

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0206a.3
(552 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
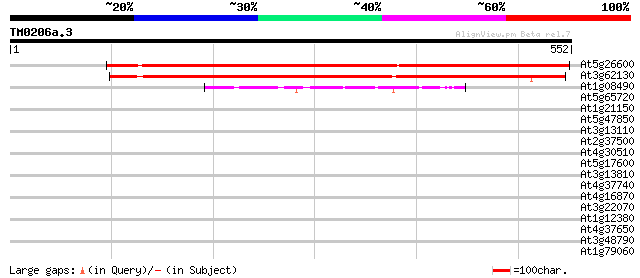
Score E
Sequences producing significant alignments: (bits) Value
At5g26600 unknown protein 659 0.0
At3g62130 unknown protein 567 e-162
At1g08490 NIFS like protein CpNifsp precursor (NIFS) 55 8e-08
At5g65720 NifS-like aminotranfserase 39 0.006
At1g21150 unknown protein 37 0.022
At5g47850 receptor kinase-like protein 34 0.24
At3g13110 serine acetyltransferase (Sat-1) 32 0.70
At2g37500 putative glutamate/ornithine acetyltransferase 32 0.92
At4g30510 unknown protein 31 2.0
At5g17600 RING-H2 zinc finger protein-like 30 2.7
At3g13810 zinc finger protein, putative 30 2.7
At4g37740 transcription activator (GRL2) 30 4.6
At4g16870 retrotransposon like protein 30 4.6
At3g22070 unknown protein 29 5.9
At1g12380 hypothetical protein 29 5.9
At4g37650 SHORT-ROOT (SHR) 29 7.8
At3g48790 serine palmitoyltransferase-like protein 29 7.8
At1g79060 unknown protein 29 7.8
>At5g26600 unknown protein
Length = 475
Score = 659 bits (1700), Expect = 0.0
Identities = 315/456 (69%), Positives = 375/456 (82%), Gaps = 4/456 (0%)
Query: 96 SSSMASTHNHNNNNNGCTTHIPKKPKLSPLSSSPFITPSEIQSEFSHHDPSVARINNGSF 155
SS+ +++ N + K+PK+S + +I+ SEI+SEFSHHDP ARINNGSF
Sbjct: 23 SSASSASSTTNGTVESSVSDFVKRPKISHPN---YISSSEIESEFSHHDPDFARINNGSF 79
Query: 156 GCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNADHLDEISIVDNAT 215
GCC +S+ A Q DWQL++LRQPD FYF+ LK I SR++IK L+NA+H DE+SIVDNAT
Sbjct: 80 GCCPSSILALQRDWQLRFLRQPDRFYFDELKPKISDSRSVIKRLINAEHDDEVSIVDNAT 139
Query: 216 TAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGGTVIEVPLPFPVK 275
TAAAIVLQ TAWAFREG F GD V+MLHYAYG+VKKS+EAYVTR+GG V EV LPFPV
Sbjct: 140 TAAAIVLQQTAWAFREGRFDKGDAVVMLHYAYGSVKKSVEAYVTRSGGHVTEVQLPFPVI 199
Query: 276 SSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDA 335
S+D+I+ FR LE GK+ G++VRLA+IDHVTSMP VVIP+KELVKICR EGVD+VFVDA
Sbjct: 200 SADEIIDRFRIGLESGKANGRRVRLALIDHVTSMPSVVIPIKELVKICRREGVDQVFVDA 259
Query: 336 AHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGSELHHPVVSHEYG 395
AH IGC DVDMKEIGADFYTSNLHKWFF PPS+AFLY RK GG ++LHHPVVS+EYG
Sbjct: 260 AHGIGCVDVDMKEIGADFYTSNLHKWFFAPPSVAFLYCRKSSNGGV-ADLHHPVVSNEYG 318
Query: 396 NGLAVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIEGIKKRNHETVVEMGDMLVKAWGTH 455
NGLAVES+W+GTRDYSAQLVVP++++FVNRFEGGI+GIKKRNHE+VVEMG MLVK+WGT
Sbjct: 319 NGLAVESSWVGTRDYSAQLVVPSILEFVNRFEGGIDGIKKRNHESVVEMGQMLVKSWGTQ 378
Query: 456 VGCPPHMCASMVMVGLPTCLGVQSDSDALKLRTHLREAFGVEVPIYYRPPRDGEVEPVTG 515
+GCPP MCASM+MVGLP CLGV S+SD LKLRT LRE F +E+PIY+RPP DGE++P+TG
Sbjct: 379 LGCPPEMCASMIMVGLPVCLGVSSESDVLKLRTFLREKFRIEIPIYFRPPGDGEIDPITG 438
Query: 516 YARISHQVYNKVDDYYKFRDAVIQLVDKGFTCALLS 551
Y RIS QVYNK +DY++ RDA+ LV GF C LS
Sbjct: 439 YVRISFQVYNKPEDYHRLRDAINGLVRDGFKCTSLS 474
>At3g62130 unknown protein
Length = 454
Score = 567 bits (1461), Expect = e-162
Identities = 272/456 (59%), Positives = 346/456 (75%), Gaps = 15/456 (3%)
Query: 99 MASTHNHNNNNNGCTTHIPKKPKLSPLSSSPFITPSEIQSEFSHHDPSVARINNGSFGCC 158
M + N ++ PKKP+L+ L +T S+I SEF+HH VARINNGSFGCC
Sbjct: 1 MEAGERRNGDSMSHNHRAPKKPRLAGL-----LTESDIDSEFAHHQTGVARINNGSFGCC 55
Query: 159 SASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNADHLDEISIVDNATTAA 218
SV AQ +WQL+YLRQPD FYFN L++G+L SRT+I DL+NAD +DE+S+VDNATTAA
Sbjct: 56 PGSVLEAQREWQLRYLRQPDEFYFNGLRRGLLASRTVISDLINADDVDEVSLVDNATTAA 115
Query: 219 AIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGGTVIEVPLPFPVKSSD 278
AIVLQ F EG ++ D V+M H A+ +VKKS++AYV+R GG+ +EV LPFPV S++
Sbjct: 116 AIVLQKVGRCFSEGKYKKEDTVVMFHCAFQSVKKSIQAYVSRVGGSTVEVRLPFPVNSNE 175
Query: 279 DIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHS 338
+I+ +FR LE+G++ G+ VRLA+IDH+TSMPCV++PV+ELVKICREEGV++VFVDAAH+
Sbjct: 176 EIISKFREGLEKGRANGRTVRLAIIDHITSMPCVLMPVRELVKICREEGVEQVFVDAAHA 235
Query: 339 IGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGSELHHPVVSHEYGNGL 398
IG VD+KEIGAD+Y SNLHKWFFCPPSIAF Y +K GS S++HHPVVSHE+GNGL
Sbjct: 236 IGSVKVDVKEIGADYYVSNLHKWFFCPPSIAFFYCKKR---GSESDVHHPVVSHEFGNGL 292
Query: 399 AVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIEGIKKRNHETVVEMGDMLVKAWGTHVGC 458
+ESAWIGTRDYS+QLVVP+V++FVNRFEGG+EGI +NH+ V MG ML AWGT++G
Sbjct: 293 PIESAWIGTRDYSSQLVVPSVMEFVNRFEGGMEGIMMKNHDEAVRMGLMLADAWGTNLGS 352
Query: 459 PPHMCASMVMVGLPTCLGVQSDSDALKLRTHLREAFGVEVPIYYRPPRDGEVEP------ 512
PP MC MVM+GLP+ L V SD DA+KLR++LR + VEVP++Y RDGE
Sbjct: 353 PPEMCVGMVMIGLPSKLCVGSDEDAIKLRSYLRVHYSVEVPVFYLGLRDGEEGVKDKDSG 412
Query: 513 -VTGYARISHQVYNKVDDYYKFRDAVIQLVDKGFTC 547
+T Y RISHQVYNK +DY + RDA+ +LV TC
Sbjct: 413 LITAYVRISHQVYNKTEDYERLRDAITELVKDQMTC 448
>At1g08490 NIFS like protein CpNifsp precursor (NIFS)
Length = 463
Score = 55.5 bits (132), Expect = 8e-08
Identities = 66/267 (24%), Positives = 116/267 (42%), Gaps = 36/267 (13%)
Query: 192 SRTIIKDLVNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVK 251
+R + +NA EI NAT A +V A+++ +PGD V++ + +
Sbjct: 121 ARKKVARFINASDSREIVFTRNATEAINLV----AYSWGLSNLKPGDEVILTVAEHHSCI 176
Query: 252 KSMEAYVTRAGGTVIEVPLPFPVKSSDDI--VREFRSALERGKSGGKKVRLAVIDHVTSM 309
+ + G L F + D++ + + R + K +L + HV+++
Sbjct: 177 VPWQIVSQKTGAV-----LKFVTLNEDEVPDINKLRELIS------PKTKLVAVHHVSNV 225
Query: 310 PCVVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFC-PPSI 368
+P++E+V + G +V VDA S+ VD++++ ADF ++ HK C P I
Sbjct: 226 LASSLPIEEIVVWAHDVGA-KVLVDACQSVPHMVVDVQKLNADFLVASSHK--MCGPTGI 282
Query: 369 AFLYSRKH------PKGGSGSELHHPVVSHE-YGNGLAVESAWIGTRDYSAQLVVPAVID 421
FLY + P G G + + H Y + A GT + + A +D
Sbjct: 283 GFLYGKSDLLHSMPPFLGGGEMISDVFLDHSTYAEPPSRFEA--GTPAIGEAIALGAAVD 340
Query: 422 FVNRFEGGIEGIKKRNHETVVEMGDML 448
+++ GI G+ K HE VE+G L
Sbjct: 341 YLS----GI-GMPK-IHEYEVEIGKYL 361
>At5g65720 NifS-like aminotranfserase
Length = 453
Score = 39.3 bits (90), Expect = 0.006
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 2/67 (2%)
Query: 312 VVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFL 371
VV P++E+ IC+E V DAA +IG VD+K+ + + HK + P + L
Sbjct: 207 VVQPMEEIGMICKEHNVP-FHTDAAQAIGKIPVDVKKWNVALMSMSAHK-IYGPKGVGAL 264
Query: 372 YSRKHPK 378
Y R+ P+
Sbjct: 265 YVRRRPR 271
>At1g21150 unknown protein
Length = 390
Score = 37.4 bits (85), Expect = 0.022
Identities = 37/127 (29%), Positives = 55/127 (43%), Gaps = 10/127 (7%)
Query: 7 LVECNKCKVFNNKHLFGFFNFGFPCGWIHVLCSLSKVKTHPLVSVPLIIYSISFSPLTHS 66
+V+C F + + N GF G I LCS+S+VK+ L + S + S L S
Sbjct: 10 VVDC----AFTSSTVRRIVNLGFLVGRIRTLCSVSQVKSENLEKQKPCLESFTVSYLVDS 65
Query: 67 ILSLCLSSSSSSSYVVVLSLVIFTHHFLISSSMASTHNHNNNNNGCTTHIPKKPKLSPLS 126
L L S+ S+S V L S +A +H N+ T+ I P++ LS
Sbjct: 66 -CGLSLESAKSNSRFVKL-----VSSKKPDSVLALFKDHGFTNDQITSVIKSFPRVLSLS 119
Query: 127 SSPFITP 133
I+P
Sbjct: 120 PEDVISP 126
>At5g47850 receptor kinase-like protein
Length = 751
Score = 33.9 bits (76), Expect = 0.24
Identities = 36/136 (26%), Positives = 57/136 (41%), Gaps = 19/136 (13%)
Query: 142 HHDPSVARINNGSFGCCSAS-VTAAQHDWQLKYLRQPDNF--------YFNHLKQ--GIL 190
+H P + + G+F C V+ WQ YL +PDN+ +F L Q G++
Sbjct: 120 YHGPELEELEAGNFRICGVERVSRRLRCWQPYYLPRPDNYRSIALGDNFFCGLSQPPGMI 179
Query: 191 RSRTIIKDLVNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAV 250
I K + + DH I+I + A AI + N + + P + L L AV
Sbjct: 180 SCEGIAK-VPSGDHY--IAIAAGSRQACAITVDNDVECWGQTQSLPREKFLAL-----AV 231
Query: 251 KKSMEAYVTRAGGTVI 266
+ V + GTV+
Sbjct: 232 GEDRGCGVRWSNGTVV 247
>At3g13110 serine acetyltransferase (Sat-1)
Length = 391
Score = 32.3 bits (72), Expect = 0.70
Identities = 29/107 (27%), Positives = 45/107 (41%), Gaps = 24/107 (22%)
Query: 126 SSSP------FITPSEIQSEFSHHD-----------PSVARINNGSFGCCSASV--TAAQ 166
SSSP F+ PS + S+F HH P A I+ G S ++
Sbjct: 19 SSSPHINHHSFLLPSFVSSKFKHHTLSPPPSPPPPPPMAACIDTCRTGKPQISPRDSSKH 78
Query: 167 HDWQ-----LKYLRQPDNFYFNHLKQGILRSRTIIKDLVNADHLDEI 208
HD + + Y R PD FN + L +R +++DL +D++
Sbjct: 79 HDDESGFRYMNYFRYPDRSSFNGTQTKTLHTRPLLEDLDRDAEVDDV 125
>At2g37500 putative glutamate/ornithine acetyltransferase
Length = 468
Score = 32.0 bits (71), Expect = 0.92
Identities = 21/77 (27%), Positives = 34/77 (43%), Gaps = 16/77 (20%)
Query: 463 CASMVMVGLP----------TCL------GVQSDSDALKLRTHLREAFGVEVPIYYRPPR 506
C VM GL TCL G +++++A K+ + + V+ +Y R P
Sbjct: 319 CLDAVMQGLAKSIAWDGEGATCLIEVTVKGTETEAEAAKIARSVASSSLVKAAVYGRDPN 378
Query: 507 DGEVEPVTGYARISHQV 523
G + GYA +S Q+
Sbjct: 379 WGRIAAAAGYAGVSFQM 395
>At4g30510 unknown protein
Length = 312
Score = 30.8 bits (68), Expect = 2.0
Identities = 19/55 (34%), Positives = 28/55 (50%)
Query: 34 IHVLCSLSKVKTHPLVSVPLIIYSISFSPLTHSILSLCLSSSSSSSYVVVLSLVI 88
+H++ +K + + P IYS+SF P T L +SSS S + LSL I
Sbjct: 153 VHLVSEATKSYSFRRGTYPSTIYSLSFGPSTQLPDILIATSSSGSIHAFSLSLAI 207
>At5g17600 RING-H2 zinc finger protein-like
Length = 362
Score = 30.4 bits (67), Expect = 2.7
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Query: 57 SISFSPLTHSILSLCLSSSSSSSYVVVLSLVIFTHHFLISSSMASTHNHNNNNNG 111
S SFSPL +++ + S+ SY ++S HH +S + T N N+N G
Sbjct: 51 SSSFSPLLIALIGILTSALILVSYYTLISKYCHRHH---QTSSSETLNLNHNGEG 102
>At3g13810 zinc finger protein, putative
Length = 513
Score = 30.4 bits (67), Expect = 2.7
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 11/76 (14%)
Query: 52 PLIIYSISFSP----LTHSILSLCLSSSSSSSYVVVLSLVIFTHHFLISSSMASTHNHNN 107
PL+I+ + P T +++ SSSSS ++ ++ SL HF ++ +T+N NN
Sbjct: 248 PLLIHQSASHPHHHHQTQPTINVSSSSSSSHNHNIINSL-----HF--DTNNGNTNNSNN 300
Query: 108 NNNGCTTHIPKKPKLS 123
+NN T KK + S
Sbjct: 301 SNNHLHTFPMKKEQQS 316
>At4g37740 transcription activator (GRL2)
Length = 535
Score = 29.6 bits (65), Expect = 4.6
Identities = 21/58 (36%), Positives = 33/58 (56%), Gaps = 9/58 (15%)
Query: 94 LISSSMASTHNHNNNNNGCTTHIPKKPKLSPLSSSPFITPSEIQSEFSHHDPSVARIN 151
+ SSS +STHN+NN +K LSPL S + S IQ++ + +P+V ++N
Sbjct: 418 IASSSPSSTHNNNN--------AQEKTTLSPLRLSRELDLS-IQTDETTIEPTVKKVN 466
>At4g16870 retrotransposon like protein
Length = 1474
Score = 29.6 bits (65), Expect = 4.6
Identities = 28/119 (23%), Positives = 51/119 (42%), Gaps = 5/119 (4%)
Query: 215 TTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRA--GGTVIEVPLPF 272
T A + N A + T L+ GA+ ++ V+RA + +
Sbjct: 65 TPAPTLTTNNVVSANPQYTLWKRQDRLIFSALIGAISPPVQPLVSRATKASQIWKTLTNT 124
Query: 273 PVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKICREEGVDRV 331
KSS D +++ R+ +++ K G K + V+ H T + + I K + EE V+R+
Sbjct: 125 YAKSSYDHIKQLRTQIKQLKKGTKTIDEYVLSHTTLLDQLAILGKPME---HEEQVERI 180
>At3g22070 unknown protein
Length = 146
Score = 29.3 bits (64), Expect = 5.9
Identities = 16/66 (24%), Positives = 34/66 (51%), Gaps = 3/66 (4%)
Query: 70 LCLSSSSSSSYVVVLSLVIFTHHFLISSSMASTHNHNNNNNGC--TTHIPKKPKLSPLSS 127
LC ++ ++ +++ V+F +++ + H+H+ + + C +T S S+
Sbjct: 2 LCYVGKATKIFIFIVT-VVFVIGLVVAFGVLRRHSHHCSGDYCSSSTDPSSSSSSSSSST 60
Query: 128 SPFITP 133
SPFITP
Sbjct: 61 SPFITP 66
>At1g12380 hypothetical protein
Length = 793
Score = 29.3 bits (64), Expect = 5.9
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query: 96 SSSMASTHNHNNNNNGC-TTHIPKKPKLSPLSSSPFITP 133
SSS TH+ N++G TT IP + P+ S +TP
Sbjct: 132 SSSSPQTHHRKRNSSGAVTTAIPSRLNPPPIGGSYHVTP 170
>At4g37650 SHORT-ROOT (SHR)
Length = 531
Score = 28.9 bits (63), Expect = 7.8
Identities = 24/66 (36%), Positives = 33/66 (49%), Gaps = 15/66 (22%)
Query: 97 SSMASTHNHNNNNNGCTTHIP--KKPKLSPLSSSPFITPSE------IQSEFS----HHD 144
SS +S HNH+N+NN T + P + P +SS TPS + S +S H+D
Sbjct: 65 SSSSSHHNHHNHNNPNTYYSPFTTPTQYHPATSS---TPSSTAAAAALASPYSSSGHHND 121
Query: 145 PSVARI 150
PS I
Sbjct: 122 PSAFSI 127
>At3g48790 serine palmitoyltransferase-like protein
Length = 386
Score = 28.9 bits (63), Expect = 7.8
Identities = 16/45 (35%), Positives = 27/45 (59%), Gaps = 2/45 (4%)
Query: 296 KKVRLAVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHSIG 340
KK+ + V++ + SM + + E+V +C E V++D AHSIG
Sbjct: 133 KKI-IVVVEGIYSMEGEICDLPEIVSVCSEYKA-YVYLDEAHSIG 175
>At1g79060 unknown protein
Length = 396
Score = 28.9 bits (63), Expect = 7.8
Identities = 15/35 (42%), Positives = 21/35 (59%), Gaps = 3/35 (8%)
Query: 87 VIFTHHFLISSSMASTHNH---NNNNNGCTTHIPK 118
V+F F SSS +S+ N+ NNNNN + HI +
Sbjct: 354 VVFGQFFSSSSSSSSSQNNRTGNNNNNRASCHISR 388
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,222,396
Number of Sequences: 26719
Number of extensions: 589079
Number of successful extensions: 1791
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1772
Number of HSP's gapped (non-prelim): 26
length of query: 552
length of database: 11,318,596
effective HSP length: 104
effective length of query: 448
effective length of database: 8,539,820
effective search space: 3825839360
effective search space used: 3825839360
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0206a.3


