34
Identities = 21/54 (38%), Positives = 22/54 (39%), Gaps = 2/54 (3%)
Query: 13 YSPPQPHSTLSPYAA--AFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHP 64
YSPP P SP +S PP P V S P PP PVH P P
Sbjct: 516 YSPPPPPPVYSPPPPPPVYSPPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPP 569
Score = 31.2 bits (69), Expect = 0.57
Identities = 19/53 (35%), Positives = 22/53 (40%), Gaps = 5/53 (9%)
Query: 13 YSPPQP-HSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHP 64
+SPP P HS P + PP + P P Y PP PVH P P
Sbjct: 550 HSPPPPVHSPPPPVHSP----PPPVHSPPPPVHSPPPPVYSPPPPPVHSPPPP 598
Score = 30.0 bits (66), Expect = 1.3
Identities = 22/58 (37%), Positives = 27/58 (45%), Gaps = 6/58 (10%)
Query: 13 YSPPQP--HSTLSP-YAAAFSVNRPP---FNDVSAPFVDSDEPAYGVPPNPVHYPVHP 64
YSPP P HS P ++ V+ PP ++ P V S P PP PVH P P
Sbjct: 585 YSPPPPPVHSPPPPVHSPPPPVHSPPPPVYSPPPPPPVHSPPPPVFSPPPPVHSPPPP 642
Score = 28.9 bits (63), Expect = 2.9
Identities = 20/53 (37%), Positives = 22/53 (40%), Gaps = 5/53 (9%)
Query: 13 YSPPQP-HSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHP 64
+SPP P HS P + PP P V S P PP PVH P P
Sbjct: 564 HSPPPPVHSPPPPVHSP----PPPVYSPPPPPVHSPPPPVHSPPPPVHSPPPP 612
Score = 27.3 bits (59), Expect = 8.3
Identities = 19/52 (36%), Positives = 22/52 (41%), Gaps = 2/52 (3%)
Query: 13 YSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHP 64
+SPP P SP +S PP P V S P PP PV+ P P
Sbjct: 600 HSPPPP--VHSPPPPVYSPPPPPPVHSPPPPVFSPPPPVHSPPPPVYSPPPP 649
>At1g23720 unknown protein
Length = 895
Score = 36.6 bits (83), Expect = 0.014
Identities = 46/158 (29%), Positives = 59/158 (37%), Gaps = 27/158 (17%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + SP + PP+ S P + S +PAY PP P Y P Y
Sbjct: 213 YSSPPPPYYSPSP-KPVYKSPPPPYVYSSPPPPYYSPSPKPAYKSPPPPYVYSSPPPPY- 270
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSF 128
Y P+ S P Y Y+ PP PS P+P + S SF
Sbjct: 271 YSPSPKPIYKSPPPPYV---------YNSPPPPYYSPS---PKPAY---KSPPPPYVYSF 315
Query: 129 GYEQWLSASSKPSLGEAQPYF-------PSYIPSTIPS 159
+ S S KP P + P Y PS P+
Sbjct: 316 PPPPYYSPSPKPVYKSPPPPYVYNSPPPPYYSPSPKPA 353
Score = 36.6 bits (83), Expect = 0.014
Identities = 35/108 (32%), Positives = 43/108 (39%), Gaps = 18/108 (16%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + SP A+ PP+ S P + S +P Y PP P Y P Y
Sbjct: 338 YNSPPPPYYSPSP-KPAYKSPPPPYVYSSPPPPYYSPSPKPTYKSPPPPYVYSSPPPPY- 395
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPP----TSSKPSLAEPQP 112
Y P+ S P Y Y+ PP S KPS P P
Sbjct: 396 YSPSPKPVYKSPPPPYI---------YNSPPPPYYSPSPKPSYKSPPP 434
Score = 36.2 bits (82), Expect = 0.018
Identities = 45/152 (29%), Positives = 61/152 (39%), Gaps = 23/152 (15%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + SP + PP+ S P + S +PAY PP P Y P Y
Sbjct: 263 YSSPPPPYYSPSP-KPIYKSPPPPYVYNSPPPPYYSPSPKPAYKSPPPPYVYSFPPPPY- 320
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSF 128
Y P+ S P Y Y+ PP PS P+P + + S
Sbjct: 321 YSPSPKPVYKSPPPPYV---------YNSPPPPYYSPS---PKPAYKSPPPPYVYSSPPP 368
Query: 129 GYEQWLSASSKPSLGEAQPYFPSYIPSTIPSP 160
Y S S KP+ ++ P P Y+ S+ P P
Sbjct: 369 PY---YSPSPKPTY-KSPP--PPYVYSSPPPP 394
Score = 33.5 bits (75), Expect = 0.12
Identities = 34/108 (31%), Positives = 43/108 (39%), Gaps = 18/108 (16%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + SP + PP+ S P + S +P Y PP P Y P Y
Sbjct: 163 YSSPPPPYYSPSP-KVDYKSPPPPYVYNSPPPPYYSPSPKPTYKSPPPPYIYSSPPPPY- 220
Query: 69 YDFFPESDSVSK----PYGYSGLASEDSFWYDQWPPTSSKPSLAEPQP 112
+ P V K PY YS S +Y P KP+ P P
Sbjct: 221 --YSPSPKPVYKSPPPPYVYS---SPPPPYYSPSP----KPAYKSPPP 259
Score = 33.1 bits (74), Expect = 0.15
Identities = 32/108 (29%), Positives = 41/108 (37%), Gaps = 18/108 (16%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + +P + PP+ S P + S +P Y PP P Y P Y
Sbjct: 640 YSSPPPPYYSPTP-KPTYKSPPPPYVYSSPPPPYYSPSPKPTYKSPPPPYVYSSPPPPY- 697
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPP----TSSKPSLAEPQP 112
Y P+ S P Y Y PP S KP+ P P
Sbjct: 698 YSPAPKPTYKSPPPPYV---------YSSPPPPYYSPSPKPTYKSPPP 736
Score = 32.7 bits (73), Expect = 0.20
Identities = 45/166 (27%), Positives = 64/166 (38%), Gaps = 27/166 (16%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + SP + PP+ S P + S +P+Y PP P Y P Y
Sbjct: 388 YSSPPPPYYSPSP-KPVYKSPPPPYIYNSPPPPYYSPSPKPSYKSPPPPYVYSSPPPPY- 445
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPPT--SSKPSLAEPQPCFPDLNSMGLASED 126
Y P+ S P Y Y PP S P + P P + S
Sbjct: 446 YSPSPKLTYKSSPPPYV---------YSSPPPPYYSPSPKVVYKSPPPPYVYSS------ 490
Query: 127 SFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSS 172
+ S S KPS ++ P P Y+ ++ P P S + + + S
Sbjct: 491 --PPPPYYSPSPKPSY-KSPP--PPYVYNSPPPPYYSPSPKVIYKS 531
Score = 32.3 bits (72), Expect = 0.26
Identities = 48/180 (26%), Positives = 62/180 (33%), Gaps = 37/180 (20%)
Query: 4 AGSYNDWGYYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVH 63
A SY + Y SPP PP D P VD Y PP P Y
Sbjct: 3 AASYEPYTYSSPP-----------------PPLYDSPTPKVD-----YKSPPPPYVYSSP 40
Query: 64 PRSYGYDFFPESD--SVSKPYGYSG-----LASEDSFWYDQWPP----TSSKPSLAEPQP 112
P Y P+ D S PY YS + Y PP +S P P P
Sbjct: 41 PPPLSYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPYYSPSP 100
Query: 113 CFPDLNSMGLASEDSFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSS 172
D S S + S S KP+ ++ P P Y+ ++ P P S + + + S
Sbjct: 101 KV-DYKSPPPPYVYSSPPPPYYSPSPKPTY-KSPP--PPYVYNSPPPPYYSPSPKVEYKS 156
Score = 32.0 bits (71), Expect = 0.34
Identities = 26/79 (32%), Positives = 33/79 (40%), Gaps = 6/79 (7%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHY--PVHPRS 66
Y SPP P+ + +P + PP+ S P + S +P Y PP P Y P P
Sbjct: 690 YSSPPPPYYSPAP-KPTYKSPPPPYVYSSPPPPYYSPSPKPTYKSPPPPYVYSSPPPPPY 748
Query: 67 YGYDFFPESDSVSKPYGYS 85
Y E S PY YS
Sbjct: 749 YSPSPKVEYKSPPPPYVYS 767
Score = 30.0 bits (66), Expect = 1.3
Identities = 47/182 (25%), Positives = 60/182 (32%), Gaps = 38/182 (20%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP-------------FVDSDEPAYGVPPNPV 58
Y SPP PH + P + P S P + S +PAY P P
Sbjct: 529 YKSPPHPHVCVCPPPPPCYSHSPKIEYKSPPTPYVYHSPPPPPYYSPSPKPAYKSSPPPY 588
Query: 59 HYPVHPRSYGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLN 118
Y P Y Y P+ S P Y Y+ PP PS P+P +
Sbjct: 589 VYSSPPPPY-YSPAPKPVYKSPPPPYV---------YNSPPPPYYSPS---PKPTYKSPP 635
Query: 119 SMGLASEDSFGYEQWLSASSKPSLGEAQPYF-------PSYIPSTIPSPTSSVAAPIHWS 171
+ S Y S + KP+ P + P Y PS P PT P +
Sbjct: 636 PPYVYSSPPPPY---YSPTPKPTYKSPPPPYVYSSPPPPYYSPS--PKPTYKSPPPPYVY 690
Query: 172 SS 173
SS
Sbjct: 691 SS 692
>At3g19020 hypothetical protein
Length = 951
Score = 35.4 bits (80), Expect = 0.030
Identities = 48/175 (27%), Positives = 58/175 (32%), Gaps = 22/175 (12%)
Query: 13 YSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDFF 72
+SPP P SP PP + P V S P PP PVH P P
Sbjct: 735 FSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPP-------- 786
Query: 73 PESDSVSKPYGYSGLASEDSFWYDQWPPTSS---KPSLAEPQPCFPDLNSMGLASEDSFG 129
V P S Y PP S KP P P N+ +S +S
Sbjct: 787 -----VHSPPPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMANAPTPSSSESGE 841
Query: 130 YEQWLSASSKPSLGEAQP----YFPSYIPSTIPSPTS--SVAAPIHWSSSSGFAP 178
+ A + S P + P + S PSP S V AP+ S AP
Sbjct: 842 ISTPVQAPTPDSEDIEAPSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSEPEVEAP 896
Score = 35.0 bits (79), Expect = 0.040
Identities = 19/52 (36%), Positives = 22/52 (41%), Gaps = 1/52 (1%)
Query: 13 YSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHP 64
+SPP P SP FS PP + P P + PP PVH P P
Sbjct: 640 HSPPPPPPVHSPPPPVFSPP-PPMHSPPPPVYSPPPPVHSPPPPPVHSPPPP 690
Score = 30.8 bits (68), Expect = 0.75
Identities = 23/56 (41%), Positives = 27/56 (48%), Gaps = 4/56 (7%)
Query: 13 YSPPQP-HSTLSP-YAAAFSVNRPPFNDVSAPF--VDSDEPAYGVPPNPVHYPVHP 64
+SPP P HS P Y+ V+ PP V +P V S P PP PVH P P
Sbjct: 656 FSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPP 711
Score = 28.1 bits (61), Expect = 4.9
Identities = 23/56 (41%), Positives = 26/56 (46%), Gaps = 4/56 (7%)
Query: 13 YSPPQP-HSTLSP--YAAAFSVNRPPFNDVSAPF-VDSDEPAYGVPPNPVHYPVHP 64
YSPP P HS P ++ V+ PP S P V S P PP PVH P P
Sbjct: 670 YSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPP 725
>At4g18670 extensin-like protein
Length = 839
Score = 34.7 bits (78), Expect = 0.052
Identities = 44/173 (25%), Positives = 66/173 (37%), Gaps = 27/173 (15%)
Query: 16 PQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDFFPES 75
P P +T SP + S + P ++ P + G PP+P P P + P +
Sbjct: 411 PSPPTTPSPGGSPPSPSISPSPPITVPSPPTTPSPGGSPPSPSIVPSPPSTTPSPGSPPT 470
Query: 76 DSVSKPYGYSGLASEDSFWYDQWPPTS-SKPSLAEPQPCFPDLNSMGLASEDSFGYEQWL 134
+ G S PP+S + P+ P P + G +
Sbjct: 471 SPTTPTPGGS-------------PPSSPTTPTPGGSPPSSPTTPTPGGSPP--------- 508
Query: 135 SASSKPSLGEAQPYFPSYIPS---TIPSPTSSVAAPIHWSSSSGFAPLDVVPS 184
S+ + PS G + P PS PS T+PSP S+ +P S S P +PS
Sbjct: 509 SSPTTPSPGGSPPS-PSISPSPPITVPSPPSTPTSPGSPPSPSSPTPSSPIPS 560
Score = 33.1 bits (74), Expect = 0.15
Identities = 35/113 (30%), Positives = 43/113 (37%), Gaps = 13/113 (11%)
Query: 12 YYS----PPQPHSTLSPYAAAFSV--NRPPFNDVSAPFVDSDEPA--YGVPPNP-VHY-P 61
YYS PP PH +L P + PP + +P S P Y PP P VHY P
Sbjct: 709 YYSSPQPPPPPHYSLPPPTPTYHYISPPPPPTPIHSPPPQSHPPCIEYSPPPPPTVHYNP 768
Query: 62 VHPRSYGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCF 114
P S + P S V Y Y+ + Y PP S P P +
Sbjct: 769 PPPPSPAHYSPPPSPPV---YYYNSPPPPPAVHYSPPPPPVIHHSQPPPPPIY 818
Score = 28.5 bits (62), Expect = 3.7
Identities = 45/165 (27%), Positives = 53/165 (31%), Gaps = 37/165 (22%)
Query: 14 SPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNP----------VHYPVH 63
SPP +T +P + S P S P + G PP+P P
Sbjct: 480 SPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSPPSPSISPSPPITVPSPPST 539
Query: 64 PRSYGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLA 123
P S G P S + S P PPT S P P P P NS +
Sbjct: 540 PTSPGSPPSPSSPTPSSPI--------------PSPPTPSTP----PTPISPGQNSPPII 581
Query: 124 SEDSFGYEQWLSASSKPSLGEAQPYFPSYIPS-TIPSPTSSVAAP 167
F + S PS P P IPS I PT S P
Sbjct: 582 PSPPF------TGPSPPS--SPSPPLPPVIPSPPIVGPTPSSPPP 618
>At3g28550 unknown protein
Length = 1018
Score = 34.7 bits (78), Expect = 0.052
Identities = 43/167 (25%), Positives = 64/167 (37%), Gaps = 12/167 (7%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + SP + PP+ S P + S + Y PP P Y P Y
Sbjct: 544 YSSPPPPYYSPSP-KVVYKSPPPPYVYSSPPPPYYSPSPKVVYKSPPPPYVYSSPPPPY- 601
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCF-PDLNSMGLASEDS 127
Y P+ S P Y + + + + PP P PC+ P + +S
Sbjct: 602 YSPSPKVYYKSPPSPYHAPSPKVLY---KSPPHPHVCVCPPPPPCYSPSPKVVYKSSPPP 658
Query: 128 FGYEQWLSA--SSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSS 172
+ Y S P + P P Y+ S+ P P S + +H+ S
Sbjct: 659 YVYSSPPPPYHSPSPKVHYKSPP-PPYVYSSPPPPYYSPSPKVHYKS 704
Score = 29.3 bits (64), Expect = 2.2
Identities = 21/62 (33%), Positives = 25/62 (39%), Gaps = 7/62 (11%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDF 71
Y SPP P+ + SP + PP+ S P P Y P VHY P Y Y
Sbjct: 661 YSSPPPPYHSPSP-KVHYKSPPPPYVYSSPP------PPYYSPSPKVHYKSPPPPYVYSS 713
Query: 72 FP 73
P
Sbjct: 714 PP 715
>At4g33970 extensin-like protein
Length = 699
Score = 33.9 bits (76), Expect = 0.089
Identities = 35/108 (32%), Positives = 38/108 (34%), Gaps = 14/108 (12%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDF 71
Y PP P SP FS PP P V S P PP PVH P P
Sbjct: 556 YSPPPPPPPVHSPPPPVFS---PP------PPVYSPPPPVHSPPPPVHSPPPPAPVHSPP 606
Query: 72 FPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNS 119
P P YS S PP+ S P + P P P +NS
Sbjct: 607 PPVHSPPPPPPVYSPPPPVFSP-----PPSQSPPVVYSPPPRPPKINS 649
>At3g54580 extensin precursor -like protein
Length = 951
Score = 33.9 bits (76), Expect = 0.089
Identities = 46/175 (26%), Positives = 65/175 (36%), Gaps = 19/175 (10%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + SP + PP+ S P + S + Y PP P Y P Y
Sbjct: 593 YSSPPPPYHSPSP-KVQYKSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPY- 650
Query: 69 YDFFPES--DSVSKPYGYSG----LASEDSFWYDQWPPTSSKPSLAEPQPCF-PDLNSMG 121
Y P+ S PY YS S Y + PP P PC+ P +
Sbjct: 651 YSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYKSPPHPHVCVCPPPPPCYSPSPKVVY 710
Query: 122 LASEDSFGYEQWLSASSKPSLGEAQPYF----PSYIPSTIPSPTSSVAAPIHWSS 172
+ + Y PS + Y+ P Y+ S+ P P S + +H+ S
Sbjct: 711 KSPPPPYVYSSPPPPHYSPS---PKVYYKSPPPPYVYSSPPPPYYSPSPKVHYKS 762
Score = 30.4 bits (67), Expect = 0.98
Identities = 23/71 (32%), Positives = 26/71 (36%), Gaps = 9/71 (12%)
Query: 12 YYSPPQPHSTLSPYAA--------AFSVNRPPFNDVSAPF-VDSDEPAYGVPPNPVHYPV 62
Y SPP PH + SP +S PP+ S S P Y P VHY
Sbjct: 719 YSSPPPPHYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYYAPTPKVHYKS 778
Query: 63 HPRSYGYDFFP 73
P Y Y P
Sbjct: 779 PPPPYVYSSPP 789
Score = 29.3 bits (64), Expect = 2.2
Identities = 35/162 (21%), Positives = 52/162 (31%), Gaps = 11/162 (6%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNP-VHYPVHPRSYGYD 70
Y +PP P+ SP + P+V S P P+P V Y P Y Y
Sbjct: 44 YKTPPLPYIDSSPPPTYSPAPEVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYS 103
Query: 71 FFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSFGY 130
P + S + + Y PP + PS P ++
Sbjct: 104 -SPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSP 162
Query: 131 EQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSS 172
+ S P P Y+ S+ P PT S + + + S
Sbjct: 163 SPKVEYKSPP---------PPYVYSSPPPPTYSPSPKVEYKS 195
Score = 28.5 bits (62), Expect = 3.7
Identities = 26/84 (30%), Positives = 30/84 (34%), Gaps = 17/84 (20%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDF 71
Y SPP P+ +P + PP+ S P P Y P VHY P Y Y
Sbjct: 760 YKSPPPPYYAPTP-KVHYKSPPPPYVYSSPP------PPYYSPSPKVHYKSPPPPYVYSS 812
Query: 72 FP----------ESDSVSKPYGYS 85
P E S PY YS
Sbjct: 813 PPPPYYSPSPKVEYKSPPPPYVYS 836
Score = 27.3 bits (59), Expect = 8.3
Identities = 34/130 (26%), Positives = 44/130 (33%), Gaps = 30/130 (23%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVS---------APFV-DSDEPAYGVPPNPVHYP 61
Y SPP P+ + SP + PP+ S P+V S P Y P V+Y
Sbjct: 527 YSSPPPPYYSPSP-KVYYKSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYK 585
Query: 62 VHPRSYGYDFFP----------ESDSVSKPYGYSG-----LASEDSFWYDQWPP----TS 102
P Y Y P + S PY YS + +Y PP +S
Sbjct: 586 SPPPPYVYSSPPPPYHSPSPKVQYKSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSS 645
Query: 103 SKPSLAEPQP 112
P P P
Sbjct: 646 PPPPYYSPSP 655
>At1g59660 nucleoporin 98-like protein
Length = 997
Score = 33.9 bits (76), Expect = 0.089
Identities = 47/174 (27%), Positives = 62/174 (35%), Gaps = 27/174 (15%)
Query: 3 GAGSYNDWGYYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP-FVDSDEPAYGVPPNPVHYP 61
G G+ N G+ P S A S N P F S P F S PA+G P P
Sbjct: 179 GFGATNTTGFGGSSTPGFGASSTPAFGSTNTPAFGASSTPLFGSSSSPAFGASPAP---- 234
Query: 62 VHPRSYGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMG 121
++G S +G + +S +F P + + A P N
Sbjct: 235 ----AFG--------SSGNAFGNNTFSSGGAFGSSSTPTFGASNTSAFGASSSPSFN--- 279
Query: 122 LASEDSFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSSSSG 175
S +FG Q SA S G Q S + ST PSP + A S+ G
Sbjct: 280 FGSSPAFG--QSTSAFGSSSFGSTQ----SSLGST-PSPFGAQGAQASTSTFGG 326
Score = 28.5 bits (62), Expect = 3.7
Identities = 34/127 (26%), Positives = 51/127 (39%), Gaps = 21/127 (16%)
Query: 100 PTSSKPSLAEPQPCFPDLNSMGLASEDSFGYEQWLSASSKPSLGEA---QPYFPSYIPST 156
P +S P A PQ + G +S SFG +S+ P G + Q F P +
Sbjct: 70 PQTSSPFGASPQAFGSSTQAFGASSTPSFG------SSNSPFGGTSTFGQKSFGLSTPQS 123
Query: 157 IP-SPTSSVAAPIHWSSSSGFAP---LDVVPSFG-----DYAAENSSEFGFS---GLGAG 204
P T+ + P +S+ G + P+FG + N+S FG + G GA
Sbjct: 124 SPFGSTTQQSQPAFGNSTFGSSTPFGASTTPAFGASSTPAFGVSNTSGFGATNTPGFGAT 183
Query: 205 SWNQFSG 211
+ F G
Sbjct: 184 NTTGFGG 190
>At2g43150 putative extensin
Length = 212
Score = 33.5 bits (75), Expect = 0.12
Identities = 43/163 (26%), Positives = 57/163 (34%), Gaps = 19/163 (11%)
Query: 14 SPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEP--AYGVPPNPVHYPVHPRSYGYDF 71
SPP P+ SP S P + P V S P Y PP PV P P Y +
Sbjct: 51 SPPPPYEYKSPPPPVKSPPPPYYYHSPPPPVKSPPPPYVYSSPPPPVKSPPPPYYY-HSP 109
Query: 72 FPESDSVSKPYGYSG-----LASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASED 126
P S PY Y + ++Y PP P P P + + S
Sbjct: 110 PPPVKSPPPPYYYHSPPPPVKSPPPPYYYHSPPPPVKSP----PPPYYYHSPPPPVKSPP 165
Query: 127 SFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIH 169
Y S P ++ P P Y+ S+ P P S P++
Sbjct: 166 PPYYYH-----SPPPPVKSPP--PPYLYSSPPPPVKSPPPPVY 201
Score = 30.4 bits (67), Expect = 0.98
Identities = 39/155 (25%), Positives = 52/155 (33%), Gaps = 22/155 (14%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSYGYDF 71
YY P P P +S PP P+ Y PP PV P P Y +
Sbjct: 73 YYHSPPPPVKSPPPPYVYSSPPPPVKSPPPPYY------YHSPPPPVKSPPPPYYY-HSP 125
Query: 72 FPESDSVSKPYGYSG-----LASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASED 126
P S PY Y + ++Y PP P P P + + S
Sbjct: 126 PPPVKSPPPPYYYHSPPPPVKSPPPPYYYHSPPPPVKSP----PPPYYYHSPPPPVKSPP 181
Query: 127 SFGYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPT 161
+L +S P + P P YI ++ P PT
Sbjct: 182 ----PPYLYSSPPPPVKSPPP--PVYIYASPPPPT 210
>At5g51300 unknown protein
Length = 804
Score = 33.1 bits (74), Expect = 0.15
Identities = 44/162 (27%), Positives = 61/162 (37%), Gaps = 15/162 (9%)
Query: 14 SPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVD---SDEPAYGVPPNPVH--YPVHPRSYG 68
+PP P + P N+PP S + S P PP P + Y + P G
Sbjct: 564 APPGPPAPQPPTQGYPPSNQPPGAYPSQQYATGGYSTAPVPWGPPVPSYSPYALPPPPPG 623
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCF-PDLNSMGLASEDS 127
+ P PYG P T+ PS +EPQ F P + + A+ S
Sbjct: 624 -SYHPVHGQHMPPYGMQYPPPPPHVTQAPPPGTTQNPSSSEPQQSFPPGVQADSGAATSS 682
Query: 128 FGYEQWLSASSKPSLGEAQPY--FPSYI----PSTIPSPTSS 163
+ SS ++ PY +PSY P T P+P SS
Sbjct: 683 IPPNVY--GSSVTAMPGQPPYMSYPSYYNAVPPPTPPAPASS 722
>At3g54590 extensin precursor -like protein
Length = 743
Score = 33.1 bits (74), Expect = 0.15
Identities = 44/164 (26%), Positives = 62/164 (36%), Gaps = 23/164 (14%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAP---FVDSDEPAYGVPPNPVHYPVHPRSYG 68
Y SPP P+ + SP + PP+ S P + S + Y PP P Y P Y
Sbjct: 277 YSSPPPPYYSPSP-KVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPY- 334
Query: 69 YDFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSF 128
Y P+ D S P Y Y PP + PS P+ + + S
Sbjct: 335 YSPSPKVDYKSPPPPYV---------YSSPPPPTYSPS---PKVDYKSPPPPYVYSSPPP 382
Query: 129 GYEQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSS 172
Y S P + P P Y+ S+ P PT S + +++ S
Sbjct: 383 PYY-----SPSPKVEYKSPP-PPYVYSSPPPPTYSPSPKVYYKS 420
Score = 31.2 bits (69), Expect = 0.57
Identities = 28/97 (28%), Positives = 38/97 (38%), Gaps = 3/97 (3%)
Query: 12 YYSPPQPHSTLSPYAAAFSVN-RPPFNDVSAPFVDSDEPAYGVPPNP-VHYPVHPRSYGY 69
Y SPP P+ SP +S + + + P+V S P P+P VHY P Y Y
Sbjct: 518 YKSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYVY 577
Query: 70 DFFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPS 106
+ P P Y + + Y PP PS
Sbjct: 578 NSPPPPYYSPSPKVYY-KSPPPPYVYSSPPPPYYSPS 613
Score = 30.0 bits (66), Expect = 1.3
Identities = 42/162 (25%), Positives = 60/162 (36%), Gaps = 19/162 (11%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNP-VHYPVHPRSYGYD 70
Y SPP P+ + SP + + PP P+V S P P+P V+Y P Y Y
Sbjct: 427 YSSPPPPYYSPSP--KVYYKSPPP------PYVYSSPPPPYYSPSPKVYYKSPPPPYVYS 478
Query: 71 FFPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPSLAEPQPCFPDLNSMGLASEDSFGY 130
P P Y + + Y PP PS P+ + + S Y
Sbjct: 479 SPPPPYYSPSPKVYYK-SPPPPYVYSSPPPPYYSPS---PKVYYKSPPPPYVYSSPPPPY 534
Query: 131 EQWLSASSKPSLGEAQPYFPSYIPSTIPSPTSSVAAPIHWSS 172
S P + P P Y+ S+ P P S + +H+ S
Sbjct: 535 Y-----SPSPKVHYKSPP-PPYVYSSPPPPYYSPSPKVHYKS 570
Score = 27.7 bits (60), Expect = 6.4
Identities = 29/102 (28%), Positives = 36/102 (34%), Gaps = 11/102 (10%)
Query: 6 SYNDWGYYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPR 65
+Y Y SPP +S+ P PP P+VDS P P V Y P
Sbjct: 22 AYEPETYASPPPLYSSPLPEV---EYKTPPL-----PYVDSSPPPTYTPAPEVEYKSPPP 73
Query: 66 SYGYDF-FPESDSVSKPYGYSGLASEDSFWYDQWPPTSSKPS 106
Y Y P + S S Y + + Y PP PS
Sbjct: 74 PYVYSSPPPPTYSPSPKVDYK--SPPPPYVYSSPPPPYYSPS 113
>At1g76930 extensin
Length = 373
Score = 33.1 bits (74), Expect = 0.15
Identities = 22/58 (37%), Positives = 25/58 (42%), Gaps = 4/58 (6%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVD--SDEPAYGVPPNPVHYPVHPRSY 67
YYSPP + + P +S PP P V S P Y PP PVHY P Y
Sbjct: 200 YYSPPPVYKSPPPPVKHYSP--PPVYKSPPPPVKYYSPPPVYKSPPPPVHYSPPPVVY 255
Score = 29.3 bits (64), Expect = 2.2
Identities = 20/56 (35%), Positives = 23/56 (40%), Gaps = 7/56 (12%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSY 67
Y+SPP P SP + PP + P V Y PP PVHY P Y
Sbjct: 271 YHSPPPPVH-YSPPPVVYHSPPPPVHYSPPPVV------YHSPPPPVHYSPPPVVY 319
Score = 29.3 bits (64), Expect = 2.2
Identities = 20/56 (35%), Positives = 23/56 (40%), Gaps = 7/56 (12%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSY 67
Y+SPP P SP + PP + P V Y PP PVHY P Y
Sbjct: 255 YHSPPPPVH-YSPPPVVYHSPPPPVHYSPPPVV------YHSPPPPVHYSPPPVVY 303
Score = 28.9 bits (63), Expect = 2.9
Identities = 20/56 (35%), Positives = 21/56 (36%), Gaps = 7/56 (12%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSY 67
Y SPP P SP + PP S P P Y PP PV Y P Y
Sbjct: 39 YKSPPPPVKHYSP-PPVYKSPPPPVKHYSPP------PVYKSPPPPVKYYSPPPVY 87
Score = 28.9 bits (63), Expect = 2.9
Identities = 20/56 (35%), Positives = 21/56 (36%), Gaps = 7/56 (12%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSY 67
Y SPP P SP + PP S P P Y PP PV Y P Y
Sbjct: 127 YKSPPPPVKHYSP-PPVYKSPPPPVKHYSPP------PVYKSPPPPVKYYSPPPVY 175
Score = 28.9 bits (63), Expect = 2.9
Identities = 20/56 (35%), Positives = 21/56 (36%), Gaps = 7/56 (12%)
Query: 12 YYSPPQPHSTLSPYAAAFSVNRPPFNDVSAPFVDSDEPAYGVPPNPVHYPVHPRSY 67
Y SPP P SP + PP S P P Y PP PV Y P Y
Sbjct: 159 YKSPPPPVKYYSP-PPVYKSPPPPVKHYSPP------PVYKSPPPPVKYYSPPPVY 207
>At1g30970 putative 11-zinc finger protein
Length = 367
Score = 33.1 bits (74), Expect = 0.15
Identities = 34/121 (28%), Positives = 51/121 (42%), Gaps = 14/121 (11%)
Query: 49 PAYGVPPNPVHYPVHPRSYGYDFFPESDSVSKPYGYSGLASEDSFWYDQWPP--TSSKPS 106
PA VPP H P+ FP + P S A + S PP +S P+
Sbjct: 171 PALSVPP-AAHLGYRPQP----LFPVQNMGMTPTPTSAPAIQPSPVTGVTPPGIPTSSPA 225
Query: 107 LAEPQPCFPDLNSMGLASEDSFGYEQWLSASSKPS----LGEAQPYFPSYIPSTIPSPTS 162
+ PQP FP +N+ + F + + +PS LG A Y P+ ++IP T+
Sbjct: 226 MPVPQPLFPVVNNSIPSQAPPFSAPLPVGGAQQPSHADALGSADAYPPN---NSIPGGTN 282
Query: 163 S 163
+
Sbjct: 283 A 283
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.132 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,054,546
Number of Sequences: 26719
Number of extensions: 357672
Number of successful extensions: 1085
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 65
Number of HSP's that attempted gapping in prelim test: 884
Number of HSP's gapped (non-prelim): 185
length of query: 258
length of database: 11,318,596
effective HSP length: 97
effective length of query: 161
effective length of database: 8,726,853
effective search space: 1405023333
effective search space used: 1405023333
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0207.8


