

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0206b.5
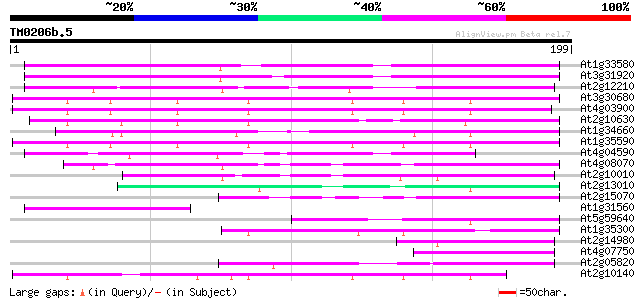
(199 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g33580 En/Spm-like transposon protein, putative 101 3e-22
At3g31920 hypothetical protein 97 6e-21
At2g12210 putative TNP2-like transposon protein 79 1e-15
At3g30680 hypothetical protein 76 1e-14
At4g03900 putative transposon protein 75 2e-14
At2g10630 En/Spm transposon hypothetical protein 1 74 5e-14
At1g34660 hypothetical protein 73 1e-13
At1g35590 CACTA-element, putative 72 2e-13
At4g04590 putative transposon protein 67 8e-12
At4g08070 65 2e-11
At2g10010 putative TNP2-like transposon protein 62 2e-10
At2g13010 pseudogene 51 4e-07
At2g15070 En/Spm-like transposon protein 49 2e-06
At1g31560 putative protein 47 9e-06
At5g59640 serine/threonine-specific protein kinase - like 45 3e-05
At1g35300 hypothetical protein 45 3e-05
At2g14980 pseudogene 44 6e-05
At4g07750 putative transposon protein 43 1e-04
At2g05820 En/Spm-like transposon protein 42 2e-04
At2g10140 putative protein 40 6e-04
>At1g33580 En/Spm-like transposon protein, putative
Length = 428
Score = 101 bits (251), Expect = 3e-22
Identities = 58/193 (30%), Positives = 91/193 (47%), Gaps = 16/193 (8%)
Query: 6 MDKSWIEMPRNTMAYEEGVKRFIDFAFERSSIEGKIICPCPRCEFRKRQTRDEVYDHLIC 65
MDKSW+ +PRN++ YE+G +F+ + R ++ CPC C Q D V +HL+
Sbjct: 11 MDKSWVWLPRNSIEYEKGATKFVYASSRRLGDPSEMFCPCTDCRNLCHQPIDTVLEHLVI 70
Query: 66 KQLPKGYT---FWFDHGETMNNSPQNVPNSVEQTMVDEDPIQNMINGAFGVDMHHGEESN 122
K + Y W HGE + +E E +I A+ H + N
Sbjct: 71 KGMNHKYKRNGCWSKHGEIR-------ADKLEAESSSEFGAYELIRTAYFDGEDHSDSQN 123
Query: 123 EDVPDGEGVIPDATQERHETRDFYDLARDGEQPLYEGCTKYSKLSFLVKLYHIKTLCGVT 182
++ D + E DF + +D E PLY GC KY+K+S +++LY IK G++
Sbjct: 124 QNENDSK------EPSTKEESDFREKLKDAETPLYYGCPKYTKVSAIMRLYRIKVKSGMS 177
Query: 183 DKAMTMILELLHD 195
+ +L L+HD
Sbjct: 178 ENYFDQLLSLVHD 190
>At3g31920 hypothetical protein
Length = 725
Score = 97.1 bits (240), Expect = 6e-21
Identities = 56/193 (29%), Positives = 90/193 (46%), Gaps = 13/193 (6%)
Query: 6 MDKSWIEMPRNTMAYEEGVKRFIDFAFERSSIEGKIICPCPRCEFRKRQTRDEVYDHLIC 65
MDKSW+ +PRN + YE+G F+ + ++ CPC C Q + + +HL+
Sbjct: 1 MDKSWVWLPRNNIEYEKGATTFVYASSRSLGDPAEMFCPCIDCRNLCHQPIETILEHLVM 60
Query: 66 KQLPKGYT---FWFDHGETMNNSPQNVPNSVEQTMVDEDPIQNMINGAFGVDMHHGEESN 122
K + Y W HGE + + P+S E +I A+ H + N
Sbjct: 61 KGMDHKYKRNRCWSKHGEIRADKLEVEPSST----TSECEAYELIRTAYFDGDVHSDSQN 116
Query: 123 EDVPDGEGVIPDATQERHETRDFYDLARDGEQPLYEGCTKYSKLSFLVKLYHIKTLCGVT 182
++ D + E E DF + ++ E PLY GC KY+K+S ++ LY IK G++
Sbjct: 117 QNEDDSK------EPETKEEADFREKLKEAETPLYHGCPKYTKVSAIMGLYRIKVKSGMS 170
Query: 183 DKAMTMILELLHD 195
+ +L L+HD
Sbjct: 171 ENYFDQLLSLVHD 183
>At2g12210 putative TNP2-like transposon protein
Length = 889
Score = 79.3 bits (194), Expect = 1e-15
Identities = 56/193 (29%), Positives = 92/193 (47%), Gaps = 26/193 (13%)
Query: 6 MDKSWIEMPRNTMAYEEGVKRFI-DFAFERSSIEGKIICPCPRCEFRKRQTRDEVYDHLI 64
+DK+W+ + R YE+G ++F+ D A I+ I+CPC C +R++ +V DHL+
Sbjct: 2 LDKAWVRLCRADPGYEKGAQKFVTDVAVALGDID-MIVCPCKDCRNVERKSGSDVVDHLV 60
Query: 65 CKQLPKGYTF---WFDHGETMNNSPQNVPNSVEQTMVDEDPIQNMINGAFGVDMHHGE-E 120
+ + + Y W+ HG+ +S + ++ + + I + A G D GE
Sbjct: 61 RRGMDEAYKLRADWYHHGDV--DSVADCESNFSRWNAE---ILELYQAAQGFDADLGEIA 115
Query: 121 SNEDVPDGEGVIPDATQERHETRDFYDLARDGEQPLYEGCTKYSKLSFLVKLYHIKTLCG 180
+ED+ + E F D E PLY C +SKLS +V L+ IKT G
Sbjct: 116 EDEDIREDE---------------FLAKLADAEIPLYPSCLNHSKLSAIVTLFRIKTKNG 160
Query: 181 VTDKAMTMILELL 193
+DK+ +L L
Sbjct: 161 WSDKSFNELLGTL 173
>At3g30680 hypothetical protein
Length = 1116
Score = 75.9 bits (185), Expect = 1e-14
Identities = 60/210 (28%), Positives = 93/210 (43%), Gaps = 16/210 (7%)
Query: 2 YLKAMDKSWIEMPRNTMA-YEEGVKRFIDFAFER---SSIEGKIICPCPRCEFRKRQTRD 57
Y M K + E+ N A Y GV+ F+ FA + S GK CPC C+ K
Sbjct: 14 YRDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHIISG 73
Query: 58 E-VYDHLICKQLPKGYTFWFDHGETMN--------NSPQNVPNSVEQTMVDEDPIQNMIN 108
V HL + Y W+ HGE +N + N E V EDP +M+N
Sbjct: 74 RRVSSHLFSQGFMPDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPYVDMVN 133
Query: 109 GAFGVDMHHGEE-SNEDVPDGEGVIPDATQE-RHETRDFYDLARDGEQPLYEGCTK-YSK 165
AF ++ + + ++D +G + + R+ + FYDL PLY+GC + S+
Sbjct: 134 DAFNFNVGYDDNVGHDDCYHHDGSYQNVEEPVRNHSNKFYDLLEGANNPLYDGCREGQSQ 193
Query: 166 LSFLVKLYHIKTLCGVTDKAMTMILELLHD 195
LS +L H K +++K + + E+ D
Sbjct: 194 LSLASRLMHNKAEYNMSEKLVDSVCEMFTD 223
>At4g03900 putative transposon protein
Length = 817
Score = 75.1 bits (183), Expect = 2e-14
Identities = 59/207 (28%), Positives = 92/207 (43%), Gaps = 16/207 (7%)
Query: 2 YLKAMDKSWIEMPRNTMA-YEEGVKRFIDFAFER---SSIEGKIICPCPRCEFRKRQTRD 57
Y M K + E+ N A Y GV+ F+ FA + S GK CPC C+ K
Sbjct: 14 YRDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFYCPCSVCKNEKHIISG 73
Query: 58 E-VYDHLICKQLPKGYTFWFDHGETMN--------NSPQNVPNSVEQTMVDEDPIQNMIN 108
V HL + Y W+ HGE +N + N E V EDP +M+N
Sbjct: 74 RRVSSHLFSHEFMPDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPYVDMVN 133
Query: 109 GAFGVDMHHGEE-SNEDVPDGEGVIPDATQE-RHETRDFYDLARDGEQPLYEGCTK-YSK 165
AF ++ + + ++D +G + + R+ + FYDL PLY+GC + S+
Sbjct: 134 DAFNFNVGYDDNVGHDDNYHHDGSYQNVEEPVRNHSNKFYDLLEGANNPLYDGCREGQSQ 193
Query: 166 LSFLVKLYHIKTLCGVTDKAMTMILEL 192
LS +L H K +++K + + E+
Sbjct: 194 LSLASRLMHNKAEYNMSEKLVDSVCEM 220
>At2g10630 En/Spm transposon hypothetical protein 1
Length = 515
Score = 73.9 bits (180), Expect = 5e-14
Identities = 58/202 (28%), Positives = 87/202 (42%), Gaps = 18/202 (8%)
Query: 8 KSWIEMPRNTMA-YEEGVKRFIDFAFERSSIE---GKIICPCPRCEFRKRQTRDE-VYDH 62
K + E+ N A Y GV++F+ FA + ++ GK CPC C+ K V H
Sbjct: 20 KRFDEVTGNLSAEYVAGVEQFMTFANSQPIVQSSRGKFHCPCSVCKNEKHIISGRRVSSH 79
Query: 63 LICKQLPKGYTFWFDHGETMN--------NSPQNVPNSVEQTMVDEDPIQNMINGAFGVD 114
L Y W+ HGE MN N N E + EDP +M+N AF +
Sbjct: 80 LFSHGFMHDYYVWYKHGEEMNMDLGTSYTNRTYFSENHEEVGNIVEDPYVDMVNDAFNFN 139
Query: 115 MHHGEESNEDVPDGEGVIPDATQERHETRDFYDLARDGEQPLYEGC-TKYSKLSFLVKLY 173
+ + + D D + + R+ + FYDL PLY+GC + S+LS +L
Sbjct: 140 VGYDDNYRHD--DSYQNVEEPV--RNHSNKFYDLLEGANNPLYDGCRERQSQLSLASRLM 195
Query: 174 HIKTLCGVTDKAMTMILELLHD 195
H K + +K + + E D
Sbjct: 196 HNKAEYNMNEKFVDSVCETFTD 217
>At1g34660 hypothetical protein
Length = 501
Score = 72.8 bits (177), Expect = 1e-13
Identities = 56/197 (28%), Positives = 90/197 (45%), Gaps = 34/197 (17%)
Query: 17 TMAYEEGVKRFIDFAFERS-SIE-GKIICPCPRCEFRKRQTRDEVYDHLICKQLPKGYTF 74
T ++EG+ F+ FA + +E GK+ CPC C K D ++ HL + GY
Sbjct: 13 TKEFKEGLVDFMRFAANQEIPLECGKMYCPCSTCGNEKFLPVDSIWKHLYSRGFMPGYYI 72
Query: 75 WFDHG---ETMNNSPQNVPNSVEQTMVDEDPIQNMINGAFGVDMHHGEESNEDVPDGEGV 131
WF HG ET+ NS ++ DE I+G D + EE + D GV
Sbjct: 73 WFSHGKDFETLANSNED----------DE------IHGLEPEDNNSNEEEENIIRDPTGV 116
Query: 132 IPDATQERHET------------RDFYDLARDGEQPLYEGCTK-YSKLSFLVKLYHIKTL 178
I T ET + F+D+ ++P+Y+GC + +S+LS +L +IKT
Sbjct: 117 IQMVTNAFRETTESGIEEPNLEAKKFFDMLDAAKKPIYDGCKEGHSRLSAATRLMNIKTE 176
Query: 179 CGVTDKAMTMILELLHD 195
+++ + I + + D
Sbjct: 177 YNLSEDCVDAIADFVKD 193
>At1g35590 CACTA-element, putative
Length = 1180
Score = 72.0 bits (175), Expect = 2e-13
Identities = 59/210 (28%), Positives = 92/210 (43%), Gaps = 16/210 (7%)
Query: 2 YLKAMDKSWIEMPRNTMA-YEEGVKRFIDFAFER---SSIEGKIICPCPRCEFRKRQTRD 57
Y M K + E+ N A Y GV+ F+ FA + S GK CPC C+ K
Sbjct: 14 YRDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHIISG 73
Query: 58 E-VYDHLICKQLPKGYTFWFDHGETMN--------NSPQNVPNSVEQTMVDEDPIQNMIN 108
V HL + Y W+ HGE +N + N E V EDP +M+N
Sbjct: 74 RRVSSHLFSQGFMPDYYVWYKHGEELNMDIGTSYTDRTYFSENHEEVGNVVEDPYVDMVN 133
Query: 109 GAFGVDMHHGEE-SNEDVPDGEGVIPDATQE-RHETRDFYDLARDGEQPLYEGCTK-YSK 165
AF ++ + + ++D +G + + R+ + FYDL LY+GC + S+
Sbjct: 134 DAFNFNVGYDDNVGHDDCYHHDGSYQNVEEPVRNHSNKFYDLLEGANNLLYDGCREGQSQ 193
Query: 166 LSFLVKLYHIKTLCGVTDKAMTMILELLHD 195
LS +L H K +++K + + E+ D
Sbjct: 194 LSLASRLMHNKAEYNMSEKLVDSVCEMFTD 223
>At4g04590 putative transposon protein
Length = 346
Score = 66.6 bits (161), Expect = 8e-12
Identities = 47/166 (28%), Positives = 72/166 (43%), Gaps = 24/166 (14%)
Query: 6 MDKSWIEMPRNTMAYEEGVKRFIDFAFERSSIEGKI---ICPCPRCEFRKRQTRDEVYDH 62
MDK+W+ +PRN + YEEG +F+ F+ +S G + +CPC Q+ + V +H
Sbjct: 1 MDKAWVWLPRNCLEYEEGATKFV---FDSASCLGNVSDMLCPCVDYRNMVHQSNETVLEH 57
Query: 63 LICKQLPKGY---TFWFDHGETMNNSPQNVPNSVEQTMVDEDPIQNMINGAFGVDMHHGE 119
L+ + + Y W HG+ N N+ EQT E ++ AF
Sbjct: 58 LVIRGMDIKYKRSKCWSKHGDRSN-------NTAEQT--SEYEAYDLFRTAFMASEEKLP 108
Query: 120 ESNEDVPDGEGVIPDATQERHETRDFYDLARDGEQPLYEGCTKYSK 165
+V + E T + E +F D E PLY C Y+K
Sbjct: 109 SQQHNVEESE------TMDSKEEVEFRKKLEDVETPLYSTCLNYTK 148
>At4g08070
Length = 767
Score = 65.1 bits (157), Expect = 2e-11
Identities = 54/180 (30%), Positives = 81/180 (45%), Gaps = 19/180 (10%)
Query: 20 YEEGVKRFI-DFAFERSSIEGKIICPCPRCEFRKRQTRDEVYDHLICKQLPKGYTF---W 75
YE G +F+ D A + I+CPC C R + V DHL+ + + + Y W
Sbjct: 32 YERGAIKFVRDVAAALG--DDMIVCPCIDCRNIDRHSGCVVVDHLVTRGMDEAYKRRCDW 89
Query: 76 FDHGETMNNSPQNVPNSVEQTMVDEDPIQNMINGAFGVDMHHGEESNEDVPDGEGVIPDA 135
+ HGE ++ + V Q D + + A +D EE + V GE D
Sbjct: 90 YHHGELVSGGESE--SKVSQW---NDEVVELYQAAEYLD----EEFDAMVQLGEIAERDD 140
Query: 136 TQERHETRDFYDLARDGEQPLYEGCTKYSKLSFLVKLYHIKTLCGVTDKAMTMILELLHD 195
+E +F D E PLY C +SKLS +V L+ +KT G +DK+ +LE L +
Sbjct: 141 KKED----EFLAKLADAETPLYPSCVNHSKLSAIVSLFRLKTKNGWSDKSFNDLLETLSE 196
>At2g10010 putative TNP2-like transposon protein
Length = 619
Score = 62.4 bits (150), Expect = 2e-10
Identities = 50/160 (31%), Positives = 77/160 (47%), Gaps = 16/160 (10%)
Query: 41 IICPCPRCEFRKRQTRDEVYDHLICKQLPKGYTF---WFDHGETMNNSPQNVPNSVEQTM 97
IICPC C RQ+ V DHL+ + + + Y W+ HG+ S V + + Q
Sbjct: 4 IICPCKDCRNVVRQSDCVVVDHLVRRGMDEAYKLHSDWYHHGD--EKSVDKVQSKLNQW- 60
Query: 98 VDEDPIQNMINGAFGVDMHHGEESNEDVPDGEGVIPDATQ--ERHETRDFYDLAR--DGE 153
+ + + A D EE GE ++ D ++ E + R+ LA+ D E
Sbjct: 61 --NEEVFELYKAAEFFD----EELACRGDLGENLVGDLSEIAECEDQREDEFLAKILDAE 114
Query: 154 QPLYEGCTKYSKLSFLVKLYHIKTLCGVTDKAMTMILELL 193
PLY C+ +SKLS +V L+ IKT G +DK+ +L+ L
Sbjct: 115 TPLYPNCSNHSKLSAIVTLFRIKTHNGWSDKSFNELLQTL 154
>At2g13010 pseudogene
Length = 1040
Score = 51.2 bits (121), Expect = 4e-07
Identities = 42/178 (23%), Positives = 69/178 (38%), Gaps = 33/178 (18%)
Query: 39 GKIICPCPRCEFRKRQTRDEVYDHLICKQLPKGYTFWFDHGETMNNSPQ----------- 87
G I+CPC +C K + V HL + Y W HG+ + + +
Sbjct: 209 GTILCPCRKCLNEKYLEFNVVKKHLYNRGFMPNYYVWLRHGDVVEIAHELVIQIMVSSNL 268
Query: 88 ---------NVPNSVEQTMVDEDPIQNMINGAFGVDMHHGEESNEDVPDGEGVIPDATQE 138
VP+ E+ +M+ AF E+ P+ P+
Sbjct: 269 VILIMMRIMQVPDQFGYGYNQENRFHDMVTNAF-------HETIASFPENISEEPNV--- 318
Query: 139 RHETRDFYDLARDGEQPLYEGCTK-YSKLSFLVKLYHIKTLCGVTDKAMTMILELLHD 195
+ + FYD+ QP+YEGC + SKLS ++ IK +++K M EL+ +
Sbjct: 319 --DAQHFYDMLDAANQPIYEGCREGNSKLSLASRMMTIKADNNLSEKCMDSWAELIKE 374
>At2g15070 En/Spm-like transposon protein
Length = 672
Score = 48.9 bits (115), Expect = 2e-06
Identities = 35/121 (28%), Positives = 56/121 (45%), Gaps = 23/121 (19%)
Query: 75 WFDHGETMNNSPQNVPNSVEQTMVDEDPIQNMINGAFGVDMHHGEESNEDVPDGEGVIPD 134
W HGE + + P+S E +I A+ GEE +++ P
Sbjct: 11 WSKHGEIWADKLEAEPSS-------EFGAYELIRTAY----FDGEEDSKE--------PV 51
Query: 135 ATQERHETRDFYDLARDGEQPLYEGCTKYSKLSFLVKLYHIKTLCGVTDKAMTMILELLH 194
+E + F + +D E PLY GC KY+K+S ++ LY IK G+++ +L L+H
Sbjct: 52 TKEESY----FREKLKDVETPLYYGCPKYTKVSAIMGLYRIKVKSGMSENYFDQLLSLVH 107
Query: 195 D 195
D
Sbjct: 108 D 108
>At1g31560 putative protein
Length = 1431
Score = 46.6 bits (109), Expect = 9e-06
Identities = 19/59 (32%), Positives = 33/59 (55%)
Query: 6 MDKSWIEMPRNTMAYEEGVKRFIDFAFERSSIEGKIICPCPRCEFRKRQTRDEVYDHLI 64
+DK+W+ + R YE+G ++F+ + IICPC C +RQ+ +V D+L+
Sbjct: 2 LDKAWVRLCRADPGYEKGAQKFVTDVAAALGVIDMIICPCKDCRSVERQSGSDVVDNLV 60
>At5g59640 serine/threonine-specific protein kinase - like
Length = 986
Score = 45.1 bits (105), Expect = 3e-05
Identities = 26/96 (27%), Positives = 46/96 (47%), Gaps = 13/96 (13%)
Query: 101 DPIQNMINGAFGVDMHHGEESNEDVPDGEGVIPDATQERHETRDFYDLARDGEQPLYEGC 160
DP M++ AFG ++ E+ P+ E + FYD+ +QP+Y+GC
Sbjct: 25 DPYVEMVSDAFGSTESEFDQMREEDPN------------FEAKKFYDILDAAKQPIYDGC 72
Query: 161 TK-YSKLSFLVKLYHIKTLCGVTDKAMTMILELLHD 195
+ SKLS +L +KT ++ M I +++ +
Sbjct: 73 KEGLSKLSLAARLMSLKTDNNLSQNCMDSIAQIMQE 108
>At1g35300 hypothetical protein
Length = 477
Score = 44.7 bits (104), Expect = 3e-05
Identities = 34/130 (26%), Positives = 56/130 (42%), Gaps = 15/130 (11%)
Query: 76 FDHGETMN--------NSPQNVPNSVEQTMVDEDPIQNMINGAFGVDMHHGEESN-EDVP 126
+ HGE MN + N E V EDP +M+N AF ++ + + +D
Sbjct: 27 YKHGEEMNMDIGTSYTDRTYFSENHEEVGNVVEDPYVDMVNDAFNFNVGYDDNVGYDDNY 86
Query: 127 DGEGVIPDATQE-RHETRDFYDLARDGEQPLYEGCTKYSKLSFLVKLYHIKTLCGVTDKA 185
+G + + R+ + FYDL D PLY+GC + +L H K + +K
Sbjct: 87 HHDGSYQNVEEPVRNHSNKFYDLLEDANNPLYDGCCEGQS-----RLMHNKAEYNMIEKL 141
Query: 186 MTMILELLHD 195
+ + E+ D
Sbjct: 142 VDSVCEMFTD 151
>At2g14980 pseudogene
Length = 665
Score = 43.9 bits (102), Expect = 6e-05
Identities = 24/58 (41%), Positives = 35/58 (59%), Gaps = 2/58 (3%)
Query: 138 ERHETRDFYDLAR--DGEQPLYEGCTKYSKLSFLVKLYHIKTLCGVTDKAMTMILELL 193
ER + ++ LA+ D E PLY C +SKLS +V L+ +KT G +DK+ +LE L
Sbjct: 9 ERDDKKEDELLAKLVDAETPLYPSCVNHSKLSAIVSLFRLKTKNGWSDKSFNDLLETL 66
>At4g07750 putative transposon protein
Length = 569
Score = 42.7 bits (99), Expect = 1e-04
Identities = 20/50 (40%), Positives = 29/50 (58%)
Query: 144 DFYDLARDGEQPLYEGCTKYSKLSFLVKLYHIKTLCGVTDKAMTMILELL 193
+F D E PLY C +SKLS +V L+ + T G +DK+ +LE+L
Sbjct: 65 EFLAKLADAETPLYPSCVNHSKLSAIVSLFRLNTKNGWSDKSFNDLLEIL 114
>At2g05820 En/Spm-like transposon protein
Length = 143
Score = 42.0 bits (97), Expect = 2e-04
Identities = 33/120 (27%), Positives = 52/120 (42%), Gaps = 15/120 (12%)
Query: 75 WFDHGETMNNSPQNVPNS-VEQTMVDEDPIQNMINGAFGVDMHHGEESNEDVPDGEGVIP 133
W+ HGE ++ S + +V+ ++ F +H GE D
Sbjct: 11 WYHHGELVSGGESESKFSQLNDEVVELYQAAEYLDEEFAGMVHLGEIVERD--------- 61
Query: 134 DATQERHETRDFYDLARDGEQPLYEGCTKYSKLSFLVKLYHIKTLCGVTDKAMTMILELL 193
++ E LA D E PLY C +SKLS ++ L+ +KT G +DK+ +LE L
Sbjct: 62 ----DKKEDEFLAKLA-DAETPLYPSCFNHSKLSAIMSLFTLKTKNGWSDKSHNDLLETL 116
>At2g10140 putative protein
Length = 663
Score = 40.4 bits (93), Expect = 6e-04
Identities = 49/191 (25%), Positives = 79/191 (40%), Gaps = 22/191 (11%)
Query: 2 YLKAMDKSWIEMPRNTMA-YEEGVKRFIDFAFERSSIEGKIICPCPRCEFRKRQTRDEVY 60
Y M K + E+ N A Y GV+ F+ FA + ++ R R + T +V
Sbjct: 14 YRDWMYKRFDEVTENLSAEYVAGVEEFMTFANSQPIVQSS------RVYARMKNTSSQVE 67
Query: 61 DHLIC---KQLPKGYTFWFD-HGETMN--------NSPQNVPNSVEQTMVDEDPIQNMIN 108
+ ++ K L F HGE +N N N E + EDP +M+N
Sbjct: 68 ELVVICLVKDLCLIIMFGISMHGEELNMDIGTSYTNRTYFRENHEEVGNIVEDPYVDMVN 127
Query: 109 GAFGVDMHHGEE-SNEDVPDGEGVIPDATQE-RHETRDFYDLARDGEQPLYEGCTK-YSK 165
AF ++ + + +D +G + + R+ + FYDL LY+ C + S+
Sbjct: 128 DAFNFNVGYDDNVGYDDNYHHDGSYKNVEEPVRNHSNKFYDLLEGANNTLYDYCREGQSQ 187
Query: 166 LSFLVKLYHIK 176
LS +L H K
Sbjct: 188 LSLASRLMHNK 198
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,892,356
Number of Sequences: 26719
Number of extensions: 217654
Number of successful extensions: 558
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 506
Number of HSP's gapped (non-prelim): 38
length of query: 199
length of database: 11,318,596
effective HSP length: 94
effective length of query: 105
effective length of database: 8,807,010
effective search space: 924736050
effective search space used: 924736050
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0206b.5


