

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0206b.3
(304 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
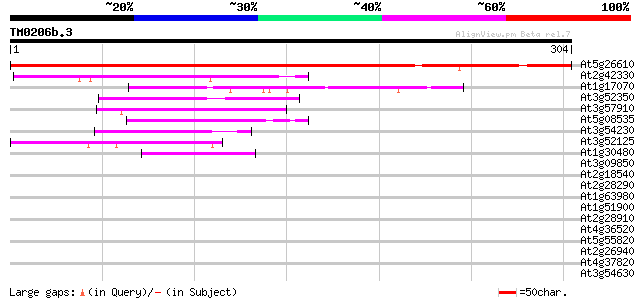
Score E
Sequences producing significant alignments: (bits) Value
At5g26610 unknown protein 386 e-108
At2g42330 hypothetical protein 62 5e-10
At1g17070 tuftelin-interacting-like protein 58 6e-09
At3g52350 putative protein 57 2e-08
At3g57910 putative protein 55 5e-08
At5g08535 unknown protein 55 6e-08
At3g54230 unknown protein 51 9e-07
At3g52125 unknown protein 47 2e-05
At1g30480 DNA damage repair protein, putative 41 7e-04
At3g09850 putative protein 39 0.003
At2g18540 putative vicilin storage protein (globulin-like) 39 0.003
At2g28290 putative SNF2 subfamily transcription regulator 39 0.003
At1g63980 unknown protein 39 0.003
At1g51900 Hypothetical protein 39 0.003
At2g28910 unknown protein 39 0.005
At4g36520 trichohyalin like protein 38 0.006
At5g55820 unknown protein 37 0.010
At2g26940 putative C2H2-type zinc finger protein 37 0.010
At4g37820 unknown protein 37 0.013
At3g54630 unknown protein 37 0.017
>At5g26610 unknown protein
Length = 301
Score = 386 bits (992), Expect = e-108
Identities = 198/308 (64%), Positives = 244/308 (78%), Gaps = 11/308 (3%)
Query: 1 MDYRRSSGRQESGGRGRRQFKKEEAHQDSLIGDLAEDFRLPINHRPTENVDLENVEQASL 60
MD R + E+ R +R ++K++ +Q+S I LAE+FRLPI HR TENVDLE+VEQASL
Sbjct: 1 MDISRRESKTEASSREKRIYEKDQMNQESFIEGLAEEFRLPITHRVTENVDLEDVEQASL 60
Query: 61 DTQLTSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEEN 120
D +++SSN+GF+LLQKMGWKGKGLGK EQGI EPIKSG+RD RLGLGKQEEDD+FTAEEN
Sbjct: 61 DVKISSSNVGFRLLQKMGWKGKGLGKQEQGITEPIKSGIRDRRLGLGKQEEDDYFTAEEN 120
Query: 121 IQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHL 180
IQRKKLD+E+EETEE +KREVLAEREQKIQ++VKEIRKVFYC+LC+KQY+ MEFE HL
Sbjct: 121 IQRKKLDIEIEETEEIAKKREVLAEREQKIQSDVKEIRKVFYCELCSKQYRTVMEFEGHL 180
Query: 181 SSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESG 240
SSYDHNH+KRFK+MKEMHG S RD+R+KREQQRQERE+ K +ADA+KQ ++Q Q+
Sbjct: 181 SSYDHNHKKRFKEMKEMHGASGRDERKKREQQRQEREMTK---MADARKQHQMQQSQQEV 237
Query: 241 SE--PVSSETR-TATPLTDQEQRNALKFGFSSK-GSASKNVIGPKRQMVAKKQNIPVSSI 296
E PVS+ + T PL Q+QR LKFGFSSK G SK+ + KK + ++S+
Sbjct: 238 PENVPVSAPAKTTVAPLAVQDQRKTLKFGFSSKSGIISKS----QPTSSIKKPKVAIASV 293
Query: 297 FNNDSDEE 304
F NDSDE+
Sbjct: 294 FGNDSDED 301
>At2g42330 hypothetical protein
Length = 752
Score = 61.6 bits (148), Expect = 5e-10
Identities = 47/181 (25%), Positives = 82/181 (44%), Gaps = 29/181 (16%)
Query: 3 YRRSSGRQESGGRGRRQFKKEEAHQDSLIGDLAE---------------DFRLPI--NHR 45
+ S +SGG R++ +K + + S G++ + D LPI +
Sbjct: 23 FESDSNSDDSGGSRRKKRRKTKPVKFSSAGNIDQVLKQNRGNCKIDENDDTILPIALGKK 82
Query: 46 PTENVDLENVEQASLDTQLTSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDPRLG 105
+ + + + S IG KLL+KMG+KG+GLGK++QGI+ PI+ +R +G
Sbjct: 83 IADKAHVREKNNKKENFEKFSGGIGMKLLEKMGYKGRGLGKNQQGIVAPIEVQLRPKNMG 142
Query: 106 LG----KQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVF 161
+G K++ F ++ KK V + +E + R L ++ K +RK
Sbjct: 143 MGYNDFKEKNAPLFPCLNKVEEKKKSVVVTVSENHGDGRRDLWKK--------KNVRKEV 194
Query: 162 Y 162
Y
Sbjct: 195 Y 195
>At1g17070 tuftelin-interacting-like protein
Length = 849
Score = 58.2 bits (139), Expect = 6e-09
Identities = 61/230 (26%), Positives = 105/230 (45%), Gaps = 54/230 (23%)
Query: 65 TSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAE----EN 120
++ IG KLL+KMG+KG GLGK++QGI+ PI++ +R +G+G +DF A+ +
Sbjct: 199 STKGIGMKLLEKMGYKGGGLGKNQQGIVAPIEAQLRPKNMGMG---YNDFKEAKLPDLKK 255
Query: 121 IQRKK-LDVELEETEEN--------------VRK-----REVLAEREQK---------IQ 151
++ KK + V + E E++ VRK E L E++Q+ I
Sbjct: 256 VEEKKIIGVSVSENEQSHGDRGGKNLWKKKKVRKAVYVTAEELLEKKQEAGFGGGQTIID 315
Query: 152 TEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKR-- 209
++R V + + + K A E + + HN R ++ RD R +R
Sbjct: 316 MRGPQVRVVTNLENLDAEEK-AKEADVPMPELQHNLRLIVDLVEHEIQKIDRDLRNERES 374
Query: 210 -------------EQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSS 246
E+++Q+R + IAD + R++L SG+ + S
Sbjct: 375 ALSLQQEKEMLINEEEKQKRHLENMEYIAD--EISRIELENTSGNLTLDS 422
>At3g52350 putative protein
Length = 180
Score = 56.6 bits (135), Expect = 2e-08
Identities = 38/110 (34%), Positives = 62/110 (55%), Gaps = 10/110 (9%)
Query: 49 NVDLENVEQASLDTQLTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLG 107
NV ++ E ++ T ++SSNIGF+LL+K GWK G GLG EQGI+ P+++ + + GLG
Sbjct: 62 NVIGDSSESSNPSTAISSSNIGFQLLKKHGWKEGTGLGITEQGILVPLQAEPKHNKQGLG 121
Query: 108 KQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEI 157
AE+ +RK + EE +K + L+++ K+ K +
Sbjct: 122 ---------AEKPAKRKPAQPQDTAFEEVSKKSKKLSKKLVKMIEHEKRL 162
>At3g57910 putative protein
Length = 265
Score = 55.1 bits (131), Expect = 5e-08
Identities = 34/107 (31%), Positives = 58/107 (53%), Gaps = 4/107 (3%)
Query: 48 ENVDLENVEQAS---LDTQLTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPR 103
E +E EQ +DT + SNIGFKLL++MG+K G LGK G EP+ +R R
Sbjct: 72 ERKQIEEDEQTLARIVDTPIGESNIGFKLLKQMGYKPGSALGKQGSGRAEPVTMDIRRSR 131
Query: 104 LGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKI 150
G+G+++ +E I+ + +++E E+ R+ R++++
Sbjct: 132 AGVGREDPHKEKKKKEEIEAENEKRKVDEMLEDFGSRQKSQWRKKRV 178
>At5g08535 unknown protein
Length = 141
Score = 54.7 bits (130), Expect = 6e-08
Identities = 32/100 (32%), Positives = 59/100 (59%), Gaps = 6/100 (6%)
Query: 64 LTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQ 122
+ SSNIGF+LL+K GWK G GLG EQGI+ P+++ + + G+G ++ A+ +
Sbjct: 39 INSSNIGFQLLKKHGWKEGTGLGIAEQGILVPLQAEPKHNKRGVGAKQPAKRKVAQPQAK 98
Query: 123 RKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFY 162
+++ + ++ + +RK + E E+ +Q KE + F+
Sbjct: 99 DEEVSKQSKKLSKKMRK---MMEHEKHLQE--KEFERAFF 133
>At3g54230 unknown protein
Length = 1007
Score = 50.8 bits (120), Expect = 9e-07
Identities = 28/86 (32%), Positives = 47/86 (54%), Gaps = 14/86 (16%)
Query: 47 TENVDLENVEQASLDTQLTSSNIGFKLLQKMGW-KGKGLGKDEQGIIEPIKSGMRDPRLG 105
T + ++ + + + + + SN+G ++L+ MGW +G GLGKD G+ EP+++ D R G
Sbjct: 909 TTSTEVSSFDVITEERAIDESNVGNRMLRNMGWHEGSGLGKDGSGMKEPVQAQGVDRRAG 968
Query: 106 LGKQEEDDFFTAEENIQRKKLDVELE 131
LG Q+ KK+D E E
Sbjct: 969 LGSQQ-------------KKVDAEFE 981
>At3g52125 unknown protein
Length = 443
Score = 46.6 bits (109), Expect = 2e-05
Identities = 39/141 (27%), Positives = 62/141 (43%), Gaps = 26/141 (18%)
Query: 1 MDYRRSSGRQESGGRGRRQFKKEEAHQDSLIGDLAEDFRLP--INHRPTENVDLENVE-- 56
M++ QE R RQ K E SL G P H + + LE ++
Sbjct: 277 MEFYMKKAAQEEKMRRPRQSKDEMPPPASLQGPSETSSTDPGKRGHHMGDYIPLEELDKF 336
Query: 57 --------------QASLDTQLTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSG-MR 100
+A+ ++ + N+G KLL KMGWK G+G+G +G+ +PI +G ++
Sbjct: 337 LSKCNDAAAQKATKEAAEKAKIQADNVGHKLLSKMGWKEGEGIGSSRKGMADPIMAGDVK 396
Query: 101 DPRLGLGK------QEEDDFF 115
LG+G + EDD +
Sbjct: 397 TNNLGVGASAPGEVKPEDDIY 417
>At1g30480 DNA damage repair protein, putative
Length = 387
Score = 41.2 bits (95), Expect = 7e-04
Identities = 22/63 (34%), Positives = 36/63 (56%), Gaps = 1/63 (1%)
Query: 72 KLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVEL 130
+++ KMGWK G+GLGK EQGI P+ + D R G+ ++ ++ E K +++
Sbjct: 220 RMMAKMGWKQGQGLGKSEQGITTPLMAKKTDRRAGVIVNASENKSSSAEKKVVKSVNING 279
Query: 131 EET 133
E T
Sbjct: 280 EPT 282
>At3g09850 putative protein
Length = 781
Score = 39.3 bits (90), Expect = 0.003
Identities = 20/46 (43%), Positives = 30/46 (64%), Gaps = 1/46 (2%)
Query: 63 QLTSSNIGFKLLQKMGW-KGKGLGKDEQGIIEPIKSGMRDPRLGLG 107
++ + G K++ KMG+ G GLGKD +GI +PI++ R LGLG
Sbjct: 632 EVHTRGFGSKMMAKMGFIDGGGLGKDGKGIAQPIEAVQRPKSLGLG 677
Score = 33.9 bits (76), Expect = 0.11
Identities = 14/45 (31%), Positives = 29/45 (64%), Gaps = 1/45 (2%)
Query: 66 SSNIGFKLLQKMGW-KGKGLGKDEQGIIEPIKSGMRDPRLGLGKQ 109
++ G +++ +MG+ +G GLG++ QGI+ P+ + R G+G +
Sbjct: 736 TTGFGSRMMARMGFVEGSGLGRESQGIVNPLVAVRRPRARGIGAE 780
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 39.3 bits (90), Expect = 0.003
Identities = 35/157 (22%), Positives = 71/157 (44%), Gaps = 13/157 (8%)
Query: 108 KQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCN 167
++EE+ EE Q +K + E E+ EE +KRE ER++K + EV+ R+
Sbjct: 493 RREEERKKREEEAEQARKREEEREKEEEMAKKRE--EERQRKEREEVERKRR-------E 543
Query: 168 KQYKLAMEFEAHLSSYDHNHR----KRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQ 223
+Q + E EA + KR +Q ++ + + + EQ+R+ E +
Sbjct: 544 EQERKRREEEARKREEERKREEEMAKRREQERQRKEREEVERKIREEQERKREEEMAKRR 603
Query: 224 IADAQKQQRLQLLQESGSEPVSSETRTATPLTDQEQR 260
+ QK++R ++ ++ E + ++E++
Sbjct: 604 EQERQKKEREEMERKKREEEARKREEEMAKIREEERQ 640
Score = 37.0 bits (84), Expect = 0.013
Identities = 37/156 (23%), Positives = 73/156 (46%), Gaps = 19/156 (12%)
Query: 108 KQEEDDFFTAEENIQRKKLDV-ELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLC 166
K+EE+ E ++RK+ + E + EE RKRE +RE+++ ++
Sbjct: 524 KREEERQRKEREEVERKRREEQERKRREEEARKREEERKREEEMAKRREQ---------- 573
Query: 167 NKQYKLAMEFEAHLSSYDHNHRKRFKQM-KEMHGGSSRDDRQKREQQRQEREIAK-FAQI 224
+Q K E E + + RKR ++M K + +R++ E++++E E K ++
Sbjct: 574 ERQRKEREEVERKIR--EEQERKREEEMAKRREQERQKKEREEMERKKREEEARKREEEM 631
Query: 225 ADAQKQQRLQLLQESGSEPVSSETRTATPLTDQEQR 260
A ++++R Q E V + R + +E+R
Sbjct: 632 AKIREEER----QRKEREDVERKRREEEAMRREEER 663
Score = 36.6 bits (83), Expect = 0.017
Identities = 32/175 (18%), Positives = 82/175 (46%), Gaps = 6/175 (3%)
Query: 86 KDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAE 145
K+E+ + ++ R+ ++EE+ E + +K + E + EE ++RE E
Sbjct: 440 KEEEEARKREEAKRREEEEAKRREEEETERKKREEEEARKREEERKREEEEAKRRE---E 496
Query: 146 REQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDD 205
+K + E ++ RK + K+ ++A + E + +R ++ ++ +
Sbjct: 497 ERKKREEEAEQARK--REEEREKEEEMAKKREEERQRKEREEVERKRREEQERKRREEEA 554
Query: 206 RQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTATPLTDQEQR 260
R++ E++++E E+AK + + Q+++R ++ ++ E +QE++
Sbjct: 555 RKREEERKREEEMAKRRE-QERQRKEREEVERKIREEQERKREEEMAKRREQERQ 608
Score = 32.3 bits (72), Expect = 0.33
Identities = 28/128 (21%), Positives = 59/128 (45%), Gaps = 11/128 (8%)
Query: 108 KQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVK---EIRKVFYCD 164
++EE + EE RK+ + + E EE R+ E ER+++ + E + E RK
Sbjct: 431 EEEEIERRRKEEEEARKREEAKRREEEEAKRREEEETERKKREEEEARKREEERK----- 485
Query: 165 LCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQI 224
++ + A E + + K+ +E +++ E+QR+ERE + +
Sbjct: 486 ---REEEEAKRREEERKKREEEAEQARKREEEREKEEEMAKKREEERQRKEREEVERKRR 542
Query: 225 ADAQKQQR 232
+ ++++R
Sbjct: 543 EEQERKRR 550
Score = 30.0 bits (66), Expect = 1.6
Identities = 19/53 (35%), Positives = 29/53 (53%), Gaps = 1/53 (1%)
Query: 108 KQEEDDFFTAEENIQRKKL-DVELEETEENVRKREVLAEREQKIQTEVKEIRK 159
K+EE+ EE QRK+ DVE + EE +RE +RE++ +E R+
Sbjct: 626 KREEEMAKIREEERQRKEREDVERKRREEEAMRREEERKREEEAAKRAEEERR 678
>At2g28290 putative SNF2 subfamily transcription regulator
Length = 1331
Score = 38.9 bits (89), Expect = 0.003
Identities = 40/185 (21%), Positives = 79/185 (42%), Gaps = 24/185 (12%)
Query: 43 NHRPTENVDLENVEQASLDTQLTSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDP 102
N T V L+ + L T L ++ + W + +E+G ++P
Sbjct: 435 NDLSTSAVQLDEFHSSVLLTNLIDNSSNMDI-----WH---IADEEEGNLQPSPKYTMSQ 486
Query: 103 RLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFY 162
+ +G+Q + +++++K D + +++ L++ V E++K+
Sbjct: 487 KWIMGRQNKRLLVDRSWSLKQQKADQAIGSRFNELKESVSLSDDISAKTKSVIELKKL-- 544
Query: 163 CDLCNKQYKLAMEFE-----------AHLSSYD-HNHRKRFKQMKEMHGGSSRDDRQKRE 210
L N Q +L EF HL SY H H +R KQ+ E + +++RQ+R
Sbjct: 545 -QLLNLQRRLRSEFVYNFFKPIATDVEHLKSYKKHKHGRRIKQL-EKYEQKMKEERQRRI 602
Query: 211 QQRQE 215
++RQ+
Sbjct: 603 RERQK 607
>At1g63980 unknown protein
Length = 391
Score = 38.9 bits (89), Expect = 0.003
Identities = 24/74 (32%), Positives = 41/74 (54%), Gaps = 3/74 (4%)
Query: 64 LTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDD--FFTAEEN 120
+ + FKL++ MGW+ G+GLGKD+QGI ++ + G+G + + F T + +
Sbjct: 13 ICKDSAAFKLMKSMGWEEGEGLGKDKQGIKGYVRVTNKQDTSGVGLDKPNPWAFDTTQFD 72
Query: 121 IQRKKLDVELEETE 134
KKL V+ T+
Sbjct: 73 NILKKLKVQAAPTK 86
>At1g51900 Hypothetical protein
Length = 765
Score = 38.9 bits (89), Expect = 0.003
Identities = 39/162 (24%), Positives = 69/162 (42%), Gaps = 5/162 (3%)
Query: 111 EDDFFTAEENIQRKKLDVELEETEENVR-KREVLAEREQKIQTEVKEIRKVFYCDLCNKQ 169
EDD+ A ++ + LD L+++ + R + E +IQ KE +KV D+ +
Sbjct: 468 EDDYIEA---LKCRMLDDILKKSGHRLEISRRQYNKPEIEIQVNEKEEKKVINTDMDIRY 524
Query: 170 YKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQK 229
+ E SS + +R K+ M + D +++R + E + A K
Sbjct: 525 DDESPEEVETYSSLTDDEEERSKEDTSMEDVNLTDIKEERSNEDTSMEDCCIEE-AQVGK 583
Query: 230 QQRLQLLQESGSEPVSSETRTATPLTDQEQRNALKFGFSSKG 271
QR+ +ES E S + +PLT+ +L + KG
Sbjct: 584 DQRVFRFRESSEEKRKSSSSPLSPLTEFRDMESLTYYMRQKG 625
>At2g28910 unknown protein
Length = 332
Score = 38.5 bits (88), Expect = 0.005
Identities = 33/137 (24%), Positives = 64/137 (46%), Gaps = 17/137 (12%)
Query: 86 KDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAE 145
KD I + SG+ R G+GK E ++ + EE + E +++ + ++AE
Sbjct: 108 KDPGAIEAAVLSGLEKIRRGVGKGEVEEVSSEEEE------ESESSDSDVDSEMERIIAE 161
Query: 146 R--EQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHG---- 199
R ++K + VK+ V K+ +++ E ++ S D R+R + + H
Sbjct: 162 RFGKKKGGSSVKKTSSV-----RKKKKRVSDESDSDSDSGDRKRRRRSMKKRSSHKRRSL 216
Query: 200 GSSRDDRQKREQQRQER 216
S D+ + R ++R+ER
Sbjct: 217 SESEDEEEGRSKRRKER 233
>At4g36520 trichohyalin like protein
Length = 1432
Score = 38.1 bits (87), Expect = 0.006
Identities = 43/208 (20%), Positives = 83/208 (39%), Gaps = 22/208 (10%)
Query: 78 GWKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENV 137
G K + + E + EP+K + R+ + E EN +R+++ VE E E+ +
Sbjct: 612 GKKMEMRSQSETKLNEPLKRMEEETRIKEARLRE-------ENDRRERVAVEKAENEKRL 664
Query: 138 RKREVLAEREQKI----QTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYD----HNHRK 189
+ E+E+KI + E R V + ++ K+ + E L + +
Sbjct: 665 KAALEQEEKERKIKEAREKAENERRAVEAREKAEQERKMKEQQELELQLKEAFEKEEENR 724
Query: 190 RFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPV----- 244
R ++ + R ++ RE++ ER I + + A+ +++ + L QE +
Sbjct: 725 RMREAFALEQEKERRIKEAREKEENERRIKEAREKAELEQRLKATLEQEEKERQIKERQE 784
Query: 245 --SSETRTATPLTDQEQRNALKFGFSSK 270
+E R L E LK K
Sbjct: 785 REENERRAKEVLEQAENERKLKEALEQK 812
Score = 31.2 bits (69), Expect = 0.73
Identities = 44/213 (20%), Positives = 88/213 (40%), Gaps = 19/213 (8%)
Query: 84 LGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVL 143
L + E+ +IE + + RL ++E+ +E +R++L E +E +EN RK+
Sbjct: 837 LEEKEKRLIEAFERAEIERRLKEDLEQEEMRMRLQEAKERERLHRENQEHQENERKQHEY 896
Query: 144 AEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSR 203
+ E ++ KE D C + EAH + + E
Sbjct: 897 SGEE----SDEKE------RDACEMEKTCETTKEAHGEQSSNESLSDTLEENESIDNDVS 946
Query: 204 DDRQKREQQRQEREIAKFAQIADAQKQQRL--QLLQESGSEPVSSETR-----TATPLTD 256
++QK+E++ + + A+ + ++ Q+ G+ + ++TR TP
Sbjct: 947 VNKQKKEEEGTRQRESMSAETCPWKVFEKTLKDASQKEGTNEMDADTRLFERNEETPRLG 1006
Query: 257 QEQRNALKFGFSSKGSAS--KNVIGPKRQMVAK 287
+ + G S + S S +N+IG K + +K
Sbjct: 1007 ENGGCNQQNGESGEESTSVTENIIGGKLEQKSK 1039
Score = 28.5 bits (62), Expect = 4.7
Identities = 27/129 (20%), Positives = 57/129 (43%), Gaps = 5/129 (3%)
Query: 118 EENIQRKKLDVELEETEENVRKREVLAEREQKI--QTEVKEIRKVFYCDLCNKQYKLAME 175
+EN +R K E EE ++ +R+ L E+E+++ E EI + DL ++ ++ ++
Sbjct: 812 KENERRLKETREKEENKKKLREAIELEEKEKRLIEAFERAEIERRLKEDLEQEEMRMRLQ 871
Query: 176 ---FEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQR 232
L + H++ ++ E G S + + + + E K A + +
Sbjct: 872 EAKERERLHRENQEHQENERKQHEYSGEESDEKERDACEMEKTCETTKEAHGEQSSNESL 931
Query: 233 LQLLQESGS 241
L+E+ S
Sbjct: 932 SDTLEENES 940
>At5g55820 unknown protein
Length = 1826
Score = 37.4 bits (85), Expect = 0.010
Identities = 41/173 (23%), Positives = 80/173 (45%), Gaps = 21/173 (12%)
Query: 97 SGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKE 156
+G RD ++ + E AE+ +KL E + E +++E L ++Q+I+ + KE
Sbjct: 1512 TGKRDVKVKALEAAEASKRIAEQKENDRKLKKEAMKLERAKQEQENL--KKQEIEKKKKE 1569
Query: 157 IRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQER 216
D K+ ++A + E + +++ +M + RQ+ E+ ++ +
Sbjct: 1570 E------DRKKKEAEMAWKQEMEKKKKEEERKRKEFEMADRK-------RQREEEDKRLK 1616
Query: 217 EIAKFAQIADAQKQQR-----LQLLQESGSEPVSSETRTATPLTDQEQRNALK 264
E K +IAD Q+QQR LQ +E + + + + L ++Q NA K
Sbjct: 1617 EAKKRQRIADFQRQQREADEKLQAEKELKRQAMDARIKAQKEL-KEDQNNAEK 1668
Score = 28.9 bits (63), Expect = 3.6
Identities = 25/143 (17%), Positives = 63/143 (43%), Gaps = 8/143 (5%)
Query: 108 KQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCN 167
++E D+ AE+ ++R+ +D ++ +E +++ + AE+ ++ + + +R
Sbjct: 1631 QREADEKLQAEKELKRQAMDARIKAQKE-LKEDQNNAEKTRQANSRIPAVRSKSNSSDDT 1689
Query: 168 KQYKLAMEFEAHLSSYDHNHRKR----FKQMKEMHGGSS---RDDRQKREQQRQEREIAK 220
+ + E + + S N + ++M+E + S DD + E + K
Sbjct: 1690 NASRSSRENDFKVISNPGNMSEEANMGIEEMEESYNISPYKCSDDEDEEEDDNDDMSNKK 1749
Query: 221 FAQIADAQKQQRLQLLQESGSEP 243
FA ++ RL ++ + +P
Sbjct: 1750 FAPTWASKSNVRLAVISQQNIDP 1772
>At2g26940 putative C2H2-type zinc finger protein
Length = 286
Score = 37.4 bits (85), Expect = 0.010
Identities = 19/86 (22%), Positives = 44/86 (51%), Gaps = 2/86 (2%)
Query: 135 ENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAH--LSSYDHNHRKRFK 192
E +R + L E + + E + ++ +C +C KQ+ + H + S ++N++ + K
Sbjct: 23 EKIRVKVKLPEERSEDEDESPQPQRKHFCVICEKQFSSGKAYGGHVRIHSIEYNNKGKMK 82
Query: 193 QMKEMHGGSSRDDRQKREQQRQEREI 218
+MK+M + + ++ +E+EI
Sbjct: 83 KMKKMKLKKKKKRKIGLVKKEKEKEI 108
>At4g37820 unknown protein
Length = 532
Score = 37.0 bits (84), Expect = 0.013
Identities = 40/195 (20%), Positives = 84/195 (42%), Gaps = 12/195 (6%)
Query: 78 GWKGKGLG----KDEQGIIEPIKSGMRDPRLG-LGKQEEDDFFTAEENIQRKKLDVELEE 132
G GK G K+E+ E ++S + ++ GK E+D + +E+ + K + EE
Sbjct: 282 GSSGKSTGYQQTKNEEDEKEKVQSSEEESKVKESGKNEKDASSSQDESKEEKPERKKKEE 341
Query: 133 TEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAME----FEAHLSSYDHNHR 188
+ +E E+ +K + +E K + K+ + E E + + +
Sbjct: 342 SSSQGEGKEEEPEKREKEDSSSQEESKEEEPENKEKEASSSQEENEIKETEIKEKEESSS 401
Query: 189 KRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQK---QQRLQLLQESGSEPVS 245
+ + KE SS R++ ++ E + ++ QK Q+ + +ESG++ +
Sbjct: 402 QEGNENKETEKKSSESQRKENTNSEKKIEQVESTDSSNTQKGDEQKTDESKRESGNDTSN 461
Query: 246 SETRTATPLTDQEQR 260
ET + T+ E++
Sbjct: 462 KETEDDSSKTESEKK 476
>At3g54630 unknown protein
Length = 560
Score = 36.6 bits (83), Expect = 0.017
Identities = 46/198 (23%), Positives = 83/198 (41%), Gaps = 18/198 (9%)
Query: 106 LGKQEEDDFFTAEENIQRKKLDVELEETEENVR----KREVLAEREQKIQTEVKEIRKVF 161
LGK E + AE +K+ ELE E++R K+E L + + ++ +V + R +
Sbjct: 203 LGKLEAEKTSVAETISGCEKISGELEAKLESLRKGPSKKESLEKVKADLENDVNKFRTIV 262
Query: 162 --YCDLCNKQYKLAMEFEAHLSSYDHN------HRKRFKQMKEMHGGSSRD-DRQKREQQ 212
Y D K+ E L + + K K+ E+ S+ D +R +RE Q
Sbjct: 263 VEYTDRNPAMEKVVEEKAKELKAKEEERERISVENKELKKSVELQNFSAADVNRMRRELQ 322
Query: 213 RQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTATPLTDQEQRNA---LKFGFSS 269
ER++A D Q+ +L + ++ + +T +Q R ++F +
Sbjct: 323 AVERDVADAEVARDGWDQKAWELNSQIRNQ--FHQIQTLAIDCNQALRRLKLDIQFAVNE 380
Query: 270 KGSASKNVIGPKRQMVAK 287
+G V+G + V K
Sbjct: 381 RGETPAAVMGVDYKSVVK 398
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.130 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,652,150
Number of Sequences: 26719
Number of extensions: 289139
Number of successful extensions: 1579
Number of sequences better than 10.0: 166
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 128
Number of HSP's that attempted gapping in prelim test: 1346
Number of HSP's gapped (non-prelim): 273
length of query: 304
length of database: 11,318,596
effective HSP length: 99
effective length of query: 205
effective length of database: 8,673,415
effective search space: 1778050075
effective search space used: 1778050075
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0206b.3


