

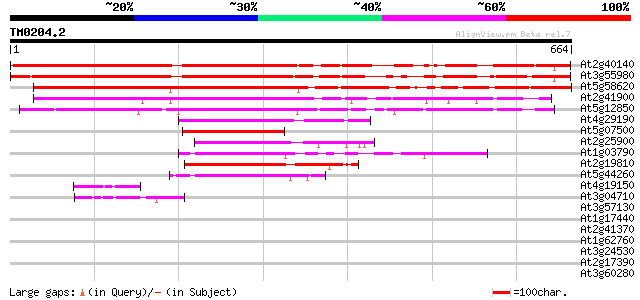
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0204.2
(664 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g40140 putative CCCH-type zinc finger protein 622 e-178
At3g55980 unknown protein 599 e-171
At5g58620 unknown protein 591 e-169
At2g41900 CCCH-type zinc finger like protein 442 e-124
At5g12850 zinc finger transcription factor -like protein 439 e-123
At4g29190 unknown protein 201 1e-51
At5g07500 zinc finger transcription factor 189 6e-48
At2g25900 Cys3His zinc finger like protein ATCTH (ATCTH) 187 2e-47
At1g03790 unknown protein 185 8e-47
At2g19810 putative CCCH-type zinc finger protein 184 1e-46
At5g44260 zinc finger like protein 171 1e-42
At4g19150 ankyrin-like protein 44 4e-04
At3g04710 ankyrin-like protein 44 4e-04
At3g57130 putative protein 39 0.012
At1g17440 unknown protein 38 0.016
At2g41370 unknown protein (At2g41370) 37 0.027
At1g62760 hypothetical protein 37 0.035
At3g24530 unknown protein 36 0.060
At2g17390 putative glucanase 36 0.079
At3g60280 uclacyanin 3 35 0.10
>At2g40140 putative CCCH-type zinc finger protein
Length = 597
Score = 622 bits (1603), Expect = e-178
Identities = 344/686 (50%), Positives = 437/686 (63%), Gaps = 114/686 (16%)
Query: 1 MCSGSKSKLSSSEQVMESQ--KENCDGLHNFSILLELSASNDFEALKREVDEK-GLDVNE 57
MC G+KS L SS+ + E + ++ + + + LLE +A +D + KRE++E ++++E
Sbjct: 1 MC-GAKSNLCSSKTLTEVEFMRQKSEDGASATCLLEFAACDDLSSFKREIEENPSVEIDE 59
Query: 58 AGFWYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCA 117
+GFWY RR+GSKKMG E+RTPLM+A+++GS V+ Y++ TGR DVN+ C +K TALHCA
Sbjct: 60 SGFWYCRRVGSKKMGFEERTPLMVAAMYGSMEVLNYIIATGRSDVNRVCSDEKVTALHCA 119
Query: 118 VAGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGES 177
V+G S +E++K+LLDA + + +DA GNKP++L+A RKA+E+LL G
Sbjct: 120 VSGCSVSIVEIIKILLDASASPNCVDANGNKPVDLLAKDSRFVPNQSRKAVEVLLTGIHG 179
Query: 178 GQLIHQEMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRA 237
+ +E +L+ + +YP D SLPDIN G+YG+D+FRM+SFKVKPCSRA
Sbjct: 180 SVMEEEEEELKSVVT-----------KYPADASLPDINEGVYGTDDFRMFSFKVKPCSRA 228
Query: 238 YSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPA 297
YSHDWTECPFVHPGENARRRDPRKYPY+CVPCPEFRKG+C KGDSCEYAHGVFESWLHPA
Sbjct: 229 YSHDWTECPFVHPGENARRRDPRKYPYTCVPCPEFRKGSCPKGDSCEYAHGVFESWLHPA 288
Query: 298 QYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALS 357
QYRTRLCKDETGC R+VCFFAHR +ELRPV ASTGSAM SP+S S+ + +MSVMSPL L
Sbjct: 289 QYRTRLCKDETGCARRVCFFAHRRDELRPVNASTGSAMVSPRS-SNQSPEMSVMSPLTLG 347
Query: 358 SSSLPMSTVSTPPMSPLAGS--LSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDF 415
SS P SP+A LSP++G LWQN++N +LTPP LQL GSRLK+ LSARD
Sbjct: 348 SS---------PMNSPMANGVPLSPRNGGLWQNRVN-SLTPPPLQLNGSRLKSTLSARDM 397
Query: 416 DLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNL 475
D+EME+ G R R+GDL P+NL++ GS DS
Sbjct: 398 DMEMELRFRGLDNR-------------------------RLGDLKPSNLEETFGSYDSAS 432
Query: 476 LSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVA 535
+ Q LQS SRH MNH PSSPVR+P GF+SS+A+A
Sbjct: 433 VMQ------------LQSPSRH----SQMNH--------YPSSPVRQPPPHGFESSAAMA 468
Query: 536 AAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVN 595
AAVMN+RS+AFAKRS SF V+S SDW SP+GKL+WG+
Sbjct: 469 AAVMNARSSAFAKRSLSF------------------KPAPVASNVSDWGSPNGKLEWGMQ 510
Query: 596 GDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKDDL------------ 643
DEL+KLR+SASFG N P A EPDVSWV+SLVK++
Sbjct: 511 RDELNKLRRSASFGIHGNNNNSVSRP-ARDYSDEPDVSWVNSLVKENAPERVNERVGNTV 569
Query: 644 ------SKEMLPPWVEQLYIEQEQMV 663
K LP W EQ+YI+ EQ +
Sbjct: 570 NGAASRDKFKLPSWAEQMYIDHEQQI 595
>At3g55980 unknown protein
Length = 580
Score = 599 bits (1545), Expect = e-171
Identities = 338/679 (49%), Positives = 429/679 (62%), Gaps = 116/679 (17%)
Query: 1 MCSGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGF 60
MCSG KS L SS + E + + +LLE +A +D ++ KREV+EKGLD++E+G
Sbjct: 1 MCSGPKSNLCSSRTLTEIESRQKE--EETMLLLEFAACDDLDSFKREVEEKGLDLDESGL 58
Query: 61 WYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAG 120
WY RR+GSKKMG E+RTPLM+A+++GS +V+ ++V TG+ DVN+ACG ++ T LHCAVAG
Sbjct: 59 WYCRRVGSKKMGLEERTPLMVAAMYGSIKVLTFIVSTGKSDVNRACGEERVTPLHCAVAG 118
Query: 121 GSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESGQL 180
S +EV+ VLLDA + +S+DA GN+P+++ + RRKA+ELLLRGG G L
Sbjct: 119 CSVNMIEVINVLLDASALVNSVDANGNQPLDVFVRVSRFVASPRRKAVELLLRGGGVGGL 178
Query: 181 IHQ--EMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAY 238
I + E +++++S +YP D SLPDIN G+YGSDEFRMYSFKVKPCSRAY
Sbjct: 179 IDEAVEEEIKIVS------------KYPADASLPDINEGVYGSDEFRMYSFKVKPCSRAY 226
Query: 239 SHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQ 298
SHDWTEC FVHPGENARRRDPRKYPY+CVPCPEFRKG+C KGDSCEYAHGVFESWLHPAQ
Sbjct: 227 SHDWTECAFVHPGENARRRDPRKYPYTCVPCPEFRKGSCPKGDSCEYAHGVFESWLHPAQ 286
Query: 299 YRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSS 358
Y+TRLCKDETGC RKVCFFAH+ EE+RPV ASTGSA+ +S S+ M +SPLA SS
Sbjct: 287 YKTRLCKDETGCARKVCFFAHKREEMRPVNASTGSAV--AQSPFSSLEMMPGLSPLAYSS 344
Query: 359 SSLPMSTVSTPPMSPLAGSL--SPKSGNLWQNKINLNLTPPSLQL-PGSRLKTALSARDF 415
VSTPP+SP+A + SP++G WQN++N LTPP+LQL GSRLK+ LSARD
Sbjct: 345 G------VSTPPVSPMANGVPSSPRNGGSWQNRVN-TLTPPALQLNGGSRLKSTLSARDI 397
Query: 416 DLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNL 475
D+EMEM + F N N+++ GS
Sbjct: 398 DMEMEM------------------------ELRLRGFGN--------NVEETFGS----- 420
Query: 476 LSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRK-PSSLGFDSSSAV 534
++ S SR+ M QNMN + PSSPVR+ PS GF+SS+A
Sbjct: 421 --------------YVSSPSRNSQMGQNMN-------QHYPSSPVRQPPSQHGFESSAAA 459
Query: 535 AAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGV 594
A AVM +RS AFAKRS SF P+ + S SDW SP+GKL+WG+
Sbjct: 460 AVAVMKARSTAFAKRSLSF--------------KPATQAA-PQSNLSDWGSPNGKLEWGM 504
Query: 595 NGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKDDL----------S 644
G+EL+K+R+S SFG N + A EPDVSWV+SLVKD
Sbjct: 505 KGEELNKMRRSVSFGIHGN----NNNNAARDYRDEPDVSWVNSLVKDSTVVSERSFGMNE 560
Query: 645 KEMLPPWVEQLYIEQEQMV 663
+ + W EQ+Y E+EQ V
Sbjct: 561 RVRIMSWAEQMYREKEQTV 579
>At5g58620 unknown protein
Length = 607
Score = 591 bits (1523), Expect = e-169
Identities = 327/647 (50%), Positives = 412/647 (63%), Gaps = 61/647 (9%)
Query: 29 FSILLELSASNDFEALKREVDEKGLD-VNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGS 87
FS+LLE SA ND K V+E+GL+ ++ +G WYGRR+GSKKMG E+RTPLMIA+LFGS
Sbjct: 11 FSLLLESSACNDLSGFKSLVEEEGLESIDGSGLWYGRRLGSKKMGFEERTPLMIAALFGS 70
Query: 88 TRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGN 147
VV Y++ TG VDVN++CGSD ATALHCAV+G S SLE+V +LL ++ DS DA GN
Sbjct: 71 KEVVDYIISTGLVDVNRSCGSDGATALHCAVSGLSANSLEIVTLLLKGSANPDSCDAYGN 130
Query: 148 KPMNLIAPAFNSSSKSRRKAMELLLRGGESGQLIHQEMDLEL---ISAPFSSKEGSDKKE 204
KP ++I P + +R K +E LL+G + ++ + + E + S GS++KE
Sbjct: 131 KPGDVIFPCLSPVFSARMKVLERLLKGNDDLNEVNGQEESEPEVEVEVEVSPPRGSERKE 190
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
YPVD +LPDI NG+YG+DEFRMY+FK+KPCSRAYSHDWTECPFVHPGENARRRDPRKY Y
Sbjct: 191 YPVDPTLPDIKNGVYGTDEFRMYAFKIKPCSRAYSHDWTECPFVHPGENARRRDPRKYHY 250
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
SCVPCPEFRKG+C +GD+CEYAHG+FE WLHPAQYRTRLCKDET C R+VCFFAH+PEEL
Sbjct: 251 SCVPCPEFRKGSCSRGDTCEYAHGIFECWLHPAQYRTRLCKDETNCSRRVCFFAHKPEEL 310
Query: 325 RPVYASTGSAMPSPKS-----YSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLS 379
RP+Y STGS +PSP+S SS A DM +SP LP+ +TPP+SP S
Sbjct: 311 RPLYPSTGSGVPSPRSSFSSCNSSTAFDMGPISP-------LPIGATTTPPLSPNGVSSP 363
Query: 380 PKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTRQQQQQQQQQQ 439
G W N N+TPP+LQLPGSRLK+AL+AR+ D EM L SPT
Sbjct: 364 IGGGKTWMNW--PNITPPALQLPGSRLKSALNAREIDFSEEMQSLTSPTTWNNTPMSSPF 421
Query: 440 LIEEIARISSPSFRNRIGDLTPTN-LDDLLGSVDSNLLSQLHGLSGPSTPTHLQSQSRHQ 498
+ + R++ G ++P N L D+ G+ D+ GL Q R
Sbjct: 422 SGKGMNRLAG-------GAMSPVNSLSDMFGTEDNT-----SGL-----------QIRRS 458
Query: 499 MMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVMNSRSAAFAK-RSQSFIDRG 557
++ + + +++ SSPV S DSS AV+ SR+A FAK RSQSFI+R
Sbjct: 459 VINPQL------HSNSLSSSPVGANSLFSMDSS-----AVLASRAAEFAKQRSQSFIERN 507
Query: 558 AAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAP 617
HH +SS ++ +DW S GKLDW V GDEL KLRKS SF R GM
Sbjct: 508 NGLNHHPAISS------MTTTCLNDWGSLDGKLDWSVQGDELQKLRKSTSFRLRAGGM-E 560
Query: 618 TGSPMAHSEHAEPDVSWVHSLVKDDLSKEMLPPWVEQLYIEQEQMVA 664
+ P + EPDVSWV LVK+ + P W+EQ Y+E EQ VA
Sbjct: 561 SRLPNEGTGLEEPDVSWVEPLVKEPQETRLAPVWMEQSYMETEQTVA 607
>At2g41900 CCCH-type zinc finger like protein
Length = 716
Score = 442 bits (1136), Expect = e-124
Identities = 275/667 (41%), Positives = 371/667 (55%), Gaps = 100/667 (14%)
Query: 29 FSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGST 88
F+ LLEL+A+ND E ++ ++ V+EAG WYGR+ GSK M ++ RTPLM+A+ +GS
Sbjct: 45 FASLLELAANNDVEGVRLSIERDPSCVDEAGLWYGRQKGSKAMVNDYRTPLMVAATYGSI 104
Query: 89 RVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNK 148
V+K +V DVN+ACG+D+ TALHCA +GG+ +++VVK+LL AG+D + LDA G +
Sbjct: 105 DVIKLIVSLTDADVNRACGNDQTTALHCAASGGAVNAIQVVKLLLAAGADLNLLDAEGQR 164
Query: 149 PMNLIAP---------------AFNSSSKSRRKAMELLLRGGESGQLIHQEMDLE----- 188
++I + + SS + R + S H
Sbjct: 165 AGDVIVVPPKLEGVKLMLQELLSADGSSTAERNLRVVTNVPNRSSSPCHSPTGENGGSGS 224
Query: 189 --LISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECP 246
+ +PF K KKEYPVD SLPDI N +Y +DEFRMYSFKV+PCSRAYSHDWTECP
Sbjct: 225 GSPLGSPFKLKSTEFKKEYPVDPSLPDIKNSIYATDEFRMYSFKVRPCSRAYSHDWTECP 284
Query: 247 FVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKD 306
FVHPGENARRRDPRK+ YSCVPCP+FRKGAC++GD CEYAHGVFE WLHPAQYRTRLCKD
Sbjct: 285 FVHPGENARRRDPRKFHYSCVPCPDFRKGACRRGDMCEYAHGVFECWLHPAQYRTRLCKD 344
Query: 307 ETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVM--SPLALSSSSLPMS 364
TGC R+VCFFAH PEELRP+YASTGSA+PSP+S + A +S++ SP +S S
Sbjct: 345 GTGCARRVCFFAHTPEELRPLYASTGSAVPSPRSNADYAAALSLLPGSPSGVSVMS---- 400
Query: 365 TVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPG-----SRLKTALSARDFDL-E 418
P+SP A N+ + N+ P+L LPG SRL+++L+ARD E
Sbjct: 401 -----PLSPSAAGNGMSHSNMAWPQPNV----PALHLPGSNLQSSRLRSSLNARDIPTDE 451
Query: 419 MEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSN--LL 476
ML +QQQL+ E + S S R+ + P+NL+DL + S+
Sbjct: 452 FNMLA----------DYEQQQLLNEYSNALSRS--GRMKSMPPSNLEDLFSAEGSSSPRF 499
Query: 477 SQLHGLSGPSTPTHL-----QSQSRHQMMQQNMNHLRSSY--PSNIPSS----------- 518
+ S +PTH Q Q + Q Q ++ + +S+ P ++ S
Sbjct: 500 TDSALASAVFSPTHKSAVFNQFQQQQQQQQSMLSPINTSFSSPKSVDHSLFSGGGRMSPR 559
Query: 519 PVRKPSSLGFDSSSAVAAAVMNSRSAAFAKRSQ----SFIDRGAAGTHHLGMSSPSNPSC 574
V +P S S +A V + ++ Q S R + SP N
Sbjct: 560 NVVEPISPMSARVSMLAQCVKQQQQQQQQQQQQHQFRSLSSRELRTNSSPIVGSPVNN-- 617
Query: 575 RVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSW 634
++ S W S +G+ DWG++ + L KLR S+SF + EPDVSW
Sbjct: 618 --NTWSSKWGSSNGQPDWGMSSEALGKLRSSSSF-----------------DGDEPDVSW 658
Query: 635 VHSLVKD 641
V SLVK+
Sbjct: 659 VQSLVKE 665
>At5g12850 zinc finger transcription factor -like protein
Length = 706
Score = 439 bits (1130), Expect = e-123
Identities = 275/678 (40%), Positives = 389/678 (56%), Gaps = 85/678 (12%)
Query: 12 SEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKM 71
+E + + + + H+FS LLE +A ND E +R++ + +N+ G WY R+ ++M
Sbjct: 22 NESLAKPMNDAAEWEHSFSALLEFAADNDVEGFRRQLSDVSC-INQMGLWYRRQRFVRRM 80
Query: 72 GSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKV 131
E+RTPLM+ASL+GS VVK+++ ++N +CG DK+TALHCA +G S SL+VVK+
Sbjct: 81 VLEQRTPLMVASLYGSLDVVKFILSFPEAELNLSCGPDKSTALHCAASGASVNSLDVVKL 140
Query: 132 LLDAGSDADSLDACGNKPMN-----------------------LIAPAFNSSSKSRRKAM 168
LL G+D + DA GN+P++ +I+ ++SS S +
Sbjct: 141 LLSVGADPNIPDAHGNRPVDVLVVSPHAPGLRTILEEILKKDEIISEDLHASSSSLGSSF 200
Query: 169 ELLLRGGESGQLIHQEMDLELISAPFSSKE-----GSDKKEYPVDVSLPDINNGLYGSDE 223
L ++G + + L+ +S+P S+KKEYP+D SLPDI +G+Y +DE
Sbjct: 201 RSLSSSPDNGSSL---LSLDSVSSPTKPHGTDVTFASEKKEYPIDPSLPDIKSGIYSTDE 257
Query: 224 FRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSC 283
FRM+SFK++PCSRAYSHDWTECPF HPGENARRRDPRK+ Y+CVPCP+F+KG+C++GD C
Sbjct: 258 FRMFSFKIRPCSRAYSHDWTECPFAHPGENARRRDPRKFHYTCVPCPDFKKGSCKQGDMC 317
Query: 284 EYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPK---S 340
EYAHGVFE WLHPAQYRTRLCKD GC R+VCFFAH EELRP+Y STGS +PSP+ +
Sbjct: 318 EYAHGVFECWLHPAQYRTRLCKDGMGCNRRVCFFAHANEELRPLYPSTGSGLPSPRASSA 377
Query: 341 YSSNAVDM-SVMSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNLW--QNKINLNLTPP 397
S++ +DM SV++ L S S+ S TPP+SP P S W QN LNL
Sbjct: 378 VSASTMDMASVLNMLPGSPSAAQHS--FTPPISPSGNGSMPHSSMGWPQQNIPALNLPGS 435
Query: 398 SLQLPGSRLKTALSARDFDLE-MEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRN-- 454
++QL SRL+++L+ARD E + ML + + Q+QL + + SP F N
Sbjct: 436 NIQL--SRLRSSLNARDIPSEQLSML---------HEFEMQRQLAGD---MHSPRFMNHS 481
Query: 455 -RIGDLTPTNLDDLLGS-VDSNLLSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYP 512
R L P+NL++L + V S S +S +P+H + +Q N + S
Sbjct: 482 ARPKTLNPSNLEELFSAEVASPRFSDQLAVSSVLSPSH--KSALLNQLQNNKQSMLSPIK 539
Query: 513 SNIPSSP--VRKPSSLGFDSSSAVAAAV--MNSR-SAAFAKRSQSFIDRGAAGTHHLGMS 567
+N+ SSP V + S L SS + MN+R RS S D G++ L +
Sbjct: 540 TNLMSSPKNVEQHSLLQQASSPRGGEPISPMNARMKQQLHSRSLSSRDFGSSLPRDLMPT 599
Query: 568 SPSNPSCRVSSGFSDWNSPSG-KLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSE 626
+P S +S W+ G K+DW V DEL +LRKS S +A++
Sbjct: 600 DSGSP----LSPWSSWDQTHGSKVDWSVQSDELGRLRKSHS--------------LANNP 641
Query: 627 HAEPDVSWVHSLVKDDLS 644
+ E DVSW ++KD S
Sbjct: 642 NREADVSWAQQMLKDSSS 659
>At4g29190 unknown protein
Length = 356
Score = 201 bits (510), Expect = 1e-51
Identities = 107/229 (46%), Positives = 138/229 (59%), Gaps = 16/229 (6%)
Query: 201 DKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPR 260
D + YP D+ PD Y D FRMY FKV+ C+R SHDWTECP+ HPGE ARRRDPR
Sbjct: 58 DPESYP-DLLGPDSPIDAYSCDHFRMYDFKVRRCARGRSHDWTECPYAHPGEKARRRDPR 116
Query: 261 KYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHR 320
KY YS CP+FRKG C+KGDSCE+AHGVFE WLHPA+YRT+ CKD C RK+CFFAH
Sbjct: 117 KYHYSGTACPDFRKGGCKKGDSCEFAHGVFECWLHPARYRTQPCKDGGNCLRKICFFAHS 176
Query: 321 PEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVS-TPPMSPLAGS-L 378
P++LR ++ + + S V SP+ + L +S VS +PPMSP A S
Sbjct: 177 PDQLRFLHTRSPDRVDS----------FDVSSPIRARAFQLSISPVSGSPPMSPRADSES 226
Query: 379 SPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSP 427
SP + +L ++ + ++ +P R S + F + G GSP
Sbjct: 227 SPMTQSLSRSLGSCSIND---VVPSFRNLQFNSVKSFPRNNPLFGFGSP 272
>At5g07500 zinc finger transcription factor
Length = 245
Score = 189 bits (479), Expect = 6e-48
Identities = 79/121 (65%), Positives = 94/121 (77%)
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
Y +D S+P+I++ +YGSDEFRMY++K+K C R SHDWTECP+ H GE A RRDPR+Y Y
Sbjct: 34 YEIDPSIPNIDDAIYGSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKATRRDPRRYTY 93
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
V CP FR GAC +GDSCE+AHGVFE WLHPA+YRTR C C RKVCFFAH PE+L
Sbjct: 94 CAVACPAFRNGACHRGDSCEFAHGVFEYWLHPARYRTRACNAGNLCQRKVCFFAHAPEQL 153
Query: 325 R 325
R
Sbjct: 154 R 154
>At2g25900 Cys3His zinc finger like protein ATCTH (ATCTH)
Length = 315
Score = 187 bits (475), Expect = 2e-47
Identities = 99/231 (42%), Positives = 135/231 (57%), Gaps = 31/231 (13%)
Query: 219 YGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQ 278
+ SDEFR+Y FK++ C+R SHDWTECPF HPGE ARRRDPRK+ YS CPEFRKG+C+
Sbjct: 86 FSSDEFRIYEFKIRRCARGRSHDWTECPFAHPGEKARRRDPRKFHYSGTACPEFRKGSCR 145
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSP 338
+GDSCE++HGVFE WLHP++YRT+ CKD T C R++CFFAH E+LR + S
Sbjct: 146 RGDSCEFSHGVFECWLHPSRYRTQPCKDGTSCRRRICFFAHTTEQLRVLPCSLDP----- 200
Query: 339 KSYSSNAVDMSVMSPLALSSSSLPMS-----TVSTPPMSPLAGSLSPKSGNLWQNKINLN 393
D+ S LA S +S+ +S +PP+SP G L + N +
Sbjct: 201 --------DLGFFSGLATSPTSILVSPSFSPPSESPPLSPSTGELIASMRKMQLNGGGCS 252
Query: 394 LTPP---SLQLPGSRLKTALSA------RDFDLE----MEMLGLGSPTRQQ 431
+ P +++LP S + A R+F++E ME + G R +
Sbjct: 253 WSSPMRSAVRLPFSSSLRPIQAATWPRIREFEIEEAPAMEFVESGKELRAE 303
>At1g03790 unknown protein
Length = 393
Score = 185 bits (469), Expect = 8e-47
Identities = 131/383 (34%), Positives = 173/383 (44%), Gaps = 78/383 (20%)
Query: 201 DKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPR 260
+ KEY D D Y SD FRM+ FK++ C+R+ SHDWT+CPF HPGE ARRRDPR
Sbjct: 64 ENKEYCYDSDSDDP----YASDHFRMFEFKIRRCTRSRSHDWTDCPFAHPGEKARRRDPR 119
Query: 261 KYPYSCVPCPEFRKGA-CQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAH 319
++ YS CPEFR+G C +GD CE+AHGVFE WLHP +YRT CKD C RKVCFFAH
Sbjct: 120 RFQYSGEVCPEFRRGGDCSRGDDCEFAHGVFECWLHPIRYRTEACKDGKHCKRKVCFFAH 179
Query: 320 RPEELR---PVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAG 376
P +LR P S SA PSP + +P L SS P ST L G
Sbjct: 180 SPRQLRVLPPENVSGVSASPSP----------AAKNPCCLFCSSSPTST--------LLG 221
Query: 377 SLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTRQQQQQQQ 436
+LS S + PSL P S A + + + +
Sbjct: 222 NLSHLSRS------------PSLSPPMSPANKAAAF------------------SRLRNR 251
Query: 437 QQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQLHGLSGPSTPT------- 489
+ A S ++++ L +L+ S+DS L++ S S T
Sbjct: 252 AASAVSAAAAAGSMNYKD--------VLSELVNSLDSMSLAEALQASSSSPVTTPVSAAA 303
Query: 490 -------HLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVMNSR 542
L +Q H QQ + L+ + + PS P + F MN
Sbjct: 304 AAFASSCGLSNQRLHLQQQQPSSPLQFALSPSTPSYLTNSPQANFFSDDFTPRRRQMNDF 363
Query: 543 SAAFAKRSQSFIDRGAAGTHHLG 565
+A A R + I+ G+ G LG
Sbjct: 364 TAMTAVRENTNIEDGSCGDPDLG 386
>At2g19810 putative CCCH-type zinc finger protein
Length = 359
Score = 184 bits (468), Expect = 1e-46
Identities = 97/212 (45%), Positives = 129/212 (60%), Gaps = 21/212 (9%)
Query: 208 DVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCV 267
D+S PD Y D FRMY FKV+ C+R SHDWTECP+ HPGE ARRRDPRK+ YS
Sbjct: 63 DLSGPDSPIDAYTCDHFRMYEFKVRRCARGRSHDWTECPYAHPGEKARRRDPRKFHYSGT 122
Query: 268 PCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPV 327
CPEFRKG C++GD+CE++HGVFE WLHPA+YRT+ CKD C R+VCFFAH P+++R +
Sbjct: 123 ACPEFRKGCCKRGDACEFSHGVFECWLHPARYRTQPCKDGGNCRRRVCFFAHSPDQIRVL 182
Query: 328 YASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVS-TPPMSPLAGS------LSP 380
P V+SP + +S S +PP+SP S LS
Sbjct: 183 ----------PNQSPDRVDSFDVLSPTIRRAFQFSISPSSNSPPVSPRGDSDSSCSLLSR 232
Query: 381 KSGNLWQNKINLNLTPPSLQLPGSRLKTALSA 412
G+ N + +L +LQL +++K++LS+
Sbjct: 233 SLGSNLGNDVVASLR--NLQL--NKVKSSLSS 260
>At5g44260 zinc finger like protein
Length = 381
Score = 171 bits (433), Expect = 1e-42
Identities = 91/198 (45%), Positives = 119/198 (59%), Gaps = 21/198 (10%)
Query: 190 ISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVH 249
+S+P ++ ++K+Y D D Y D FRMY FK++ C+R+ SHDWT+CPF H
Sbjct: 33 VSSPL--RDYKEQKDYCYDSDSEDP----YAGDHFRMYEFKIRRCTRSRSHDWTDCPFSH 86
Query: 250 PGENARRRDPRKYPYSCVPCPEF-RKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDET 308
PGE ARRRDPR++ Y+ CPEF R G C +GD C +AHGVFE WLHP++YRT CKD
Sbjct: 87 PGEKARRRDPRRFHYTGEVCPEFSRHGDCSRGDECGFAHGVFECWLHPSRYRTEACKDGK 146
Query: 309 GCGRKVCFFAHRPEELRPVYAST---------GSAMPSPKSYSSNAVDMSVM----SPLA 355
C RKVCFFAH P +LR + S GS SP S SN + + SP +
Sbjct: 147 HCKRKVCFFAHSPRQLRVLPPSPENHISGGCGGSPSSSPASVLSNKNNRCCLFCSHSPTS 206
Query: 356 LSSSSLPMSTVSTPPMSP 373
+ +L S S+PP+SP
Sbjct: 207 -TLLNLSRSPSSSPPLSP 223
>At4g19150 ankyrin-like protein
Length = 243
Score = 43.5 bits (101), Expect = 4e-04
Identities = 31/79 (39%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query: 76 RTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDA 135
RTPL +A+ G VV YL + + DV A G D A+H A G LEVV+ LL A
Sbjct: 51 RTPLHLAAWAGHNEVVSYLCK-NKADVGAAAGDDMG-AIHFASQKG---HLEVVRTLLSA 105
Query: 136 GSDADSLDACGNKPMNLIA 154
G S+ G P++ A
Sbjct: 106 GGSVKSITRKGLTPLHYAA 124
>At3g04710 ankyrin-like protein
Length = 456
Score = 43.5 bits (101), Expect = 4e-04
Identities = 45/143 (31%), Positives = 64/143 (44%), Gaps = 24/143 (16%)
Query: 77 TPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAG 136
TPL+ A GS ++ LV+ G N G AT LH A G+ LE++ LL AG
Sbjct: 189 TPLLSAVAAGSLSCLELLVKAG-AKANVFAGG--ATPLHIAADIGN---LELINCLLKAG 242
Query: 137 SDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLL----------RGGESGQLIHQE-- 184
+D + D GN+P+ + A ++ RK +E+L G L H E
Sbjct: 243 ADPNQKDEEGNRPLEVAA------ARDNRKVVEILFPLTTKPETVSDWTVDGILAHMESN 296
Query: 185 MDLELISAPFSSKEGSDKKEYPV 207
+ E S S+E KK+ PV
Sbjct: 297 KEQEENSNKAKSQETGIKKDLPV 319
Score = 38.5 bits (88), Expect = 0.012
Identities = 37/116 (31%), Positives = 53/116 (44%), Gaps = 12/116 (10%)
Query: 77 TPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAG 136
TPL+ A+ G VKYL+E G D N A ATALH A G +E++K LL G
Sbjct: 91 TPLVHAARQGQIETVKYLLEQG-ADPNIA-SELGATALHHAAGTG---EIELLKELLSRG 145
Query: 137 SDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESGQLIHQEMDLELISA 192
DS G P ++ ++ A+E+LL + ++ L+SA
Sbjct: 146 VPVDSESESG-------TPLIWAAGHDQKNAVEVLLEHNANPNAETEDNITPLLSA 194
Score = 31.6 bits (70), Expect = 1.5
Identities = 20/74 (27%), Positives = 35/74 (47%), Gaps = 4/74 (5%)
Query: 73 SEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVL 132
+ KR L A+ G T + +YL+E +++ + + +H A G +E VK L
Sbjct: 53 ANKRGALHFAAREGQTEICRYLLEELKLNADAKDETGDTPLVHAARQG----QIETVKYL 108
Query: 133 LDAGSDADSLDACG 146
L+ G+D + G
Sbjct: 109 LEQGADPNIASELG 122
>At3g57130 putative protein
Length = 467
Score = 38.5 bits (88), Expect = 0.012
Identities = 35/116 (30%), Positives = 53/116 (45%), Gaps = 21/116 (18%)
Query: 38 SNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVET 97
S+D E +K V +GL+++E+ L+ A S VVK L+E
Sbjct: 258 SSDVELVKLMVMGEGLNLDES------------------LALIYAVENCSREVVKALLEL 299
Query: 98 GRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLI 153
G DVN G TALH A +S ++V VLLD +D + G P++++
Sbjct: 300 GAADVNYPAGPTGKTALHIA---AEMVSPDMVAVLLDHHADPNVQTVDGITPLDIL 352
>At1g17440 unknown protein
Length = 683
Score = 38.1 bits (87), Expect = 0.016
Identities = 76/324 (23%), Positives = 121/324 (36%), Gaps = 48/324 (14%)
Query: 333 SAMPSPKSYSS-NAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGS-------LSPKSGN 384
S+ SPKS S N ++ S S L SSS + T P+S S ++ G
Sbjct: 7 SSSLSPKSLQSPNPMEPSPASSTPLPSSSSQQQQLMTAPISNSVNSAASPAMTVTTTEGI 66
Query: 385 LWQNKINLNLTPPS---------LQLPG-SRLKTALSARDFDLEMEMLGLGSPTRQQQQQ 434
+ QN N++ P+ Q+P S L S+ D + + L + QQ
Sbjct: 67 VIQNNSQPNISSPNPTSSNPPIGAQIPSPSPLSHPSSSLDQQTQTQQL----VQQTQQLP 122
Query: 435 QQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQLHGLSGPSTPTHLQ-- 492
QQQQQ++++I+ SSP I L+P L ++ SQ +S LQ
Sbjct: 123 QQQQQIMQQIS--SSP-----IPQLSPQQQQIL---QQQHMTSQQIPMSSYQIAQSLQRS 172
Query: 493 -SQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVMN--------SRS 543
S SR +QQ N+ ++ F S +V + N SR+
Sbjct: 173 PSLSRLSQIQQQQQQQHQGQYGNVLRQQAGLYGTMNFGGSGSVQQSQQNQQMVNPNMSRA 232
Query: 544 AAFAKRSQSFIDRGAAGTHHLGMSSP-SNPSCRVSSGFSDWNSPSGKLDWGVNGDELSKL 602
+ + GAAG + + S R SG + G +G +L +
Sbjct: 233 GLVGQSGHLPMLNGAAGAAQMNIQPQLLAASPRQKSGMVQ----GSQFHPGSSGQQLQGM 288
Query: 603 RKSASFGFRNNGMAPTGSPMAHSE 626
+ G N G+P +++
Sbjct: 289 QAMGMMGSLNLTSQMRGNPALYAQ 312
>At2g41370 unknown protein (At2g41370)
Length = 491
Score = 37.4 bits (85), Expect = 0.027
Identities = 51/194 (26%), Positives = 80/194 (40%), Gaps = 28/194 (14%)
Query: 34 ELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKY 93
+LS + D E K + LD ++ +G + + ++ L A S VVK
Sbjct: 244 DLSVAQDLEDQKIRRMRRALDSSDVELVKLMVMG-EGLNLDESLALHYAVESCSREVVKA 302
Query: 94 LVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLI 153
L+E G DVN G T LH A +S ++V VLLD +D + G P++++
Sbjct: 303 LLELGAADVNYPAGPAGKTPLHIA---AEMVSPDMVAVLLDHHADPNVRTVGGITPLDIL 359
Query: 154 APAFNSSSKSRRKAMELLLRGGESGQLIHQE-----MDLELISAP---FSSKEGSDKKEY 205
R + L +G G L H E + LEL+ + S +EG++
Sbjct: 360 ----------RTLTSDFLFKGAVPG-LTHIEPNKLRLCLELVQSAAMVISREEGNNSNN- 407
Query: 206 PVDVSLPDINNGLY 219
D N G+Y
Sbjct: 408 ----QNNDNNTGIY 417
>At1g62760 hypothetical protein
Length = 312
Score = 37.0 bits (84), Expect = 0.035
Identities = 31/88 (35%), Positives = 44/88 (49%), Gaps = 10/88 (11%)
Query: 329 ASTGSAMPSPKSYSSNAVDMSVMSP-----LALSSSSLPMSTVSTPPMSPLAGSLSPKSG 383
+S+ S PSP S S ++ S +SP L+LS SS P S+ P+S L+ SLSP
Sbjct: 31 SSSPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSLSPSPP 90
Query: 384 NLWQNKINLNLTPPSLQLPGSRLKTALS 411
+ + + PPS P S +LS
Sbjct: 91 S-----SSPSSAPPSSLSPSSPPPLSLS 113
Score = 28.9 bits (63), Expect = 9.6
Identities = 29/95 (30%), Positives = 35/95 (36%)
Query: 321 PEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLSP 380
P P S S P S SS SPL+ S SL S S+ P S SLSP
Sbjct: 45 PSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSLSPSPPSSSPSSAPPSSLSP 104
Query: 381 KSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDF 415
S + PP P S L + S+ +
Sbjct: 105 SSPPPLSLSPSSPPPPPPSSSPLSSLSPSSSSSTY 139
>At3g24530 unknown protein
Length = 481
Score = 36.2 bits (82), Expect = 0.060
Identities = 33/106 (31%), Positives = 49/106 (46%), Gaps = 5/106 (4%)
Query: 61 WYGR-RIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAV- 118
W G ++ + M + TPL +A+ G K L+E+G KA S+ T LH AV
Sbjct: 72 WTGSDKVELEAMNTYGETPLHMAAKNGCNEAAKLLLESGAFIEAKA--SNGMTPLHLAVW 129
Query: 119 AGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSR 164
+ + VK LLD +D + D G P++ + P S K R
Sbjct: 130 YSITAKEISTVKTLLDHNADCSAKDNEGMTPLDHL-PQGQGSEKLR 174
>At2g17390 putative glucanase
Length = 344
Score = 35.8 bits (81), Expect = 0.079
Identities = 26/79 (32%), Positives = 41/79 (50%), Gaps = 11/79 (13%)
Query: 108 SDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKA 167
S+ TALH A G + +VLLDAG++A+++D N P++ A R++
Sbjct: 251 SEGRTALHFACGYGE---VRCAQVLLDAGANANAIDKNKNTPLHYAA------GYGRKEC 301
Query: 168 MELLLRGGESGQLIHQEMD 186
+ LLL G + + Q MD
Sbjct: 302 VSLLLENGAA--VTQQNMD 318
>At3g60280 uclacyanin 3
Length = 222
Score = 35.4 bits (80), Expect = 0.10
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Query: 326 PVYASTGSAMPSPKSYSSNAVDMSVMSPLALS-SSSLPMSTVSTPPMSPLAGSLSPKSGN 384
P ST S+ PSP S S ++ S + P A ++ P S TPP +PL SLSP + +
Sbjct: 142 PSTPSTPSSPPSPPSPPSPSLPPSSLPPSASPPTNGTPDSETLTPPPAPLPPSLSPNAAS 201
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.129 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,720,348
Number of Sequences: 26719
Number of extensions: 725992
Number of successful extensions: 2811
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 82
Number of HSP's that attempted gapping in prelim test: 2518
Number of HSP's gapped (non-prelim): 206
length of query: 664
length of database: 11,318,596
effective HSP length: 106
effective length of query: 558
effective length of database: 8,486,382
effective search space: 4735401156
effective search space used: 4735401156
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0204.2


