

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.4
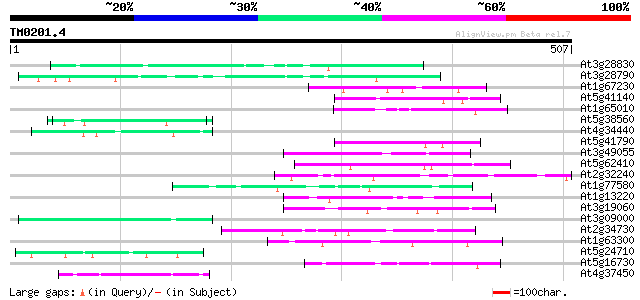
(507 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g28830 hypothetical protein 51 1e-06
At3g28790 unknown protein 51 2e-06
At1g67230 unknown protein 51 2e-06
At5g41140 putative protein 50 2e-06
At1g65010 hypothetical protein 50 4e-06
At5g38560 putative protein 49 5e-06
At4g34440 serine/threonine protein kinase - like 49 7e-06
At5g41790 myosin heavy chain-like protein 48 1e-05
At3g49055 putative protein 48 1e-05
At5g62410 chromosomal protein - like 47 3e-05
At2g32240 putative myosin heavy chain 47 3e-05
At1g77580 unknown protein 46 4e-05
At1g13220 putative nuclear matrix constituent protein 46 4e-05
At3g19060 hypothetical protein 46 6e-05
At3g09000 unknown protein 46 6e-05
At2g34730 putative myosin heavy chain 46 6e-05
At1g63300 hypothetical protein 45 7e-05
At5g24710 unknown protein 45 9e-05
At5g16730 putative protein 45 9e-05
At4g37450 arabinogalactan protein AGP18 45 9e-05
>At3g28830 hypothetical protein
Length = 536
Score = 51.2 bits (121), Expect = 1e-06
Identities = 70/342 (20%), Positives = 138/342 (39%), Gaps = 27/342 (7%)
Query: 38 ASSDAGTSNPGAQPTTAA-APKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSI 96
ASS++ ++ G+ ++ + + K K ESS + ++ +V S S+G +AA ++ S
Sbjct: 197 ASSESSSTKSGSVSSSGSVSTKSK---ESSSSGSSASGSVATKSKESSGGSAATKSKESS 253
Query: 97 GATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQG 156
G +AA+ S ++ +A++ G + +PK+ P S A + S QG
Sbjct: 254 GGSAATKSKESSGGSATTGKTSGSP---SGSPKASPSGSVSGKSSSKGSASAQGSASAQG 310
Query: 157 EKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFFLAGLNRDVIEKEVLSRG 216
S G+A+ +A ++ + + S ++ S+ + V +
Sbjct: 311 SASAQGSASAQGSASAQRRESGAMAMSKSRETKTSSQRQSKSSSESSSSSTTTTTV-KQV 369
Query: 217 LNDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKVESAKHQEAAVAWEKRFDKLATQ 276
++T +E ++ +++ EK AA E ++ ES K A A + ++
Sbjct: 370 ESETSKEVMSFIMQ--------LEKKYAAKAEL-KVFFESLKSSMQASA------SVGSK 414
Query: 277 AGKDKVYADK----MIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIA 332
KD V A K + A + + A MK D + C K+ + +++
Sbjct: 415 TAKDYVSASKAATGKLSEAMASVSSKNVKSAKMKSNLDTSKDEMLKCVKQIQDINGKMVS 474
Query: 333 RGEALIAKESELAVLCAELELVKKALAEQEKKSAESLALAKS 374
++SEL + E V E S+ S + + S
Sbjct: 475 GKTVSSTQQSELKQTITKWEKVTTQFVETAASSSSSSSSSSS 516
Score = 35.0 bits (79), Expect = 0.097
Identities = 49/217 (22%), Positives = 85/217 (38%), Gaps = 41/217 (18%)
Query: 18 GGAENQAESTKAPKRRRL------TKASSDAGTSNPGAQPTTAAAPKGKNVA-------- 63
G A +++ STK+ +K SS +G+S G+ T + G + A
Sbjct: 195 GAASSESSSTKSGSVSSSGSVSTKSKESSSSGSSASGSVATKSKESSGGSAATKSKESSG 254
Query: 64 --------ESSVAAAT------EPTAVPASSP--------ASAGATAAVAAESSIGATAA 101
ESS +AT P+ P +SP +S G+ +A + S+ G+ +A
Sbjct: 255 GSAATKSKESSGGSATTGKTSGSPSGSPKASPSGSVSGKSSSKGSASAQGSASAQGSASA 314
Query: 102 SASVNATKAAASSDTPIG--DKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKS 159
S +A +A++ G K ET S RQ S S+ + + E S
Sbjct: 315 QGSASAQGSASAQRRESGAMAMSKSRETKTSSQRQSKSSSESSSSSTTTTTVKQVESETS 374
Query: 160 CPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPS 196
S Q+E+ A + ++ L ++++ S
Sbjct: 375 ---KEVMSFIMQLEKKYAAKAELKVFFESLKSSMQAS 408
>At3g28790 unknown protein
Length = 608
Score = 50.8 bits (120), Expect = 2e-06
Identities = 91/415 (21%), Positives = 161/415 (37%), Gaps = 67/415 (16%)
Query: 9 NTEGDAKKRGGAENQA--ESTKAPKRRRLTKASS-----DAGTSNPGAQPT--------- 52
+T G + G++ +A +S+ + K + ++ SS D S+ GA P+
Sbjct: 226 STAGSIETNTGSKTEAGSKSSSSAKTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPST 285
Query: 53 -TAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAES----------SIGATAA 101
T + P S+ +T + PA S +AG T+ +ES S +AA
Sbjct: 286 PTPSTPTPSTPTPSTPTPSTPTPSTPAPSTPAAGKTSEKGSESASMKKESNSKSESESAA 345
Query: 102 SASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCP 161
S SV+ TK + GD K+ S +P P+ PS G+ S
Sbjct: 346 SGSVSKTKETNKGSS--GDTYKDTTGTSSGSPSGSPSGSPT-------PSTSTDGKASSK 396
Query: 162 GAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFFLAGLNRDVIEKEVLSRGLNDTK 221
G+A+ S A + + G+S+ +E A ++ K S +
Sbjct: 397 GSASASAGA----SASASAGASA----------SAEESAASQKKESNSKSSSSSSSTTSV 442
Query: 222 EETLACLLRAGCIFAHTFEKFNAANVEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDK 281
+E F EK N E ++ E K +A A KL+T K+
Sbjct: 443 KEVETQTSSEVNSFISNLEKKYTGNSEL-KVFFEKLKTSMSASA------KLSTSNAKEL 495
Query: 282 VYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDL------IARGE 335
V + +A KI E ++ +++E S+ +C++E Q+ K+L I G+
Sbjct: 496 VTG---MRSAASKIAEAMMFVSSRFSKSEETKTSMASCQQEVMQSLKELQDINSQIVSGK 552
Query: 336 ALIA-KESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKK 389
+ + +++EL + E V E S+ S + + S + Q +++ K
Sbjct: 553 TVTSTQQTELKQTITKWEQVTTQFVETAASSSSSSSSSSSSSSSSSQGSAKMAMK 607
Score = 37.4 bits (85), Expect = 0.020
Identities = 75/399 (18%), Positives = 143/399 (35%), Gaps = 43/399 (10%)
Query: 53 TAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAA 112
+ A +GK++ ESS + A SS S G++++ SS + ++S + +++ A
Sbjct: 169 SVAEKRGKSIDESSYGLDVDVNASIGSSSGSDGSSSSDNESSSNTKSQGTSSKSGSESTA 228
Query: 113 SS-DTPIGDKEKENETPKSPPR-------------QDAPPSPPSTHDAGSMPSPPHQGEK 158
S +T G K + S + +D S +GS P
Sbjct: 229 GSIETNTGSKTEAGSKSSSSAKTKEVSGGSSGNTYKDTTGSSSGASPSGSPTPTPSTPTP 288
Query: 159 SCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFFLAGLNRDVIEKEVLSRGLN 218
S P +T + + P P + S P A + SE G ++KE +
Sbjct: 289 STPTPSTPTPSTPTPSTPTPSTPAPS----TPAAGKTSE---KGSESASMKKE------S 335
Query: 219 DTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKVESAK---HQEAAVAWEKRFDKLAT 275
++K E+ + + T K ++ + + S + D A+
Sbjct: 336 NSKSESESAASGSVSKTKET-NKGSSGDTYKDTTGTSSGSPSGSPSGSPTPSTSTDGKAS 394
Query: 276 QAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAE-------K 328
G A + E+ A K+E++ +S + ++ E
Sbjct: 395 SKGSASASAGASASASAGASASAEESAASQKKESNSKSSSSSSSTTSVKEVETQTSSEVN 454
Query: 329 DLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLAL----AKSDMEAVMQATS 384
I+ E SEL V +L+ A A+ +A+ L A S + M S
Sbjct: 455 SFISNLEKKYTGNSELKVFFEKLKTSMSASAKLSTSNAKELVTGMRSAASKIAEAMMFVS 514
Query: 385 EEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEK 423
K+ ET ++A+ E+ L +++ + ++ + K
Sbjct: 515 SRFSKSEETKT-SMASCQQEVMQSLKELQDINSQIVSGK 552
Score = 35.8 bits (81), Expect = 0.057
Identities = 32/126 (25%), Positives = 57/126 (44%), Gaps = 13/126 (10%)
Query: 5 SPKFNTEGDAKKRGGAENQAESTKA---PKRRRLTKASSDAGTSNPGAQPTTAAAPKGKN 61
S K + KK +++++ES + K + K SS G + T++ +P G
Sbjct: 322 SEKGSESASMKKESNSKSESESAASGSVSKTKETNKGSS--GDTYKDTTGTSSGSPSGSP 379
Query: 62 VAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDK 121
+ + +T+ A SS SA A+A +A +S GA+A+ A ++AAS K
Sbjct: 380 SGSPTPSTSTDGKA---SSKGSASASAGASASASAGASAS-----AEESAASQKKESNSK 431
Query: 122 EKENET 127
+ +
Sbjct: 432 SSSSSS 437
Score = 30.4 bits (67), Expect = 2.4
Identities = 40/170 (23%), Positives = 67/170 (38%), Gaps = 14/170 (8%)
Query: 18 GGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVP 77
G + + ES+ K + SS +G+ + T K + ++SS +A T+
Sbjct: 201 GSSSSDNESSSNTKSQG---TSSKSGSESTAGSIETNTGSKTEAGSKSSSSAKTKEV--- 254
Query: 78 ASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPI-GDKEKENETPKSPPRQDA 136
S S+G T SS GA+ S S T + + TP TP +P
Sbjct: 255 --SGGSSGNTYKDTTGSSSGASP-SGSPTPTPSTPTPSTPTPSTPTPSTPTPSTPTPSTP 311
Query: 137 PPSPPS---THDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 183
PS P+ T + GS S + E + + ++ + + + GSS
Sbjct: 312 APSTPAAGKTSEKGS-ESASMKKESNSKSESESAASGSVSKTKETNKGSS 360
>At1g67230 unknown protein
Length = 1132
Score = 50.8 bits (120), Expect = 2e-06
Identities = 51/188 (27%), Positives = 82/188 (43%), Gaps = 32/188 (17%)
Query: 271 DKLATQAGKDKVYADKMIGTAGIKIGELED-------QLALMKEEADELDASLQACKKEK 323
DK+ Q GK+ A K I A + + +LED LAL ++E D L S++ +E
Sbjct: 264 DKIIKQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALREQETDVLKKSIETKAREL 323
Query: 324 EQAEKDLIARGEALIAK-----ESELAVLCAELEL------------VKKALAEQEKKSA 366
+ ++ L AR + + + +++L E EL +K +AE EK+ A
Sbjct: 324 QALQEKLEAREKMAVQQLVDEHQAKLDSTQREFELEMEQKRKSIDDSLKSKVAEVEKREA 383
Query: 367 ESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAE---IASQLAKIKSLEDELATEK 423
E ME + + + + E H E D I+ + +KS E L TEK
Sbjct: 384 E-----WKHMEEKVAKREQALDRKLEKHKEKENDFDLRLKGISGREKALKSEEKALETEK 438
Query: 424 AKAIEARE 431
K +E +E
Sbjct: 439 KKLLEDKE 446
Score = 35.8 bits (81), Expect = 0.057
Identities = 32/146 (21%), Positives = 69/146 (46%), Gaps = 10/146 (6%)
Query: 294 KIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEAL--IAKESE-----LAV 346
+I + Q L+++EA++L A ++ +KE E+ ++ G L I + E + +
Sbjct: 496 QIEKCRSQQELLQKEAEDLKAQRESFEKEWEELDERKAKIGNELKNITDQKEKLERHIHL 555
Query: 347 LCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIA 406
L+ K+A E ++ E+L +AK+ M+ + K E+ L +I
Sbjct: 556 EEERLKKEKQAANENMERELETLEVAKASFAETMEYERSMLSKKAESERSQLL---HDIE 612
Query: 407 SQLAKIKSLEDELATEKAKAIEAREQ 432
+ K++S + EK + ++A+++
Sbjct: 613 MRKRKLESDMQTILEEKERELQAKKK 638
Score = 28.9 bits (63), Expect = 7.0
Identities = 51/276 (18%), Positives = 112/276 (40%), Gaps = 50/276 (18%)
Query: 207 VIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKVESAKHQEAAVAW 266
V + +++ + D E+ + + G +K +AAN+ ++L+ + + +
Sbjct: 246 VAKSQMIVKQREDRANESDKIIKQKGKELEEAQKKIDAANLAVKKLEDDVSSRIKDLALR 305
Query: 267 EKRFDKLA----TQA-----------GKDKVYADKMIGTAGIKIGELEDQLAL-MKEEAD 310
E+ D L T+A ++K+ +++ K+ + + L M+++
Sbjct: 306 EQETDVLKKSIETKARELQALQEKLEAREKMAVQQLVDEHQAKLDSTQREFELEMEQKRK 365
Query: 311 ELDASLQA--CKKEKEQAE----KDLIARGEALI--------AKESELAVLCAELELVKK 356
+D SL++ + EK +AE ++ +A+ E + KE++ + + +K
Sbjct: 366 SIDDSLKSKVAEVEKREAEWKHMEEKVAKREQALDRKLEKHKEKENDFDLRLKGISGREK 425
Query: 357 ALAEQEKKSAESLALAKSDMEAVM-------------QATSEEIKK------ATETHAEA 397
AL +EK D E ++ QA EI K TE
Sbjct: 426 ALKSEEKALETEKKKLLEDKEIILNLKALVEKVSGENQAQLSEINKEKDELRVTEEERSE 485
Query: 398 LATKDAEIASQLAKIKSLEDELATEKAKAIEAREQA 433
E+ Q+ K +S + EL ++A+ ++A+ ++
Sbjct: 486 YLRLQTELKEQIEKCRS-QQELLQKEAEDLKAQRES 520
>At5g41140 putative protein
Length = 983
Score = 50.4 bits (119), Expect = 2e-06
Identities = 40/156 (25%), Positives = 75/156 (47%), Gaps = 10/156 (6%)
Query: 294 KIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELEL 353
K+ EL + L +E + A L+ K++KE DL + ++ E+ +L +LE
Sbjct: 693 KLNELSGKTDLKTKEMKRMSADLEYQKRQKEDVNADLT---HEITRRKDEIEILRLDLEE 749
Query: 354 VKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKA---TETHAEALATKDAEIAS--- 407
+K+ E E +E L + EAV+ A +++ A + +L+ ++EI +
Sbjct: 750 TRKSSMETEASLSEELQRIIDEKEAVITALKSQLETAIAPCDNLKHSLSNNESEIENLRK 809
Query: 408 QLAKIKSLEDELATEKAKAIEAREQAADIALNNRER 443
Q+ +++S E E E+ +E RE +AD +R
Sbjct: 810 QVVQVRS-ELEKKEEEMANLENREASADNITKTEQR 844
Score = 32.7 bits (73), Expect = 0.48
Identities = 51/220 (23%), Positives = 89/220 (40%), Gaps = 17/220 (7%)
Query: 203 LNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKVESAKHQEA 262
L R + EKE + L E +A ++ + + +++ E K +E
Sbjct: 765 LQRIIDEKEAVITALKSQLETAIAPCDNLKHSLSNNESEIENLRKQVVQVRSELEKKEEE 824
Query: 263 AVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKE 322
E R +A D + + +I +LE Q+ L KE A L+A K
Sbjct: 825 MANLENR------EASADNITKTEQRSNED-RIKQLEGQIKL-KENA------LEASSKI 870
Query: 323 KEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLALAKSD--MEAVM 380
+ EKDL R E L K +E++ E + + + E L L+KSD + V
Sbjct: 871 FIEKEKDLKNRIEELQTKLNEVSQNSQETDETLQGPEAIAMQYTEVLPLSKSDNLQDLVN 930
Query: 381 QATS-EEIKKATETHAEALATKDAEIASQLAKIKSLEDEL 419
+ S E ET + + + +EI+ + A+++ +L
Sbjct: 931 EVASLREQNGLMETELKEMQERYSEISLRFAEVEGERQQL 970
>At1g65010 hypothetical protein
Length = 1318
Score = 49.7 bits (117), Expect = 4e-06
Identities = 50/168 (29%), Positives = 80/168 (46%), Gaps = 21/168 (12%)
Query: 293 IKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELE 352
+K EL+ QL ++E+ + D ++ KK+K +A DL KESE V A E
Sbjct: 51 VKGTELQTQLNQIQEDLKKADEQIELLKKDKAKAIDDL---------KESEKLVEEAN-E 100
Query: 353 LVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKI 412
+K+ALA Q K++ ES + K + QA E ++K T L + ++ A ++ +
Sbjct: 101 KLKEALAAQ-KRAEESFEVEKFRAVELEQAGLEAVQKKDVTSKNELESIRSQHALDISAL 159
Query: 413 KSLEDEL----------ATEKAKAIEAREQAADIALNNRERGFYLAKD 450
S +EL A K KA+ E+A IA + E+ LA +
Sbjct: 160 LSTTEELQRVKHELSMTADAKNKALSHAEEATKIAEIHAEKAEILASE 207
Score = 34.3 bits (77), Expect = 0.17
Identities = 30/118 (25%), Positives = 54/118 (45%), Gaps = 15/118 (12%)
Query: 307 EEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSA 366
++ EL L+ CK E+E+++KD+ + AL +E + A L + ++ L E +
Sbjct: 406 DQRTELSIELERCKVEEEKSKKDMESLTLALQEASTESSEAKATLLVCQEELKNCESQ-V 464
Query: 367 ESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKA 424
+SL LA K+ E + + L EI S + + S+++E KA
Sbjct: 465 DSLKLAS--------------KETNEKYEKMLEDARNEIDSLKSTVDSIQNEFENSKA 508
Score = 33.5 bits (75), Expect = 0.28
Identities = 52/196 (26%), Positives = 83/196 (41%), Gaps = 36/196 (18%)
Query: 240 EKFNAANVEAERLKVESAKHQEAAVAWEKRFDKLATQAG---KDKVYADKMIGTAGIKIG 296
E A+ + E LK + AK A+ K +KL +A K+ + A K
Sbjct: 65 EDLKKADEQIELLKKDKAK----AIDDLKESEKLVEEANEKLKEALAAQK---------- 110
Query: 297 ELEDQLALMKEEADELD-ASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVK 355
E+ + K A EL+ A L+A +K+ ++ +L + S L EL+ VK
Sbjct: 111 RAEESFEVEKFRAVELEQAGLEAVQKKDVTSKNELESIRSQHALDISALLSTTEELQRVK 170
Query: 356 KALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSL 415
L+ ++L+ A EE K E HAE K +AS+L ++K+L
Sbjct: 171 HELSMTADAKNKALSHA------------EEATKIAEIHAE----KAEILASELGRLKAL 214
Query: 416 EDELATEKAKAIEARE 431
+ E+ +AIE E
Sbjct: 215 LG--SKEEKEAIEGNE 228
>At5g38560 putative protein
Length = 681
Score = 49.3 bits (116), Expect = 5e-06
Identities = 44/161 (27%), Positives = 62/161 (38%), Gaps = 12/161 (7%)
Query: 35 LTKASSDAGTSNPG---AQPTTAAAPKGKNVAESS-------VAAATEPTAVPASSPASA 84
L+ SS++ T+ P QPTT +AP S V+++ P V + P+S+
Sbjct: 10 LSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSSS 69
Query: 85 GATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSP--PS 142
+ S A+S A+ TP +T PP DA PSP P+
Sbjct: 70 PPPSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDASPSPPAPT 129
Query: 143 THDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 183
T + PSP GE P T S P P +S
Sbjct: 130 TTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTS 170
Score = 42.4 bits (98), Expect = 6e-04
Identities = 36/143 (25%), Positives = 56/143 (38%), Gaps = 9/143 (6%)
Query: 39 SSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGA 98
SS +S+P P +P +VA++ P V AS P S AT A ++
Sbjct: 62 SSPPPSSSPPPSPPVITSPP------PTVASSPPPPVVIASPPPSTPATTPPAPPQTVSP 115
Query: 99 TAA-SASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSP-PSTHDAGSMPSPPHQ- 155
AS + ++ P ETP P +PP P PST + SPP
Sbjct: 116 PPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPP 175
Query: 156 GEKSCPGAATTSEAAQIEQAPAP 178
+ P ++ ++ + + P P
Sbjct: 176 ATSASPPSSNPTDPSTLAPPPTP 198
Score = 28.9 bits (63), Expect = 7.0
Identities = 30/129 (23%), Positives = 40/129 (30%), Gaps = 11/129 (8%)
Query: 76 VPASSPASAGATAAVAAESSIGATAASAS--VNATKAAASSDTPIGDKEKENETPKSPPR 133
+P SP S+ ++ T SA V + S P+ SPP
Sbjct: 7 LPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPP 66
Query: 134 QDAPP-------SPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYY 186
+PP SPP T S P PP P T+ A + P +S
Sbjct: 67 SSSPPPSPPVITSPPPT--VASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDASPS 124
Query: 187 NMLPNAIEP 195
P P
Sbjct: 125 PPAPTTTNP 133
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 48.9 bits (115), Expect = 7e-06
Identities = 42/181 (23%), Positives = 67/181 (36%), Gaps = 24/181 (13%)
Query: 20 AENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAES--SVAAATEPTAVP 77
A++ +S+ AP+ T S+ SN + PT ++P +++ ++A+ P P
Sbjct: 2 ADSPVDSSPAPETSNGTPPSNGTSPSNESSPPTPPSSPPPSSISAPPPDISASFSPPPAP 61
Query: 78 ----ASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPR 133
S P S ++ V A S N + A TP+ P
Sbjct: 62 PTQETSPPTSPSSSPPVVANPS-----PQTPENPSPPAPEGSTPVTPPAPPQTPSNQSPE 116
Query: 134 QDAPPSPPSTHDA-----------GSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGS 182
+ PPSP + D GS PSPP G ++ + S I +P G
Sbjct: 117 RPTPPSPGANDDRNRTNGGNNNRDGSTPSPPSSGNRTSGDGGSPSPPRSI--SPPQNSGD 174
Query: 183 S 183
S
Sbjct: 175 S 175
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 47.8 bits (112), Expect = 1e-05
Identities = 32/148 (21%), Positives = 78/148 (52%), Gaps = 16/148 (10%)
Query: 294 KIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELEL 353
++ ELE QL L+++ +L ASL A ++EK+ ++ + L +S++ L EL
Sbjct: 486 RLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSKVQELVTELAE 545
Query: 354 VKKALAEQEKKSAESLALAKS----------DMEAVMQATSEEIKK------ATETHAEA 397
K L ++E + + + + ++ ++EA +++ E++K+ ++E +
Sbjct: 546 SKDTLTQKENELSSFVEVHEAHKRDSSSQVKELEARVESAEEQVKELNQNLNSSEEEKKI 605
Query: 398 LATKDAEIASQLAKIKSLEDELATEKAK 425
L+ + +E++ ++ + +S EL++E +
Sbjct: 606 LSQQISEMSIKIKRAESTIQELSSESER 633
Score = 41.6 bits (96), Expect = 0.001
Identities = 49/219 (22%), Positives = 91/219 (41%), Gaps = 17/219 (7%)
Query: 247 VEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKV-YADKMIGTAGIKIGELEDQLALM 305
++ L++E+ Q + + E R K DK+ A I I L+++L +
Sbjct: 937 IKGRELELETLGKQRSELDEELRTKKEENVQMHDKINVASSEIMALTELINNLKNELDSL 996
Query: 306 KEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEK-- 363
+ + E +A L+ K+EK + + +AL+ +E+ L E + + + E E
Sbjct: 997 QVQKSETEAELEREKQEKSELSNQITDVQKALVEQEAAYNTLEEEHKQINELFKETEATL 1056
Query: 364 -------KSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLE 416
K A+ L + +T ++ E+ L K EI + + KI ++E
Sbjct: 1057 NKVTVDYKEAQRLLEERGKEVTSRDSTIGVHEETMESLRNELEMKGDEIETLMEKISNIE 1116
Query: 417 DELATEKAKAIEAREQAADIALNNRERGFYLAKDQAQHL 455
+L K + EQ L +E F K++A+HL
Sbjct: 1117 VKLRLSNQK-LRVTEQ----VLTEKEEAF--RKEEAKHL 1148
Score = 41.2 bits (95), Expect = 0.001
Identities = 34/158 (21%), Positives = 75/158 (46%), Gaps = 19/158 (12%)
Query: 294 KIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELEL 353
++ ELE ++ +E+ EL+ +L + ++EK+ + + + ES + L +E E
Sbjct: 574 QVKELEARVESAEEQVKELNQNLNSSEEEKKILSQQISEMSIKIKRAESTIQELSSESER 633
Query: 354 VKKALAEQEKK----------SAESLALAKSDMEAVMQATS------EEIKKATETHAEA 397
+K + AE++ + L+ +EA ++++ E KA E +
Sbjct: 634 LKGSHAEKDNELFSLRDIHETHQRELSTQLRGLEAQLESSEHRVLELSESLKAAEEESRT 693
Query: 398 LATKDAEIASQLAKIKSLEDELATEKAKAIEAREQAAD 435
++TK +E + +L + + + EL + +K +EQ A+
Sbjct: 694 MSTKISETSDELERTQIMVQELTADSSK---LKEQLAE 728
Score = 39.7 bits (91), Expect = 0.004
Identities = 30/152 (19%), Positives = 74/152 (47%), Gaps = 16/152 (10%)
Query: 294 KIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELEL 353
+ ELE QL K++ +L ASL+A ++E + + L ++ + L AEL
Sbjct: 155 RASELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMNKLEQTQNTIQELMAELGK 214
Query: 354 VKKALAEQEKKSAESLALAKS----------DMEAVMQATSEEIKKATET------HAEA 397
+K + E+E + + + + ++ ++E ++++ + + + +T +
Sbjct: 215 LKDSHREKESELSSLVEVHETHQRDSSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKV 274
Query: 398 LATKDAEIASQLAKIKSLEDELATEKAKAIEA 429
L+ K AE+++++ + ++ EL +E + E+
Sbjct: 275 LSQKIAELSNEIKEAQNTIQELVSESGQLKES 306
Score = 36.6 bits (83), Expect = 0.034
Identities = 34/166 (20%), Positives = 75/166 (44%), Gaps = 20/166 (12%)
Query: 291 AGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAE 350
+ I + ELE+Q+ K+ EL+ +L ++EK+ + + + ++ + L +E
Sbjct: 240 SSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSE 299
Query: 351 LELVKK----------ALAEQEKKSAESLALAKSDMEAVMQATSEEIK------KATETH 394
+K+ +L + + + S++EA ++++ + I K E
Sbjct: 300 SGQLKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEE 359
Query: 395 AEALATKDAEIASQLAK----IKSLEDELATEKAKAIEAREQAADI 436
+A+++K+ EI +L + IK L DEL K + E + + +
Sbjct: 360 NKAISSKNLEIMDKLEQAQNTIKELMDELGELKDRHKEKESELSSL 405
Score = 31.6 bits (70), Expect = 1.1
Identities = 39/172 (22%), Positives = 78/172 (44%), Gaps = 29/172 (16%)
Query: 291 AGIKIGELEDQLALMKEEADELDA-----SLQACKKEKEQAE---------KDLIAR--- 333
A +KI L+D++ ++++ LD+ +Q KK +E +E +++I +
Sbjct: 863 ASVKIKRLDDEVNGLRQQVASLDSQRAELEIQLEKKSEEISEYLSQITNLKEEIINKVKV 922
Query: 334 GEALIAKESELAVLCAELELVKKALAEQEKKSAESLALAKSD---MEAVMQATSEEIKKA 390
E+++ + + L+ EL + L +Q + E L K + M + S EI
Sbjct: 923 HESILEEINGLSEKIKGRELELETLGKQRSELDEELRTKKEENVQMHDKINVASSEIMAL 982
Query: 391 TE------THAEALATKDAEIASQLAKIKSLEDELA---TEKAKAIEAREQA 433
TE ++L + +E ++L + K + EL+ T+ KA+ +E A
Sbjct: 983 TELINNLKNELDSLQVQKSETEAELEREKQEKSELSNQITDVQKALVEQEAA 1034
Score = 30.4 bits (67), Expect = 2.4
Identities = 44/210 (20%), Positives = 87/210 (40%), Gaps = 23/210 (10%)
Query: 245 ANVEAER--LKVESAKHQEAAVAWEKRFDKLATQA-GKDKVYADKMIGTAGI--KIGELE 299
A+++++R L+++ K E + + L + K KV+ + G+ KI E
Sbjct: 882 ASLDSQRAELEIQLEKKSEEISEYLSQITNLKEEIINKVKVHESILEEINGLSEKIKGRE 941
Query: 300 DQLALMKEEADELDASLQACKKEKEQ-------AEKDLIARGEALIAKESELAVLCAELE 352
+L + ++ ELD L+ K+E Q A +++A E + ++EL L +
Sbjct: 942 LELETLGKQRSELDEELRTKKEENVQMHDKINVASSEIMALTELINNLKNELDSLQVQKS 1001
Query: 353 LVKKALAEQEKKSAE------SLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIA 406
+ L ++++ +E + A + EA EE K+ E E AT +
Sbjct: 1002 ETEAELEREKQEKSELSNQITDVQKALVEQEAAYNTLEEEHKQINELFKETEATLNKVTV 1061
Query: 407 SQLAKIKSLEDELATEKAKAIEAREQAADI 436
+ LE E+ K + +R+ +
Sbjct: 1062 DYKEAQRLLE-----ERGKEVTSRDSTIGV 1086
>At3g49055 putative protein
Length = 480
Score = 47.8 bits (112), Expect = 1e-05
Identities = 39/169 (23%), Positives = 78/169 (46%), Gaps = 9/169 (5%)
Query: 248 EAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKE 307
E +LK+ + + V K ++ A + ++ VY +K+ +E+ + ++E
Sbjct: 277 EVSQLKISLEESRLEEVGLRKVTEEQAQKLAENTVYINKLQNQEKFLAQNVEELVKAIRE 336
Query: 308 EADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAE 367
E+ +AC+ E E ++++ R + + AVL +E+E ++ ALA E K
Sbjct: 337 AESEVSRWREACELEVEAGQREVEVRDQLI-------AVLKSEVEKLRSALARSEGKLKL 389
Query: 368 SLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLE 416
LAK+ M V + +E+ + E L ++ + QL + +S E
Sbjct: 390 KEELAKAAM--VAEEAAEKSLRLAERRIAQLLSRIEHLYRQLEEAESTE 436
Score = 39.3 bits (90), Expect = 0.005
Identities = 38/165 (23%), Positives = 78/165 (47%), Gaps = 11/165 (6%)
Query: 298 LEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKA 357
+E + +++E +L SL+ + E+ K + + L +E V +L+ +K
Sbjct: 267 IEKTMKKLRQEVSQLKISLEESRLEEVGLRKVTEEQAQKL----AENTVYINKLQNQEKF 322
Query: 358 LAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSL-- 415
LA+ ++ +++ A+S++ +A E++ A + E A + S++ K++S
Sbjct: 323 LAQNVEELVKAIREAESEVSRWREACELEVE-AGQREVEVRDQLIAVLKSEVEKLRSALA 381
Query: 416 ----EDELATEKAKAIEAREQAADIALNNRERGFYLAKDQAQHLY 456
+ +L E AKA E+AA+ +L ER + +HLY
Sbjct: 382 RSEGKLKLKEELAKAAMVAEEAAEKSLRLAERRIAQLLSRIEHLY 426
>At5g62410 chromosomal protein - like
Length = 1175
Score = 46.6 bits (109), Expect = 3e-05
Identities = 50/214 (23%), Positives = 105/214 (48%), Gaps = 20/214 (9%)
Query: 258 KHQEAAVAWEKRFDKLATQAGK-DKVYADKMIG-TAGIKIGELEDQLALMK-------EE 308
K +E A ++RF +L+T + +K + + G ++G + LEDQL K E
Sbjct: 354 KSEEGAADLKQRFQELSTTLEECEKEHQGVLAGKSSGDEEKCLEDQLRDAKIAVGTAGTE 413
Query: 309 ADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAES 368
+L ++ C+KE ++ + L+++ E I E+EL ++E VKKAL +
Sbjct: 414 LKQLKTKIEHCEKELKERKSQLMSKLEEAIEVENELGARKNDVEHVKKALESIPYNEGQM 473
Query: 369 LALAK---SDMEAV------MQATSEEIKKATETHAEALATKD-AEIASQLAKIKSLEDE 418
AL K +++E V ++ S ++ T+++ + D +++ +AK+ ++D
Sbjct: 474 EALEKDRGAELEVVQRLEDKVRGLSAQLANFQFTYSDPVRNFDRSKVKGVVAKLIKVKDR 533
Query: 419 LATEKAKAIEAREQAADIALNNRERGFYLAKDQA 452
++ A + A + D+ +++ + G L ++ A
Sbjct: 534 -SSMTALEVTAGGKLYDVVVDSEDTGKQLLQNGA 566
Score = 33.1 bits (74), Expect = 0.37
Identities = 44/209 (21%), Positives = 83/209 (39%), Gaps = 52/209 (24%)
Query: 276 QAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKE----------- 324
QA K + A +G K+G+++ + +EE E + ++A + KE
Sbjct: 241 QAEKIRDNAVLGVGEMKAKLGKIDAETEKTQEEIQEFEKQIKALTQAKEASMGGEVKTLS 300
Query: 325 -----------QAEKDLIARGEALIAKESELAVLCAELELVKKALAEQE---KKSAESLA 370
+ L + + L+ ++ + + +E +KK++ E+ KKS E A
Sbjct: 301 EKVDSLAQEMTRESSKLNNKEDTLLGEKENVEKIVHSIEDLKKSVKERAAAVKKSEEGAA 360
Query: 371 LAKSDMEAVMQATSEEIKKATETHAEALATK--------------DAEIASQLA------ 410
D++ Q S +++ + H LA K DA+IA A
Sbjct: 361 ----DLKQRFQELSTTLEECEKEHQGVLAGKSSGDEEKCLEDQLRDAKIAVGTAGTELKQ 416
Query: 411 ---KIKSLEDELATEKAKAIEAREQAADI 436
KI+ E EL K++ + E+A ++
Sbjct: 417 LKTKIEHCEKELKERKSQLMSKLEEAIEV 445
Score = 32.0 bits (71), Expect = 0.83
Identities = 29/142 (20%), Positives = 68/142 (47%), Gaps = 12/142 (8%)
Query: 282 VYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAK- 340
V A+K+ A + +GE++ +L ++DA + ++E ++ EK + A +A A
Sbjct: 240 VQAEKIRDNAVLGVGEMKAKLG-------KIDAETEKTQEEIQEFEKQIKALTQAKEASM 292
Query: 341 ESELAVLCAELELVKKALAEQEKK---SAESLALAKSDMEAVMQATSEEIKKATETHAEA 397
E+ L +++ + + + + K ++L K ++E ++ + E++KK+ + A A
Sbjct: 293 GGEVKTLSEKVDSLAQEMTRESSKLNNKEDTLLGEKENVEKIVHSI-EDLKKSVKERAAA 351
Query: 398 LATKDAEIASQLAKIKSLEDEL 419
+ + A + + L L
Sbjct: 352 VKKSEEGAADLKQRFQELSTTL 373
Score = 31.6 bits (70), Expect = 1.1
Identities = 38/174 (21%), Positives = 75/174 (42%), Gaps = 12/174 (6%)
Query: 253 KVESAKHQ--EAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEAD 310
++E AK Q E +A++ FD ++ K + G ++ +LE + +K +
Sbjct: 749 ELEEAKSQIKEKELAYKNCFDAVSKLENSIKDHDKNREG----RLKDLEKNIKTIKAQMQ 804
Query: 311 ELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELEL-VKKALAEQEKKSAESL 369
L++ + EKE+ L+ EA+ ++S L LE + +E +++ A+
Sbjct: 805 AASKDLKSHENEKEK----LVMEEEAMKQEQSSLESHLTSLETQISTLTSEVDEQRAKVD 860
Query: 370 ALAKSDMEAVMQATSEEIK-KATETHAEALATKDAEIASQLAKIKSLEDELATE 422
AL K E++ + K K +T T + +L+ +K +L E
Sbjct: 861 ALQKIHDESLAELKLIHAKMKECDTQISGFVTDQEKCLQKLSDMKLERKKLENE 914
Score = 28.5 bits (62), Expect = 9.1
Identities = 46/199 (23%), Positives = 78/199 (39%), Gaps = 28/199 (14%)
Query: 311 ELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLA 370
E+ +L+ +KEK Q + A G A + + L C E V Q +K ++
Sbjct: 202 EILPALEKLRKEKSQYMQ--WANGNAELDR---LRRFCIAFEYV------QAEKIRDNAV 250
Query: 371 LAKSDMEAVM-------QATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEK 423
L +M+A + + T EEI++ + K+A + ++ + D LA E
Sbjct: 251 LGVGEMKAKLGKIDAETEKTQEEIQEFEKQIKALTQAKEASMGGEVKTLSEKVDSLAQEM 310
Query: 424 AKAIEAREQAADIALNNRERGFYLAKDQAQHLYPNFDFSAMGVMKEITAA--GLVGPDDP 481
RE + LNN+E K+ + + + + V + A G D
Sbjct: 311 -----TRESS---KLNNKEDTLLGEKENVEKIVHSIEDLKKSVKERAAAVKKSEEGAADL 362
Query: 482 PLIDQNLWTATEEEEEEEE 500
Q L T EE E+E +
Sbjct: 363 KQRFQELSTTLEECEKEHQ 381
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 46.6 bits (109), Expect = 3e-05
Identities = 67/279 (24%), Positives = 117/279 (41%), Gaps = 22/279 (7%)
Query: 240 EKFNAANVEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 299
E+F + + EA L + + ++E ++LA +GK +K+ T G ++ E
Sbjct: 813 EEFTSRDSEASSLTEKLRDLEGKIKSYE---EQLAEASGKSSSLKEKLEQTLG-RLAAAE 868
Query: 300 DQLALMKEEADEL-DASLQACKKEKEQAE-----KDLIARGEALIAKES-ELAVLCAELE 352
+K+E D+ + SLQ+ + + AE K I E LI S E LE
Sbjct: 869 SVNEKLKQEFDQAQEKSLQSSSESELLAETNNQLKIKIQELEGLIGSGSVEKETALKRLE 928
Query: 353 LVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKI 412
+ ++E +S++ + K+ + EE KK + T+ E+ L+K+
Sbjct: 929 EAIERFNQKETESSDLVEKLKTHENQI-----EEYKKLAHEASGVADTRKVELEDALSKL 983
Query: 413 KSLEDELATEKAKAIEAREQAADIALNNRERGFYLAKDQAQHLYPNFDFSAMGVMKEITA 472
K+LE + AK +++ D+A N + LA ++ SA+ KE TA
Sbjct: 984 KNLESTIEELGAKCQGLEKESGDLAEVNLKLNLELANHGSEANELQTKLSALEAEKEQTA 1043
Query: 473 AGLVGPDDPPLIDQNLWTATEEEEEEEEQ----EKENNE 507
L I+ T E E+ + Q +ENN+
Sbjct: 1044 NELEA--SKTTIEDLTKQLTSEGEKLQSQISSHTEENNQ 1080
Score = 42.4 bits (98), Expect = 6e-04
Identities = 70/274 (25%), Positives = 121/274 (43%), Gaps = 32/274 (11%)
Query: 240 EKFNAANVEAERLK---VESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIG 296
E F+A +E E + +E + + + ++F++L Q+ +AD A ++
Sbjct: 184 EAFDALGIELESSRKKLIELEEGLKRSAEEAQKFEELHKQSAS---HADSESQKA-LEFS 239
Query: 297 ELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKK 356
EL +E +E ASLQ KE + + AL + ELA + EL L K
Sbjct: 240 ELLKSTKESAKEMEEKMASLQQEIKELNEKMSENEKVEAALKSSAGELAAVQEELALSKS 299
Query: 357 ALAEQEKKSAESLALAKSDMEAVMQATSE-EIKKATETHAEALATKDAEIASQL-AKIKS 414
L E E+K + + AL + + T E E KKA+E+ + ++ + L A+ K
Sbjct: 300 RLLETEQKVSSTEAL-------IDELTQELEQKKASESRFK----EELSVLQDLDAQTKG 348
Query: 415 LEDELATEKAKAIEAREQAADIALNNRERGFYLAKDQAQHL-YPNFDFSAMGVMKEITAA 473
L+ +L+ ++ + E+ L +E L+KDQ + L N + + KE A
Sbjct: 349 LQAKLSEQEGINSKLAEE-----LKEKELLESLSKDQEEKLRTANEKLAEVLKEKEALEA 403
Query: 474 GLVGPDDPPLIDQNLWTATEEEEEEEEQEKENNE 507
+ + N+ T TE E EE+ K ++E
Sbjct: 404 NVAE------VTSNVATVTEVCNELEEKLKTSDE 431
Score = 41.6 bits (96), Expect = 0.001
Identities = 49/201 (24%), Positives = 89/201 (43%), Gaps = 22/201 (10%)
Query: 255 ESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEA----- 309
ESAK E +A ++ K + + + + ++ ++ ++++LAL K
Sbjct: 247 ESAKEMEEKMASLQQEIKELNEKMSENEKVEAALKSSAGELAAVQEELALSKSRLLETEQ 306
Query: 310 ---------DELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAE 360
DEL L+ KK E K+ ++ + L A+ L +E E + LAE
Sbjct: 307 KVSSTEALIDELTQELEQ-KKASESRFKEELSVLQDLDAQTKGLQAKLSEQEGINSKLAE 365
Query: 361 Q--EKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDE 418
+ EK+ ESL+ D E ++ +E++ + + EAL AE+ S +A + + +E
Sbjct: 366 ELKEKELLESLS---KDQEEKLRTANEKLAEVLK-EKEALEANVAEVTSNVATVTEVCNE 421
Query: 419 LATEKAKAIEAREQAADIALN 439
L EK K + D L+
Sbjct: 422 L-EEKLKTSDENFSKTDALLS 441
Score = 41.6 bits (96), Expect = 0.001
Identities = 54/275 (19%), Positives = 110/275 (39%), Gaps = 53/275 (19%)
Query: 246 NVEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALM 305
N+E E + S++ E A + K + T A + ++ + +K + E +L +
Sbjct: 475 NLELEDVVRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKEL 534
Query: 306 KEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKS 365
E++ EL +++ ++EK+QA + E ++K+
Sbjct: 535 SEKSSELQTAIEVAEEEKKQATTQM----------------------------QEYKQKA 566
Query: 366 AE-SLALAKS---------DMEAVMQATSEEIKKATETHAEALATKDAEIASQL------ 409
+E L+L +S D+ +Q +E +A TH ++ + +SQ
Sbjct: 567 SELELSLTQSSARNSELEEDLRIALQKGAEHEDRANTTHQRSIELEGLCQSSQSKHEDAE 626
Query: 410 AKIKSLEDELATEKAKAIEAREQAADIALNNRE-----RGFYLAKDQAQHLYPNFDFSAM 464
++K LE L TEK + E EQ + + + E +G+ + Q F +
Sbjct: 627 GRLKDLELLLQTEKYRIQELEEQVSSLEKKHGETEADSKGYLGQVAELQSTLEAFQVKS- 685
Query: 465 GVMKEITAAGLVGPDDPPLIDQNLWTATEEEEEEE 499
+ AA + ++ + +NL T E+++ E
Sbjct: 686 ---SSLEAALNIATENEKELTENLNAVTSEKKKLE 717
Score = 41.6 bits (96), Expect = 0.001
Identities = 41/190 (21%), Positives = 87/190 (45%), Gaps = 18/190 (9%)
Query: 250 ERLKVESAKHQEA---AVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMK 306
E +V+S+ + A A EK + +K + + +KI E E+ L ++
Sbjct: 679 EAFQVKSSSLEAALNIATENEKELTENLNAVTSEKKKLEATVDEYSVKISESENLLESIR 738
Query: 307 EEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSA 366
E + L E E DL A G +ESE+ +L+ +++L EQ+ +
Sbjct: 739 NELNVTQGKL-------ESIENDLKAAG----LQESEVM---EKLKSAEESL-EQKGREI 783
Query: 367 ESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKAKA 426
+ + ++EA+ Q+ S + + + E ++D+E +S K++ LE ++ + + +
Sbjct: 784 DEATTKRMELEALHQSLSIDSEHRLQKAMEEFTSRDSEASSLTEKLRDLEGKIKSYEEQL 843
Query: 427 IEAREQAADI 436
EA +++ +
Sbjct: 844 AEASGKSSSL 853
Score = 39.7 bits (91), Expect = 0.004
Identities = 40/200 (20%), Positives = 91/200 (45%), Gaps = 11/200 (5%)
Query: 246 NVEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALM 305
N+E E+ + Q A E ++ A + K + + + +L+ Q++
Sbjct: 1015 NLELANHGSEANELQTKLSALEAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSH 1074
Query: 306 KEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKS 365
EE ++++A Q+ K+E + L E L + S+ L +E+E ++ AE+
Sbjct: 1075 TEENNQVNAMFQSTKEELQSVIAKL---EEQLTVESSKADTLVSEIEKLRAVAAEKSVLE 1131
Query: 366 AESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSL--EDELATEK 423
+ ++E + ++K+ E A A + K AE+ S+L + + + E ++ E+
Sbjct: 1132 SHF-----EELEKTLSEVKAQLKENVENAATA-SVKVAELTSKLQEHEHIAGERDVLNEQ 1185
Query: 424 AKAIEAREQAADIALNNRER 443
++ QAA +++ +++
Sbjct: 1186 VLQLQKELQAAQSSIDEQKQ 1205
Score = 39.3 bits (90), Expect = 0.005
Identities = 37/144 (25%), Positives = 61/144 (41%), Gaps = 18/144 (12%)
Query: 298 LEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKA 357
+E+Q +++ + L +++ ++ E +L L ESE L EL K+
Sbjct: 70 VEEQKEVIERSSSGSQRELHESQEKAKELELELERVAGELKRYESENTHLKDELLSAKEK 129
Query: 358 LAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLED 417
L E EKK D+E V + E+I + E H+ L + + + S AK K L +
Sbjct: 130 LEETEKK--------HGDLEVVQKKQQEKIVEGEERHSSQLKSLEDALQSHDAKDKELTE 181
Query: 418 ----------ELATEKAKAIEARE 431
EL + + K IE E
Sbjct: 182 VKEAFDALGIELESSRKKLIELEE 205
Score = 35.8 bits (81), Expect = 0.057
Identities = 42/201 (20%), Positives = 85/201 (41%), Gaps = 22/201 (10%)
Query: 311 ELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLA 370
EL+ +++ + E+A+ + A E + A L +L L++ ++ E++ E L+
Sbjct: 477 ELEDVVRSSSQAAEEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKE-LS 535
Query: 371 LAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKAKAIEAR 430
S+++ ++ EE K+AT T + K +E+ L + + EL + A++
Sbjct: 536 EKSSELQTAIEVAEEEKKQAT-TQMQEYKQKASELELSLTQSSARNSELEEDLRIALQKG 594
Query: 431 EQAADIALNNRERGFYL----AKDQAQHLYPNFDFSAMGVMKEITAAGLVGPDDPPLIDQ 486
+ D A +R L Q++H A G +K++ L+
Sbjct: 595 AEHEDRANTTHQRSIELEGLCQSSQSKH------EDAEGRLKDLEL----------LLQT 638
Query: 487 NLWTATEEEEEEEEQEKENNE 507
+ E EE+ EK++ E
Sbjct: 639 EKYRIQELEEQVSSLEKKHGE 659
Score = 35.0 bits (79), Expect = 0.097
Identities = 53/246 (21%), Positives = 97/246 (38%), Gaps = 37/246 (15%)
Query: 209 EKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKVESAKHQEAAVAWEK 268
+KEV+ R + ++ E + EK +E ER+ E K
Sbjct: 73 QKEVIERSSSGSQRE-----------LHESQEKAKELELELERVAGEL-----------K 110
Query: 269 RFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEK 328
R++ T + + A + + K G+LE +E+ E + + K E A +
Sbjct: 111 RYESENTHLKDELLSAKEKLEETEKKHGDLEVVQKKQQEKIVEGEERHSSQLKSLEDALQ 170
Query: 329 DLIARGEALIAKESELAVLCAELELVKKALAEQE---KKSAESL---------ALAKSDM 376
A+ + L + L ELE +K L E E K+SAE + + +D
Sbjct: 171 SHDAKDKELTEVKEAFDALGIELESSRKKLIELEEGLKRSAEEAQKFEELHKQSASHADS 230
Query: 377 EAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADI 436
E+ E+ K+T+ A+ + K +AS +IK L ++++ + + A ++
Sbjct: 231 ESQKALEFSELLKSTKESAKEMEEK---MASLQQEIKELNEKMSENEKVEAALKSSAGEL 287
Query: 437 ALNNRE 442
A E
Sbjct: 288 AAVQEE 293
Score = 33.9 bits (76), Expect = 0.22
Identities = 50/247 (20%), Positives = 95/247 (38%), Gaps = 23/247 (9%)
Query: 168 EAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFFLAGLNRDVIEKEVLSRGLNDTKEETLAC 227
EA + + A E ++ ++ E + ++ E ++ TKEE +
Sbjct: 1036 EAEKEQTANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQVNAMFQSTKEELQSV 1095
Query: 228 LLRAGCIFAHTFEKFNAANVEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKM 287
+ + K + E E+L+ +A+ E F++L + K +
Sbjct: 1096 IAKLEEQLTVESSKADTLVSEIEKLRAVAAEKS----VLESHFEELEKTLSEVKAQLKEN 1151
Query: 288 I---GTAGIKIGELEDQLALMKE---EADELDASLQACKKEKEQAEKDLIARGEALIAKE 341
+ TA +K+ EL +L + E D L+ + +KE + A+ + + +A K+
Sbjct: 1152 VENAATASVKVAELTSKLQEHEHIAGERDVLNEQVLQLQKELQAAQSSIDEQKQAHSQKQ 1211
Query: 342 SELAVLCAELELVKKALAEQ---EKKSAESLALAKSDMEAVMQATSEEIKKATETHAEAL 398
SEL E K E+ +KK+ D+E +Q + K ET A +
Sbjct: 1212 SEL-------ESALKKSQEEIEAKKKAVTEFESMVKDLEQKVQLADAKTK---ETEAMDV 1261
Query: 399 ATKDAEI 405
K +I
Sbjct: 1262 GVKSRDI 1268
>At1g77580 unknown protein
Length = 779
Score = 46.2 bits (108), Expect = 4e-05
Identities = 66/276 (23%), Positives = 112/276 (39%), Gaps = 33/276 (11%)
Query: 148 SMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFFLAGLNRDV 207
++P P KS P + T E + A E+ +L + I+ E L L
Sbjct: 313 ALPETPDGNGKSGPESVTEEVVVPSENSLASEI------EVLTSRIKELEEKLEKLEA-- 364
Query: 208 IEKEVLSRGLNDTKEETLACLLRAGCIFAHTFE---KFNAANVEAERLKVESAKHQEAAV 264
EK L + +EE + + + + + T E K E E LK E ++E AV
Sbjct: 365 -EKHELENEVKCNREEAVVHIENSEVLTSRTKELEEKLEKLEAEKEELKSEVKCNREKAV 423
Query: 265 AWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEK- 323
+ + + A+ + T+ K ELE+QL ++ E EL++ ++ C +E+
Sbjct: 424 VHVE-----------NSLAAEIEVLTSRTK--ELEEQLEKLEAEKVELESEVK-CNREEA 469
Query: 324 -EQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQA 382
Q E L E L + +L +LE+ K L + K + E + + ++EA+
Sbjct: 470 VAQVENSLATEIEVLTCRIKQLEEKLEKLEVEKDELKSEVKCNREVESTLRFELEAIACE 529
Query: 383 TSEEIKKATETHAEALATKDAEIASQLAKIKSLEDE 418
K E E L + AE+ IK +E
Sbjct: 530 -----KMELENKLEKLEVEKAELQISFDIIKDKYEE 560
Score = 35.4 bits (80), Expect = 0.075
Identities = 49/217 (22%), Positives = 91/217 (41%), Gaps = 43/217 (19%)
Query: 220 TKEETLACLLRAGCIFAHTFEKFNAANVEAERLKVESAKHQEAAVAWEKRFDKLATQAGK 279
T+ E L C ++ EK VE + LK E ++E + +A
Sbjct: 479 TEIEVLTCRIK------QLEEKLEKLEVEKDELKSEVKCNREVESTLRFELEAIA----- 527
Query: 280 DKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIA 339
+KM ELE++L ++ E EL S K + E+++ L
Sbjct: 528 ----CEKM---------ELENKLEKLEVEKAELQISFDIIKDKYEESQV-------CLQE 567
Query: 340 KESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALA 399
E++L + E++LV + AE E ++ A AK+ A +++ E+++K E A
Sbjct: 568 IETKLGEIQTEMKLVNELKAEVESQTIAMEADAKTK-SAKIESLEEDMRK------ERFA 620
Query: 400 TKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADI 436
+ K ++LE+E++ K +I++ + I
Sbjct: 621 FDELR-----RKCEALEEEISLHKENSIKSENKEPKI 652
Score = 32.3 bits (72), Expect = 0.63
Identities = 27/131 (20%), Positives = 52/131 (39%)
Query: 298 LEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKA 357
LE+Q+ +++EL ++ KE +L+AR E L + E + E K
Sbjct: 168 LENQIFETATKSEELSQMAESVAKENVMLRHELLARCEELEIRTIERDLSTQAAETASKQ 227
Query: 358 LAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLED 417
+ KK A+ A + + S ++T++H++ D + A +E
Sbjct: 228 QLDSIKKVAKLEAECRKFRMLAKSSASFNDHRSTDSHSDGGERMDVSCSDSWASSTLIEK 287
Query: 418 ELATEKAKAIE 428
+ +IE
Sbjct: 288 RSLQGTSSSIE 298
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 46.2 bits (108), Expect = 4e-05
Identities = 43/190 (22%), Positives = 82/190 (42%), Gaps = 22/190 (11%)
Query: 248 EAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKM--IGTAGIKIGELEDQLALM 305
E E + K +E WEK+ GK++ ++ + K+ E+E +L L
Sbjct: 250 ERESYEGTFQKQREYLNEWEKKLQ------GKEESITEQKRNLNQREEKVNEIEKKLKLK 303
Query: 306 KEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKS 365
++E +E + + + ++ E+D+ R E L KE E L ++ L+ K E E ++
Sbjct: 304 EKELEEWNRKVDLSMSKSKETEEDITKRLEELTTKEKEAHTL--QITLLAK---ENELRA 358
Query: 366 AESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATEKAK 425
E +A+ EI+K + E L +K E + +I+ D+ K +
Sbjct: 359 FEEKLIARE---------GTEIQKLIDDQKEVLGSKMLEFELECEEIRKSLDKELQRKIE 409
Query: 426 AIEAREQAAD 435
+E ++ D
Sbjct: 410 ELERQKVEID 419
Score = 30.4 bits (67), Expect = 2.4
Identities = 39/202 (19%), Positives = 92/202 (45%), Gaps = 29/202 (14%)
Query: 240 EKFNAANVEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELE 299
E+ EA L++ + A+E++ +A + + + D G K+ E E
Sbjct: 333 EELTTKEKEAHTLQITLLAKENELRAFEEKL--IAREGTEIQKLIDDQKEVLGSKMLEFE 390
Query: 300 DQLALMKEEAD-ELDASLQACKKEK---EQAEKDLIARGEALIAKESELAVLCAELELVK 355
+ +++ D EL ++ +++K + +E+ L R +A+ K + +LE
Sbjct: 391 LECEEIRKSLDKELQRKIEELERQKVEIDHSEEKLEKRNQAMNKKFDRVNEKEMDLEAKL 450
Query: 356 KALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDA--EIASQLAKIK 413
K + E+EK ++QA E K+ + + L+ K++ ++ ++ KI+
Sbjct: 451 KTIKEREK---------------IIQA---EEKRLSLEKQQLLSDKESLEDLQQEIEKIR 492
Query: 414 ---SLEDELATEKAKAIEAREQ 432
+ ++E+ E+ K++E +++
Sbjct: 493 AEMTKKEEMIEEECKSLEIKKE 514
Score = 30.0 bits (66), Expect = 3.1
Identities = 45/205 (21%), Positives = 84/205 (40%), Gaps = 21/205 (10%)
Query: 240 EKFNAANVEAERLKVESAK---HQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIG 296
E+ E +RL +E + +E+ ++ +K+ + K + ++ + IK
Sbjct: 455 EREKIIQAEEKRLSLEKQQLLSDKESLEDLQQEIEKIRAEMTKKEEMIEEECKSLEIKKE 514
Query: 297 ELEDQLALMKEEADELDAS----------LQACKKEKEQAEKDLIARGEALIAKESELAV 346
E E+ L L E +++ S ++ K+EKE+ EK+ E E
Sbjct: 515 EREEYLRLQSELKSQIEKSRVHEEFLSKEVENLKQEKERFEKEWEILDEKQAVYNKERIR 574
Query: 347 LCAELELVKK-ALAEQEKKSAESLALAKSDMEAV----MQATSEEIKKATETHA--EALA 399
+ E E ++ L E E+ E AL M+ + +Q S E E A E +
Sbjct: 575 ISEEKEKFERFQLLEGERLKKEESALRVQIMQELDDIRLQRESFEANMEHERSALQEKVK 634
Query: 400 TKDAEIASQLAKI-KSLEDELATEK 423
+ +++ L + ++LE EL K
Sbjct: 635 LEQSKVIDDLEMMRRNLEIELQERK 659
>At3g19060 hypothetical protein
Length = 1647
Score = 45.8 bits (107), Expect = 6e-05
Identities = 52/202 (25%), Positives = 91/202 (44%), Gaps = 27/202 (13%)
Query: 248 EAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKE 307
E LK E K + A E R+ + A K YAD E E+++ L++
Sbjct: 1388 EVLNLKEEFGKMKSEAKEMEARYIEAQQIAESRKTYAD-----------EREEEVKLLEG 1436
Query: 308 EADELDASLQACKKE----KEQAEKDLIARGEALIAKESELAVLCAELELVKKALAEQEK 363
+EL+ ++ + + K++AE+ + R E E EL + ++E + A E ++
Sbjct: 1437 SVEELEYTINVLENKVNVVKDEAERQRLQREEL----EMELHTIRQQMESARNADEEMKR 1492
Query: 364 KSAE---SLALAKSDMEAVMQATSE---EIKKATETHAEALATKDAEIASQLAKIKSLED 417
E LA AK +EA+ + T++ EI + +E +E +A+ + + K K L
Sbjct: 1493 ILDEKHMDLAQAKKHIEALERNTADQKTEITQLSEHISELNLHAEAQASEYMHKFKEL-- 1550
Query: 418 ELATEKAKAIEAREQAADIALN 439
E E+ K QA D +L+
Sbjct: 1551 EAMAEQVKPEIHVSQAIDSSLS 1572
Score = 33.1 bits (74), Expect = 0.37
Identities = 52/230 (22%), Positives = 96/230 (41%), Gaps = 27/230 (11%)
Query: 205 RDVIEKEVLSRGLNDTKEETLACLLRA----GCIFAHTFEKFNAANVEAERLKVESAK-- 258
R+V+ + + N+ E++ + G + ++ +A + ++VE A
Sbjct: 551 RNVVVSSCIEKTPNNNHTESMRLSSKVSSERGKVIILLKQEMESALASLKEVQVEMANLK 610
Query: 259 -HQEAAVAWEKR----FDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELD 313
+E A EKR + LA Q M K+ LE ++A MK EAD+
Sbjct: 611 GEKEELKASEKRSLSNLNDLAAQICNLNTVMSNMEEQYEHKMETLEHEIAKMKIEADQEY 670
Query: 314 ASLQACKKEKEQAEKDLIARGEALIAKESELAV--LCAELELVKKALAEQEKKSAESLAL 371
K+ E+A+ + +E+++ V L E ++ L +Q+K+ +
Sbjct: 671 VENLCILKKFEEAQGTI---------READITVNELVIANEKMRFDLEKQKKRGISLVGE 721
Query: 372 AKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELAT 421
K+ +E + + S +K+ E LA + S L I +L +ELAT
Sbjct: 722 KKALVEKLQELESINVKE-----NEKLAYLEKLFESSLMGIGNLVEELAT 766
>At3g09000 unknown protein
Length = 541
Score = 45.8 bits (107), Expect = 6e-05
Identities = 37/176 (21%), Positives = 63/176 (35%), Gaps = 5/176 (2%)
Query: 9 NTEGDAKKRGGAENQAESTKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVA 68
N D + Q S+ RR + + S TS P + P +
Sbjct: 121 NCREDIVSGNNNKPQTSSSSVAGLRRPSSSGSSRSTSRPATPTRRSTTPTTSTSRPVTTR 180
Query: 69 AATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTP-IGDKEKENET 127
A+ ++ P S A A + + T +S S + S+ P +K
Sbjct: 181 ASNSRSSTPTSRATLTAARATTSTAAPRTTTTSSGSARSATPTRSNPRPSSASSKKPVSR 240
Query: 128 PKSPPRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSS 183
P +P R+ + P+ PS + P +G P + S+A +P+P + SS
Sbjct: 241 PATPTRRPSTPTGPSIVSS----KAPSRGTSPSPTVNSLSKAPSRGTSPSPTLNSS 292
Score = 38.9 bits (89), Expect = 0.007
Identities = 42/180 (23%), Positives = 65/180 (35%), Gaps = 22/180 (12%)
Query: 26 STKAPKRRRLTKASSDAGTSNP---GAQPTTA-AAPKGKNVAESSVAAAT------EPTA 75
ST P R + + S TS A+ TT+ AAP+ + S +AT P++
Sbjct: 172 STSRPVTTRASNSRSSTPTSRATLTAARATTSTAAPRTTTTSSGSARSATPTRSNPRPSS 231
Query: 76 VPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQD 135
+ P S AT + G + S+ + + S K T SP
Sbjct: 232 ASSKKPVSRPATPTRRPSTPTGPSIVSSKAPSRGTSPSPTVNSLSKAPSRGTSPSPTLNS 291
Query: 136 APPSPPSTHDAGSMPSPPH----------QGEKSCPGAATT--SEAAQIEQAPAPEVGSS 183
+ P P S+ +PP+ + PG A+ S + IE+ P G S
Sbjct: 292 SRPWKPPEMPGFSLEAPPNLRTTLADRPVSASRGRPGVASAPGSRSGSIERGGGPTSGGS 351
>At2g34730 putative myosin heavy chain
Length = 829
Score = 45.8 bits (107), Expect = 6e-05
Identities = 55/247 (22%), Positives = 104/247 (41%), Gaps = 27/247 (10%)
Query: 192 AIEPSEFFLAGLNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFNAA--NVEA 249
A++ + + LN V EKE R KE + R GC+ A N+
Sbjct: 544 AVKEAHKKIVELNLHVTEKEGTLRSEMVDKERLKEEIHRLGCLVKEKENLVQTAENNLAT 603
Query: 250 ERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGI--------KIGELEDQ 301
ER K+E Q + + + ++ T+ +DK+ A ++ + KI L ++
Sbjct: 604 ERKKIEVVSQQINDL--QSQVERQETEI-QDKIEALSVVSARELEKVKGYETKISSLREE 660
Query: 302 LAL-------MKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELV 354
L L MK+E + + L K EKE +K L++ + + L +++
Sbjct: 661 LELARESLKEMKDEKRKTEEKLSETKAEKETLKKQLVSLDLVVPPQ------LIKGFDIL 714
Query: 355 KKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKS 414
+ +AE+ +K+ L +S + + + E+K T+ + L K ++ A++
Sbjct: 715 EGLIAEKTQKTNSRLKNMQSQLSDLSHQIN-EVKGKASTYKQRLEKKCCDLKKAEAEVDL 773
Query: 415 LEDELAT 421
L DE+ T
Sbjct: 774 LGDEVET 780
Score = 37.7 bits (86), Expect = 0.015
Identities = 53/243 (21%), Positives = 97/243 (39%), Gaps = 34/243 (13%)
Query: 247 VEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMK 306
V+ ERLK E H+ + EK + L A + K I +I +L+ Q+ +
Sbjct: 571 VDKERLKEEI--HRLGCLVKEK--ENLVQTAENNLATERKKIEVVSQQINDLQSQVERQE 626
Query: 307 EEA-DELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAELELVKKALAE---QE 362
E D+++A +E E+ + E++++ L ELEL +++L E ++
Sbjct: 627 TEIQDKIEALSVVSARELEKVK-----------GYETKISSLREELELARESLKEMKDEK 675
Query: 363 KKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDELATE 422
+K+ E L+ E K ET + L + D + QL K + + L E
Sbjct: 676 RKTEEKLS---------------ETKAEKETLKKQLVSLDLVVPPQLIKGFDILEGLIAE 720
Query: 423 KAKAIEAREQAADIALNNRERGFYLAKDQAQHLYPNFDFSAMGVMKEITAAGLVGPDDPP 482
K + +R + L++ K +A + + K L+G +
Sbjct: 721 KTQKTNSRLKNMQSQLSDLSHQINEVKGKASTYKQRLEKKCCDLKKAEAEVDLLGDEVET 780
Query: 483 LID 485
L+D
Sbjct: 781 LLD 783
>At1g63300 hypothetical protein
Length = 1029
Score = 45.4 bits (106), Expect = 7e-05
Identities = 47/222 (21%), Positives = 102/222 (45%), Gaps = 21/222 (9%)
Query: 234 IFAHTFEKFNAANVEAERLKVESAKHQEAAVAWEKRFDKLATQAGKDK---VYADKMIGT 290
+ A+ ++ E E LK K+Q++ + ++ + L K K + A+ +
Sbjct: 722 VTANLNQEIKILKEEIENLK----KNQDSLMLQAEQAENLRVDLEKTKKSVMEAEASLQR 777
Query: 291 AGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEALIAKESELAVLCAE 350
+K ELE +++LM++E++ L A LQ K K++ E A+ ++EL + ++
Sbjct: 778 ENMKKIELESKISLMRKESESLAAELQVIKLAKDEKE-------TAISLLQTELETVRSQ 830
Query: 351 LELVKKALAEQE---KKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIAS 407
+ +K +L+E + +K + +A KS+++ + + KK E+ T +
Sbjct: 831 CDDLKHSLSENDLEMEKHKKQVAHVKSELKKKEETMANLEKKLKESRTAITKTAQRNNIN 890
Query: 408 QLAKI----KSLEDELATEKAKAIEAREQAADIALNNRERGF 445
+ + + S E + +K K +E + + + AL + F
Sbjct: 891 KGSPVGAHGGSKEVAVMKDKIKLLEGQIKLKETALESSSNMF 932
Score = 34.7 bits (78), Expect = 0.13
Identities = 42/212 (19%), Positives = 88/212 (40%), Gaps = 41/212 (19%)
Query: 266 WEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQ 325
+E + +L+ + ++M+ K E+++Q ++ L+ ++ K+E E
Sbjct: 680 YEAKLHELSEKLSFKTSQMERMLENLDEKSNEIDNQKRHEEDVTANLNQEIKILKEEIEN 739
Query: 326 AEKDLIARGEALIAKESELAVLCAELELVKKALAEQE--------------------KKS 365
+K+ ++L+ + + L +LE KK++ E E +K
Sbjct: 740 LKKNQ----DSLMLQAEQAENLRVDLEKTKKSVMEAEASLQRENMKKIELESKISLMRKE 795
Query: 366 AESLA-------LAKSDME---AVMQATSEEIKKATETHAEALATKDAE-------IASQ 408
+ESLA LAK + E +++Q E ++ + +L+ D E +A
Sbjct: 796 SESLAAELQVIKLAKDEKETAISLLQTELETVRSQCDDLKHSLSENDLEMEKHKKQVAHV 855
Query: 409 LAKIKSLEDELATEKAKAIEAREQAADIALNN 440
+++K E+ +A + K E+R A N
Sbjct: 856 KSELKKKEETMANLEKKLKESRTAITKTAQRN 887
>At5g24710 unknown protein
Length = 1003
Score = 45.1 bits (105), Expect = 9e-05
Identities = 51/199 (25%), Positives = 76/199 (37%), Gaps = 33/199 (16%)
Query: 6 PKFNTEGDAKKRG-------GAENQAESTKAPKRRRLTKASSDAGTSNPGA--------- 49
PK + DA ++ G + S APK+ LT+ ++ + P A
Sbjct: 785 PKLTSLSDASRKPPIEILPPGMSSIFASITAPKKPLLTQKTAQPEVAKPLALEEPTKPLA 844
Query: 50 --QPTTAAAPKGKNVAESSVAAATEP---TAVPASSPASAGATAAVAAESSIGATAASAS 104
P ++ AP+ ++ E++ AAA P TA A SPA A A A S A
Sbjct: 845 IEAPPSSEAPQTESAPETA-AAAESPAPETAAVAESPAPGTAAVAEAPASETAAAPVDGP 903
Query: 105 VNATKAAASSDTPIGDKE---KENETPKSPPRQDAPPSPPSTHDAGSMP-----SPPHQG 156
V T S P+ +E +E P S P + S +T + P +PP
Sbjct: 904 VTET---VSEPPPVEKEETSLEEKSDPSSTPNTETATSTENTSQTTTTPESVTTAPPEPI 960
Query: 157 EKSCPGAATTSEAAQIEQA 175
+ P TT+ E A
Sbjct: 961 TTAPPETVTTTAVKPTENA 979
>At5g16730 putative protein
Length = 853
Score = 45.1 bits (105), Expect = 9e-05
Identities = 43/180 (23%), Positives = 83/180 (45%), Gaps = 13/180 (7%)
Query: 267 EKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQA 326
E+R KL T K + ++ G + + +L+ +KE+ + + + + +K+K +A
Sbjct: 68 ERRSPKLPTPPEKSQA---RVAAVKGTESPQTTTRLSQIKEDLKKANERISSLEKDKAKA 124
Query: 327 EKDLIARGEALIAKESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEE 386
+L KE+E L + + +K +E E ++ +EAV Q EE
Sbjct: 125 LDEL-----KQAKKEAEQVTLKLD-DALKAQKHVEENSEIEKFQAVEAGIEAV-QNNEEE 177
Query: 387 IKKATETHAEALATKDAEIASQLAKIKSLEDELAT---EKAKAIEAREQAADIALNNRER 443
+KK ET A+ A + + +++ + +ELA K+KA+ E A+ A + E+
Sbjct: 178 LKKELETVKNQHASDSAALVAVRQELEKINEELAAAFDAKSKALSQAEDASKTAEIHAEK 237
Score = 33.1 bits (74), Expect = 0.37
Identities = 54/218 (24%), Positives = 89/218 (40%), Gaps = 53/218 (24%)
Query: 307 EEADELDASLQACKKEKEQAEKDL-------IARGEALIAKESELAVLCAELELVKKALA 359
E+ ++A ++A + +E+ +K+L + AL+A EL
Sbjct: 159 EKFQAVEAGIEAVQNNEEELKKELETVKNQHASDSAALVAVRQEL--------------- 203
Query: 360 EQEKKSAESLALAKSDMEAVMQATSEEIKKATETHAEALATKDAEIASQLAKIKSLEDEL 419
EK + E A + +A+ QA E+ K E HAE K ++S+L ++K+L D
Sbjct: 204 --EKINEELAAAFDAKSKALSQA--EDASKTAEIHAE----KVDILSSELTRLKALLD-- 253
Query: 420 ATEKAKAIEAREQAA----DIALNNRE----RGFYLAKDQAQHLYP--NFDFSAMGVMKE 469
+T + AI E A +I + R+ RGF + + + N D A M E
Sbjct: 254 STREKTAISDNEMVAKLEDEIVVLKRDLESARGFEAEVKEKEMIVEKLNVDLEA-AKMAE 312
Query: 470 ITAAGLVGPDDPPLIDQNLWTATEEEEEEEEQEKENNE 507
A L N W + +E EE+ +E E
Sbjct: 313 SNAHSL----------SNEWQSKAKELEEQLEEANKLE 340
>At4g37450 arabinogalactan protein AGP18
Length = 209
Score = 45.1 bits (105), Expect = 9e-05
Identities = 39/137 (28%), Positives = 60/137 (43%), Gaps = 9/137 (6%)
Query: 45 SNPGAQPTTAAAPKGKNVAESSVAAATEPTAVPASSPASAGATAAVAAESSIGATAASAS 104
S+P PTT +AP + + A T PT PA +P +A A++ V + S + S+
Sbjct: 26 SSPTKSPTTPSAP---TTSPTKSPAVTSPTTAPAKTP-TASASSPVESPKSPAPVSESSP 81
Query: 105 VNATKAAASSDTPIGDKEKENETPKSP-PRQDAPPSPPSTHDAGSMPSPPHQGEKSCPGA 163
+S P +P P P D+PP+P + A +P+P K
Sbjct: 82 PPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVAAPVA-DVPAPAPSKHKKTTKK 140
Query: 164 ATTSEAAQIEQAPAPEV 180
+ +AA APAPE+
Sbjct: 141 SKKHQAAP---APAPEL 154
Score = 37.4 bits (85), Expect = 0.020
Identities = 39/147 (26%), Positives = 54/147 (36%), Gaps = 16/147 (10%)
Query: 18 GGAENQAESTKAPKRRRLTKASSDAGTSNPGA-QPTTAAAPKGKNVAESSVAA------- 69
GG + TK+P A + + T +P PTTA A A S V +
Sbjct: 20 GGQSPISSPTKSPTT---PSAPTTSPTKSPAVTSPTTAPAKTPTASASSPVESPKSPAPV 76
Query: 70 ---ATEPTAVPASSPASAGATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENE 126
+ PT VP SSP + S + S A AA +D P K +
Sbjct: 77 SESSPPPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVAAPVADVPAPAPSKHKK 136
Query: 127 TPKSPPRQDAPPSPPSTHDAGSMPSPP 153
T K + A P+P + P+PP
Sbjct: 137 TTKKSKKHQAAPAP--APELLGPPAPP 161
Score = 36.6 bits (83), Expect = 0.034
Identities = 39/145 (26%), Positives = 51/145 (34%), Gaps = 12/145 (8%)
Query: 27 TKAPKRRRLTKASSDAGTSNPGAQPTTAAAPKGKNVAESSVAAATEPTAVP-ASSPASAG 85
T AP + ASS + A P + ++P V ESS P P SSP S+
Sbjct: 52 TTAPAKTPTASASSPVESPKSPA-PVSESSPPPTPVPESS-----PPVPAPMVSSPVSSP 105
Query: 86 ATAAVAAESSIGATAASASVNATKAAASSDTPIGDKEKENETPKSPPRQDAPPSPPSTHD 145
A A+S AA + A + +K P P PP+PP+
Sbjct: 106 PVPAPVADSPPAPVAAPVADVPAPAPSKHKKTTKKSKKHQAAPAPAPELLGPPAPPT--- 162
Query: 146 AGSMPSPPHQGEKSCPGAATTSEAA 170
P P P A S AA
Sbjct: 163 --ESPGPNSDAFSPGPSADDQSGAA 185
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.304 0.121 0.318
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,713,249
Number of Sequences: 26719
Number of extensions: 471010
Number of successful extensions: 5712
Number of sequences better than 10.0: 506
Number of HSP's better than 10.0 without gapping: 133
Number of HSP's successfully gapped in prelim test: 381
Number of HSP's that attempted gapping in prelim test: 4001
Number of HSP's gapped (non-prelim): 1405
length of query: 507
length of database: 11,318,596
effective HSP length: 104
effective length of query: 403
effective length of database: 8,539,820
effective search space: 3441547460
effective search space used: 3441547460
T: 11
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0201.4


