

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
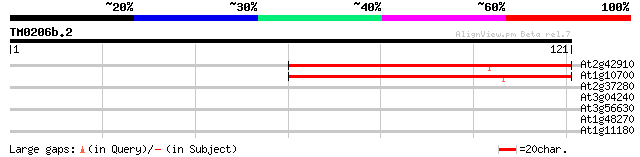
Query= TM0206b.2
(121 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g42910 putative ribose phosphate pyrophosphokinase 77 1e-15
At1g10700 putative phosphoribosyl diphosphate synthase 75 7e-15
At2g37280 putative ABC transporter 26 2.8
At3g04240 O-linked GlcNAc transferase like protein 25 4.7
At3g56630 cytochrome P450-like protein 25 6.1
At1g48270 putative G-protein-coupled receptor (GCR1) 25 8.0
At1g11180 secretory carrier membrane protein, putative 25 8.0
>At2g42910 putative ribose phosphate pyrophosphokinase
Length = 337
Score = 77.0 bits (188), Expect = 1e-15
Identities = 42/62 (67%), Positives = 47/62 (75%), Gaps = 1/62 (1%)
Query: 61 FLAQLSSPTQGFEQFTAIYALHCLFVASFALVPPFFPTGSFE-*KEEGDVTTAFTLARML 119
FLA SSP FEQ + IY L LFVASF LV PFFPTGSFE +EEGDV TAFT+AR++
Sbjct: 82 FLASFSSPAVIFEQISVIYLLPRLFVASFTLVLPFFPTGSFERMEEEGDVATAFTMARIV 141
Query: 120 SN 121
SN
Sbjct: 142 SN 143
>At1g10700 putative phosphoribosyl diphosphate synthase
Length = 411
Score = 74.7 bits (182), Expect = 7e-15
Identities = 42/62 (67%), Positives = 46/62 (73%), Gaps = 1/62 (1%)
Query: 61 FLAQLSSPTQGFEQFTAIYALHCLFVASFALVPPFFPTGSFE*KE-EGDVTTAFTLARML 119
FLA SSP FEQ + IYAL LFV+SF LV PFFPTG+ E E EGDV TAFTLAR+L
Sbjct: 155 FLASFSSPAVIFEQLSVIYALPKLFVSSFTLVLPFFPTGTSERMEDEGDVATAFTLARIL 214
Query: 120 SN 121
SN
Sbjct: 215 SN 216
>At2g37280 putative ABC transporter
Length = 1413
Score = 26.2 bits (56), Expect = 2.8
Identities = 14/49 (28%), Positives = 23/49 (46%), Gaps = 4/49 (8%)
Query: 54 VVW*TCC---FLAQLSSPTQGFEQFTAIYALHCLFVASFALVPPFFPTG 99
+VW TC + P + F QF ++A+H ++ F + F TG
Sbjct: 603 LVW-TCLTYYVIGYTPEPYRFFRQFMILFAVHFTSISMFRCIAAIFQTG 650
>At3g04240 O-linked GlcNAc transferase like protein
Length = 977
Score = 25.4 bits (54), Expect = 4.7
Identities = 12/29 (41%), Positives = 14/29 (47%)
Query: 79 YALHCLFVASFALVPPFFPTGSFE*KEEG 107
YA HC +AS +PPF K EG
Sbjct: 559 YAAHCSIIASRFGLPPFTHPAGLPVKREG 587
>At3g56630 cytochrome P450-like protein
Length = 499
Score = 25.0 bits (53), Expect = 6.1
Identities = 18/62 (29%), Positives = 23/62 (37%), Gaps = 12/62 (19%)
Query: 71 GFEQFTAIYALHCLFVASFALVPP------------FFPTGSFE*KEEGDVTTAFTLARM 118
GFE + LH S L PP P G+F K+ G A+ + RM
Sbjct: 344 GFEDLKLMNYLHAAITESLRLYPPVPVDTMSCAEDNVLPDGTFIGKDWGISYNAYAMGRM 403
Query: 119 LS 120
S
Sbjct: 404 ES 405
>At1g48270 putative G-protein-coupled receptor (GCR1)
Length = 326
Score = 24.6 bits (52), Expect = 8.0
Identities = 11/31 (35%), Positives = 15/31 (47%)
Query: 59 CCFLAQLSSPTQGFEQFTAIYALHCLFVASF 89
C F + P++GF + Y H VASF
Sbjct: 65 CSFFLIVGDPSKGFICYAQGYTTHFFCVASF 95
>At1g11180 secretory carrier membrane protein, putative
Length = 291
Score = 24.6 bits (52), Expect = 8.0
Identities = 14/41 (34%), Positives = 17/41 (41%)
Query: 54 VVW*TCCFLAQLSSPTQGFEQFTAIYALHCLFVASFALVPP 94
V+W + A S F F Y LH LF A+ PP
Sbjct: 176 VLWYRPLYRAFRSDSAFNFGWFFLFYMLHILFCLFAAVAPP 216
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.370 0.164 0.657
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,188,077
Number of Sequences: 26719
Number of extensions: 66324
Number of successful extensions: 294
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 286
Number of HSP's gapped (non-prelim): 7
length of query: 121
length of database: 11,318,596
effective HSP length: 97
effective length of query: 24
effective length of database: 8,726,853
effective search space: 209444472
effective search space used: 209444472
T: 11
A: 40
X1: 14 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0206b.2


