

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0206b.6
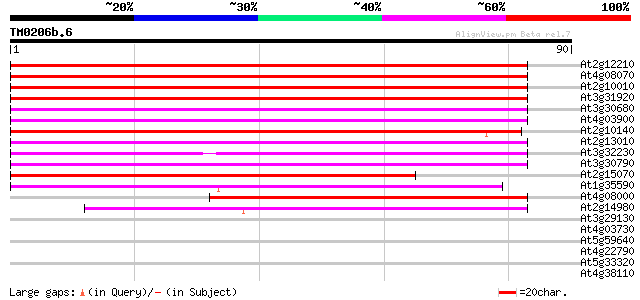
(90 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g12210 putative TNP2-like transposon protein 80 1e-16
At4g08070 77 1e-15
At2g10010 putative TNP2-like transposon protein 77 1e-15
At3g31920 hypothetical protein 69 4e-13
At3g30680 hypothetical protein 69 4e-13
At4g03900 putative transposon protein 69 5e-13
At2g10140 putative protein 63 2e-11
At2g13010 pseudogene 63 3e-11
At3g32230 hypothetical protein 61 1e-10
At3g30790 hypothetical protein 60 1e-10
At2g15070 En/Spm-like transposon protein 59 3e-10
At1g35590 CACTA-element, putative 55 5e-09
At4g08000 putative transposon protein 47 1e-06
At2g14980 pseudogene 41 1e-04
At3g29130 hypothetical protein 30 0.27
At4g03730 putative transposon protein 29 0.47
At5g59640 serine/threonine-specific protein kinase - like 27 1.4
At4g22790 predicted protein 27 1.4
At5g33320 phosphate/phosphoenolpyruvate translocator precursor 27 2.3
At4g38110 unknown protein 27 2.3
>At2g12210 putative TNP2-like transposon protein
Length = 889
Score = 80.5 bits (197), Expect = 1e-16
Identities = 36/83 (43%), Positives = 54/83 (64%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
LKSHD+H+LM+Q LP+A+ G LP + L SFF LC +V++++ + ++ +IV T
Sbjct: 663 LKSHDYHVLMQQLLPVALMGLLPKGPRTAIIRLCSFFNHLCQRVIDIEVISVMEAEIVET 722
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
LC E FP +FF + VHL + L
Sbjct: 723 LCMFERFFPPTFFDIMVHLTVHL 745
>At4g08070
Length = 767
Score = 77.4 bits (189), Expect = 1e-15
Identities = 36/83 (43%), Positives = 53/83 (63%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
LKSHD+H+LM+Q LP+A+ G LP + L FF LC +V++++ L +++ +IV T
Sbjct: 570 LKSHDYHVLMQQLLPIALRGLLPKGPRVAIGRLCKFFNHLCQRVIDMEQLLEMEAEIVET 629
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
LC E FP SFF + HL + L
Sbjct: 630 LCLFERFFPPSFFDIMEHLTVHL 652
>At2g10010 putative TNP2-like transposon protein
Length = 619
Score = 77.0 bits (188), Expect = 1e-15
Identities = 34/83 (40%), Positives = 54/83 (64%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
LKSHD+H++M+Q LP+A+ G LP + L +FF LC +V++++ + L+ +IV T
Sbjct: 517 LKSHDYHVMMQQLLPVALRGLLPKGPRTAILRLCAFFNYLCQRVIDIEVISVLETEIVET 576
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
LC + FP +FF + VHL + L
Sbjct: 577 LCMFKRFFPPTFFDIMVHLTIHL 599
>At3g31920 hypothetical protein
Length = 725
Score = 68.9 bits (167), Expect = 4e-13
Identities = 31/83 (37%), Positives = 51/83 (61%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
LKSHD H+L++ LP+A+ G LP ++ L +F +LC +V++ + L L+ ++V T
Sbjct: 620 LKSHDHHVLLQNLLPVALRGLLPRVPRVAISRLCHYFNRLCQRVIDPEKLMSLESELVET 679
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
+C +E FP S F + HL + L
Sbjct: 680 MCQLERYFPPSLFDIMFHLPIHL 702
>At3g30680 hypothetical protein
Length = 1116
Score = 68.9 bits (167), Expect = 4e-13
Identities = 33/83 (39%), Positives = 48/83 (57%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
+KSHD H+ ME+ LP L V L+ + +FFR LC + L + L+ +IVL
Sbjct: 640 MKSHDCHVFMERLLPFIFAELLDRNVHLALSGIGAFFRDLCSRTLQTSRVQILKQNIVLI 699
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
+C++E +FP SFF V HL + L
Sbjct: 700 ICNLEKIFPPSFFDVMEHLPIHL 722
>At4g03900 putative transposon protein
Length = 817
Score = 68.6 bits (166), Expect = 5e-13
Identities = 33/83 (39%), Positives = 48/83 (57%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
+KSHD H+ ME+ LP L V L+ + +FFR LC + L + L+ +IVL
Sbjct: 670 MKSHDCHVFMERLLPFIFAELLDRNVHLALSGIRAFFRDLCSRTLQTSRVQILKQNIVLI 729
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
+C++E +FP SFF V HL + L
Sbjct: 730 ICNLEKIFPPSFFDVMEHLPIHL 752
>At2g10140 putative protein
Length = 663
Score = 63.2 bits (152), Expect = 2e-11
Identities = 30/83 (36%), Positives = 50/83 (60%), Gaps = 1/83 (1%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
+KSHD H+ ME+ LP L V L+ + +FFR LC + L + + L+ +IVL
Sbjct: 557 MKSHDCHVFMERLLPFIFAELLDRNVHLALSGIGAFFRDLCSRTLQISRVQILKQNIVLI 616
Query: 61 LCHMEMLFPLSFFTV-SVHLILS 82
+C++E +FP SF + +++L +S
Sbjct: 617 ICNLEKIFPPSFLMLWNIYLSIS 639
>At2g13010 pseudogene
Length = 1040
Score = 62.8 bits (151), Expect = 3e-11
Identities = 28/83 (33%), Positives = 47/83 (55%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
+KSHD H+ M++ LP + LP + + + FF+ LC + L +++L +I +
Sbjct: 826 MKSHDCHVFMQRLLPFVLVELLPTNIHEAIAGIGVFFKDLCTRTLTTDTIEQLDKNIPVL 885
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
LC++E LF +FF V HL + L
Sbjct: 886 LCNLEKLFSPAFFDVMEHLPIHL 908
>At3g32230 hypothetical protein
Length = 1169
Score = 60.8 bits (146), Expect = 1e-10
Identities = 29/83 (34%), Positives = 46/83 (54%), Gaps = 2/83 (2%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
+KSHD H+ +++ LP LP V + +FFR LC + L + +L +I +
Sbjct: 690 MKSHDCHVFIQRLLPFVFLELLPANVHDAIE--GAFFRDLCTRTLTTDGIQQLDENIPML 747
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
LC++E +FP +FF V HL + L
Sbjct: 748 LCNLEKVFPPTFFDVMEHLSIHL 770
>At3g30790 hypothetical protein
Length = 953
Score = 60.5 bits (145), Expect = 1e-10
Identities = 28/83 (33%), Positives = 46/83 (54%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
+KSHD H+ M++ LP + LP + + + FF+ LC + L +++L +I +
Sbjct: 587 MKSHDCHVFMQRLLPFVLVELLPTNIHEAIAGIGVFFKDLCTRTLTTYTIEQLDKNIPVL 646
Query: 61 LCHMEMLFPLSFFTVSVHLILSL 83
LC++E LF FF V HL + L
Sbjct: 647 LCNLEKLFLPVFFDVMEHLPIHL 669
>At2g15070 En/Spm-like transposon protein
Length = 672
Score = 59.3 bits (142), Expect = 3e-10
Identities = 24/65 (36%), Positives = 43/65 (65%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLT 60
LKSHD H+L++ LP+A+ G LP ++ L +F +LC +V++ + L L++++V T
Sbjct: 511 LKSHDHHVLLQNLLPVALRGLLPKRPRVAISRLGHYFNRLCQRVIDPEKLISLEYELVET 570
Query: 61 LCHME 65
+C +E
Sbjct: 571 MCQLE 575
>At1g35590 CACTA-element, putative
Length = 1180
Score = 55.5 bits (132), Expect = 5e-09
Identities = 33/102 (32%), Positives = 46/102 (44%), Gaps = 23/102 (22%)
Query: 1 LKSHDFHILMEQFLPLAMWGSLPDEVSAVLNE-----------------------LSSFF 37
+KSHD HI ME+ LP L V L+ + +FF
Sbjct: 688 MKSHDCHIFMERLLPFIFAELLDRNVHLALSGTILTIFYGEIFLCNYLTSTYILGIGAFF 747
Query: 38 RQLCGKVLNVKALDKLQHDIVLTLCHMEMLFPLSFFTVSVHL 79
R LC + L + L+ +IVL +C++E +FP SFF V HL
Sbjct: 748 RDLCSRTLQTSRVQILKQNIVLIICNLEKIFPPSFFDVMEHL 789
>At4g08000 putative transposon protein
Length = 609
Score = 47.4 bits (111), Expect = 1e-06
Identities = 21/51 (41%), Positives = 32/51 (62%)
Query: 33 LSSFFRQLCGKVLNVKALDKLQHDIVLTLCHMEMLFPLSFFTVSVHLILSL 83
+ +FFR LC + L + L+ +IVL +C++E +FP SFF V HL + L
Sbjct: 494 IGAFFRDLCSRTLQKSRVQILKQNIVLIICNLEKIFPPSFFDVMEHLPIHL 544
>At2g14980 pseudogene
Length = 665
Score = 40.8 bits (94), Expect = 1e-04
Identities = 25/85 (29%), Positives = 39/85 (45%), Gaps = 14/85 (16%)
Query: 13 FLPLAMWGSLPDEVSAVLNELSSF--------------FRQLCGKVLNVKALDKLQHDIV 58
+LP A+W E L F + C +V++++ L +++ +IV
Sbjct: 466 YLPAALWSLSKSEKKLFCKRLFDFKGPDGYCSDIARGVSLEDCKRVIDMEQLLEIEAEIV 525
Query: 59 LTLCHMEMLFPLSFFTVSVHLILSL 83
TLC E FP SFF + VHL + L
Sbjct: 526 ETLCLFESFFPPSFFDIMVHLTVHL 550
>At3g29130 hypothetical protein
Length = 185
Score = 29.6 bits (65), Expect = 0.27
Identities = 19/57 (33%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query: 22 LPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLTLCHMEMLFPLSFFTVSVH 78
+ + + A E+S+ F+ L KV ++ +L LQH IVLTL ++ S + V+V+
Sbjct: 43 MEETIRAASQEVSNEFKTLV-KVEDLNSLRHLQHLIVLTLITLDSTDVSSLYAVAVN 98
>At4g03730 putative transposon protein
Length = 696
Score = 28.9 bits (63), Expect = 0.47
Identities = 12/21 (57%), Positives = 15/21 (71%)
Query: 59 LTLCHMEMLFPLSFFTVSVHL 79
L LC++E +FP SFF V HL
Sbjct: 316 LKLCNLEKIFPPSFFDVMEHL 336
>At5g59640 serine/threonine-specific protein kinase - like
Length = 986
Score = 27.3 bits (59), Expect = 1.4
Identities = 9/17 (52%), Positives = 14/17 (81%)
Query: 1 LKSHDFHILMEQFLPLA 17
+KSHD H+LM++ P+A
Sbjct: 568 MKSHDCHVLMQRLFPIA 584
>At4g22790 predicted protein
Length = 491
Score = 27.3 bits (59), Expect = 1.4
Identities = 15/58 (25%), Positives = 30/58 (50%), Gaps = 1/58 (1%)
Query: 28 AVLNELSSFFRQLCGKVLNVKALDKLQHDIVLTLCHMEML-FPLSFFTVSVHLILSLF 84
+VL +S+ +CG+ K L + + + + ++ P+SF ++VH IL+ F
Sbjct: 83 SVLYGISAAMEPICGQAFGAKNFKLLHKTLFMAVLLLLLISVPISFLWLNVHKILTGF 140
>At5g33320 phosphate/phosphoenolpyruvate translocator precursor
Length = 408
Score = 26.6 bits (57), Expect = 2.3
Identities = 15/54 (27%), Positives = 31/54 (56%), Gaps = 3/54 (5%)
Query: 27 SAVLNELSSFFRQLCGKVLNVK---ALDKLQHDIVLTLCHMEMLFPLSFFTVSV 77
SA+ + L++ R + K + VK +LD + ++TL + ++ P++FFT +
Sbjct: 254 SAMASNLTNQSRNVLSKKVMVKKDDSLDNITLFSIITLMSLVLMAPVTFFTEGI 307
>At4g38110 unknown protein
Length = 549
Score = 26.6 bits (57), Expect = 2.3
Identities = 23/77 (29%), Positives = 32/77 (40%), Gaps = 3/77 (3%)
Query: 15 PLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLTLCH-MEMLFPLSFF 73
PLA S PD V V + L S N K L++ + T+ H + ++ F
Sbjct: 411 PLAYGRSFPDVVKGVEHTLQSLHSDRETTPANFKYKRSLENQLTSTMLHLLSLVSSCHFE 470
Query: 74 TVSVHLILSLFFL--WL 88
+S LI FL WL
Sbjct: 471 ALSEFLIRKASFLEEWL 487
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.145 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,867,600
Number of Sequences: 26719
Number of extensions: 57974
Number of successful extensions: 337
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 311
Number of HSP's gapped (non-prelim): 33
length of query: 90
length of database: 11,318,596
effective HSP length: 66
effective length of query: 24
effective length of database: 9,555,142
effective search space: 229323408
effective search space used: 229323408
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0206b.6


