

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.15
(324 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
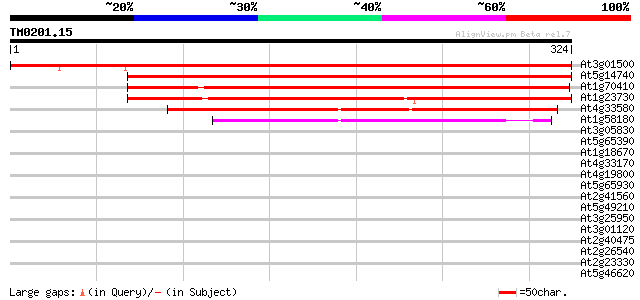
At3g01500 carbonic anhydrase, chloroplast precursor 431 e-121
At5g14740 CARBONIC ANHYDRASE 2 367 e-102
At1g70410 unknown protein 305 2e-83
At1g23730 putative carbonic anhydrase 286 8e-78
At4g33580 carbonate dehydratase - like protein 189 2e-48
At1g58180 carbonic anhydrase like protein 145 3e-35
At3g05830 unknown protein 35 0.055
At5g65390 arabinogalactan protein AGP7 32 0.61
At1g18670 hypothetical protein 32 0.61
At4g33170 putative protein 31 1.0
At4g19800 chitinase - like protein 30 1.4
At5g65930 kinesin-like calmodulin-binding protein 30 2.3
At2g41560 putative Ca2+-ATPase 30 2.3
At5g49210 unknown protein 29 3.0
At3g25950 hypothetical protein 29 3.0
At3g01120 cystathionine gamma-synthase like protein 29 3.0
At2g40475 unknown protein 29 3.0
At2g26540 unknown protein 29 3.0
At2g23330 putative retroelement pol polyprotein 29 3.0
At5g46620 unknown protein 29 4.0
>At3g01500 carbonic anhydrase, chloroplast precursor
Length = 347
Score = 431 bits (1108), Expect = e-121
Identities = 223/336 (66%), Positives = 267/336 (79%), Gaps = 12/336 (3%)
Query: 1 MSTSTINGFYLSSISPAKTSLKRVTLR--------PLVVASASSPSSSSSSSFPSLIQDR 52
MST+ ++GF+L+S+SP+++SL++++LR P +S+SS SSSSS S P+LI++
Sbjct: 1 MSTAPLSGFFLTSLSPSQSSLQKLSLRTSSTVACLPPASSSSSSSSSSSSRSVPTLIRNE 60
Query: 53 PVFAAPAPIIPTL---DMGKN-YEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSS 108
PVFAAPAPII +MG Y+EAIE L+KLL EK ELK AA KVEQITA L T +S
Sbjct: 61 PVFAAPAPIIAPYWSEEMGTEAYDEAIEALKKLLIEKEELKTVAAAKVEQITAALQTGTS 120
Query: 109 DGIPSSEASDRIKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDF 168
+ + + IK GF+ FKKEKY+ NPALYGELAKGQSP YMVFACSDSRVCPSHVLDF
Sbjct: 121 SDKKAFDPVETIKQGFIKFKKEKYETNPALYGELAKGQSPKYMVFACSDSRVCPSHVLDF 180
Query: 169 QPGEAFVVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFP 228
QPG+AFVVRN+ANMVPP+D+ K+ G GAAIEYAVLHLKV NIVVIGHSACGGIKGL+SFP
Sbjct: 181 QPGDAFVVRNIANMVPPFDKVKYGGVGAAIEYAVLHLKVENIVVIGHSACGGIKGLMSFP 240
Query: 229 FDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVRE 288
DG STDFIEDWV I LPAK+KV ++ GD+ F + C CE+EAVNVSL NLL+YPFVRE
Sbjct: 241 LDGNNSTDFIEDWVKICLPAKSKVISELGDSAFEDQCGRCEREAVNVSLANLLTYPFVRE 300
Query: 289 GLVNKTLALKGGYYDFVKGSFELWSLQFDLASSFSV 324
GLV TLALKGGYYDFVKG+FELW L+F L+ + SV
Sbjct: 301 GLVKGTLALKGGYYDFVKGAFELWGLEFGLSETSSV 336
>At5g14740 CARBONIC ANHYDRASE 2
Length = 259
Score = 367 bits (941), Expect = e-102
Identities = 180/256 (70%), Positives = 210/256 (81%)
Query: 69 KNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYFK 128
++YE+AIE L+KLL EK++LK AA KV++ITA+L SS S + +RIK GF+ FK
Sbjct: 4 ESYEDAIEALKKLLIEKDDLKDVAAAKVKKITAELQAASSSDSKSFDPVERIKEGFVTFK 63
Query: 129 KEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQ 188
KEKY+ NPALYGELAKGQSP YMVFACSDSRVCPSHVLDF PG+AFVVRN+ANMVPP+D+
Sbjct: 64 KEKYETNPALYGELAKGQSPKYMVFACSDSRVCPSHVLDFHPGDAFVVRNIANMVPPFDK 123
Query: 189 AKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPA 248
K++G GAAIEYAVLHLKV NIVVIGHSACGGIKGL+SFP DG STDFIEDWV I LPA
Sbjct: 124 VKYAGVGAAIEYAVLHLKVENIVVIGHSACGGIKGLMSFPLDGNNSTDFIEDWVKICLPA 183
Query: 249 KAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGS 308
K+KV A+ + F + C CE+EAVNVSL NLL+YPFVREG+V TLALKGGYYDFV GS
Sbjct: 184 KSKVLAESESSAFEDQCGRCEREAVNVSLANLLTYPFVREGVVKGTLALKGGYYDFVNGS 243
Query: 309 FELWSLQFDLASSFSV 324
FELW LQF ++ S+
Sbjct: 244 FELWELQFGISPVHSI 259
>At1g70410 unknown protein
Length = 280
Score = 305 bits (782), Expect = 2e-83
Identities = 151/256 (58%), Positives = 191/256 (73%), Gaps = 4/256 (1%)
Query: 69 KNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYFK 128
++YE AI+ L LL K +L AA K++ +TA+L S +S+A +RIKTGF FK
Sbjct: 26 ESYEAAIKGLNDLLSTKADLGNVAAAKIKALTAELKELDSS---NSDAIERIKTGFTQFK 82
Query: 129 KEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQ 188
EKY KN L+ LAK Q+P ++VFACSDSRVCPSH+L+FQPGEAFVVRN+ANMVPP+DQ
Sbjct: 83 TEKYLKNSTLFNHLAKTQTPKFLVFACSDSRVCPSHILNFQPGEAFVVRNIANMVPPFDQ 142
Query: 189 AKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGT-TSTDFIEDWVSIGLP 247
+ SG GAA+EYAV+HLKV NI+VIGHS CGGIKGL+S D T +DFIE+WV IG
Sbjct: 143 KRHSGVGAAVEYAVVHLKVENILVIGHSCCGGIKGLMSIEDDAAPTQSDFIENWVKIGAS 202
Query: 248 AKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKG 307
A+ K+K +H D + + C CEKEAVNVSLGNLLSYPFVR +V TLA++GG+Y+FVKG
Sbjct: 203 ARNKIKEEHKDLSYDDQCNKCEKEAVNVSLGNLLSYPFVRAEVVKNTLAIRGGHYNFVKG 262
Query: 308 SFELWSLQFDLASSFS 323
+F+LW L F +F+
Sbjct: 263 TFDLWELDFKTTPAFA 278
>At1g23730 putative carbonic anhydrase
Length = 258
Score = 286 bits (733), Expect = 8e-78
Identities = 141/258 (54%), Positives = 191/258 (73%), Gaps = 6/258 (2%)
Query: 69 KNYEEAIEELQKLLREKNELKATAAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYFK 128
++YE+AI+ L +LL +K++L AA K++++T +L S+ + +A +RIK+GFL+FK
Sbjct: 4 ESYEDAIKRLGELLSKKSDLGNVAAAKIKKLTDELEELDSNKL---DAVERIKSGFLHFK 60
Query: 129 KEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQ 188
Y+KNP LY LAK Q+P ++VFAC+DSRV PSH+L+FQ GEAF+VRN+ANMVPPYD+
Sbjct: 61 TNNYEKNPTLYNSLAKSQTPKFLVFACADSRVSPSHILNFQLGEAFIVRNIANMVPPYDK 120
Query: 189 AKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGT--TSTDFIEDWVSIGL 246
K S GAA+EY + L V NI+VIGHS CGGIKGL++ D T T T+FIE+W+ I
Sbjct: 121 TKHSNVGAALEYPITVLNVENILVIGHSCCGGIKGLMAIE-DNTAPTKTEFIENWIQICA 179
Query: 247 PAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVK 306
PAK ++K D F + CT+CEKEAVNVSLGNLLSYPFVRE +V LA++G +YDFVK
Sbjct: 180 PAKNRIKQDCKDLSFEDQCTNCEKEAVNVSLGNLLSYPFVRERVVKNKLAIRGAHYDFVK 239
Query: 307 GSFELWSLQFDLASSFSV 324
G+F+LW L F +F++
Sbjct: 240 GTFDLWELDFKTTPAFAL 257
>At4g33580 carbonate dehydratase - like protein
Length = 246
Score = 189 bits (480), Expect = 2e-48
Identities = 99/225 (44%), Positives = 140/225 (62%), Gaps = 2/225 (0%)
Query: 92 AAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFLYFKKEKYDKNPALYGELAKGQSPPYM 151
A+ K +T + + D +++ D +K FL FKK KY + Y LA Q+P ++
Sbjct: 2 ASGKTPGLTQEANGVAIDRQNNTDVFDDMKQRFLAFKKLKYMDDFEHYKNLADAQAPKFL 61
Query: 152 VFACSDSRVCPSHVLDFQPGEAFVVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIV 211
V AC+DSRVCPS VL FQPG+AF VRN+AN+VPPY+ + + AA+E++V L V NI+
Sbjct: 62 VIACADSRVCPSAVLGFQPGDAFTVRNIANLVPPYESGP-TETKAALEFSVNTLNVENIL 120
Query: 212 VIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKE 271
VIGHS CGGI+ L+ +G S FI +WV +G AK KA + F C HCEK
Sbjct: 121 VIGHSRCGGIQALMKMEDEG-DSRSFIHNWVVVGKKAKESTKAVASNLHFDHQCQHCEKA 179
Query: 272 AVNVSLGNLLSYPFVREGLVNKTLALKGGYYDFVKGSFELWSLQF 316
++N SL LL YP++ E + +L+L GGYY+FV +FE W++ +
Sbjct: 180 SINHSLERLLGYPWIEEKVRQGSLSLHGGYYNFVDCTFEKWTVDY 224
>At1g58180 carbonic anhydrase like protein
Length = 256
Score = 145 bits (366), Expect = 3e-35
Identities = 80/196 (40%), Positives = 111/196 (55%), Gaps = 16/196 (8%)
Query: 118 DRIKTGFLYFKKEKYDKNPALYGELAKGQSPPYMVFACSDSRVCPSHVLDFQPGEAFVVR 177
D ++ FL FK++KY + LA QSP MV C+DSRVCPS+VL FQPGEAF +R
Sbjct: 77 DEMRHRFLKFKRQKYLPEIEKFKALAIAQSPKVMVIGCADSRVCPSYVLGFQPGEAFTIR 136
Query: 178 NVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIGHSACGGIKGLLSFPFDGTTSTDF 237
NVAN+V P + + +A+E+AV L+V NI+V+GHS CGGI L+S +
Sbjct: 137 NVANLVTPVQNGP-TETNSALEFAVTTLQVENIIVMGHSNCGGIAALMSHQNHQGQHSSL 195
Query: 238 IEDWVSIGLPAKAKVKAKHGDAPFGELCTHCEKEAVNVSLGNLLSYPFVREGLVNKTLAL 297
+E WV G AK + + F E C +CEKE++ S+ NL++Y ++R
Sbjct: 196 VERWVMNGKAAKLRTQLASSHLSFDEQCRNCEKESIKDSVMNLITYSWIR---------- 245
Query: 298 KGGYYDFVKGSFELWS 313
D VK E+WS
Sbjct: 246 -----DRVKRDREIWS 256
>At3g05830 unknown protein
Length = 336
Score = 35.0 bits (79), Expect = 0.055
Identities = 26/78 (33%), Positives = 40/78 (50%), Gaps = 7/78 (8%)
Query: 68 GKNYEEAIEELQKLLREKN---ELKATAAEK----VEQITAQLGTTSSDGIPSSEASDRI 120
GKN E I +LQK L E+N E A+AA+K +E+ +L TT S++++
Sbjct: 62 GKNMEMEICKLQKRLEERNCQLEASASAADKFIKELEEFRLKLDTTKQTAEASADSAQST 121
Query: 121 KTGFLYFKKEKYDKNPAL 138
K K++ DK +L
Sbjct: 122 KIQCSMLKQQLDDKTRSL 139
>At5g65390 arabinogalactan protein AGP7
Length = 130
Score = 31.6 bits (70), Expect = 0.61
Identities = 22/67 (32%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Query: 3 TSTINGFYLSSISPAKTSLKRVTLRPLVVASASSPSSSSSSSFPSLIQDRPVFAAPAPII 62
T+T+ +++ PA T T P V + +P+SS SS PS D P + PAP
Sbjct: 29 TTTVTPPPVATPPPAATPAPTTTPPPAV---SPAPTSSPPSSAPSPSSDAPTASPPAPEG 85
Query: 63 PTLDMGK 69
P + G+
Sbjct: 86 PGVSPGE 92
>At1g18670 hypothetical protein
Length = 662
Score = 31.6 bits (70), Expect = 0.61
Identities = 20/78 (25%), Positives = 31/78 (39%), Gaps = 6/78 (7%)
Query: 212 VIGHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKVKAKHGDAPFGELC------ 265
V + C G ++ T T + W G + +K + FGEL
Sbjct: 21 VFRDNVCSGSGRIVVEDLPPVTETKLLSWWSKSGKKSSSKKSGSELGSDFGELSESGRAS 80
Query: 266 THCEKEAVNVSLGNLLSY 283
++C E+V+ LGNL Y
Sbjct: 81 SNCRSESVSFRLGNLSKY 98
>At4g33170 putative protein
Length = 990
Score = 30.8 bits (68), Expect = 1.0
Identities = 18/26 (69%), Positives = 19/26 (72%)
Query: 19 TSLKRVTLRPLVVASASSPSSSSSSS 44
TSL LRPL SA+SPSSSSSSS
Sbjct: 14 TSLIVQCLRPLRFTSAASPSSSSSSS 39
>At4g19800 chitinase - like protein
Length = 398
Score = 30.4 bits (67), Expect = 1.4
Identities = 15/39 (38%), Positives = 17/39 (43%)
Query: 214 GHSACGGIKGLLSFPFDGTTSTDFIEDWVSIGLPAKAKV 252
G S G L P DG + + DW GLPAK V
Sbjct: 193 GWSTVTGPPASLYLPTDGRSGDSGVRDWTEAGLPAKKAV 231
>At5g65930 kinesin-like calmodulin-binding protein
Length = 1260
Score = 29.6 bits (65), Expect = 2.3
Identities = 14/37 (37%), Positives = 26/37 (69%), Gaps = 3/37 (8%)
Query: 66 DMGKNYEEAIEELQKLL---REKNELKATAAEKVEQI 99
D+ K YEE+ ++++KL+ +EKN+ + T E++E I
Sbjct: 626 DLSKAYEESQKKIEKLMDEQQEKNQQEVTLREELEAI 662
>At2g41560 putative Ca2+-ATPase
Length = 1030
Score = 29.6 bits (65), Expect = 2.3
Identities = 21/63 (33%), Positives = 36/63 (56%), Gaps = 6/63 (9%)
Query: 76 EELQKLLREKNELKATAAEK-VEQITAQLGTTSSDGIPSSEASDRIKTGFLYFKKEKYDK 134
+EL ++R KN+ K+ A + VE++ ++ + S+GI SSE R K F + +Y +
Sbjct: 98 DELASMVR-KNDTKSLAQKGGVEELAKKVSVSLSEGIRSSEVPIREK----IFGENRYTE 152
Query: 135 NPA 137
PA
Sbjct: 153 KPA 155
>At5g49210 unknown protein
Length = 195
Score = 29.3 bits (64), Expect = 3.0
Identities = 16/37 (43%), Positives = 22/37 (59%)
Query: 67 MGKNYEEAIEELQKLLREKNELKATAAEKVEQITAQL 103
MG+ EE I+E +KLLRE++ L E +IT L
Sbjct: 153 MGQKREEKIKEREKLLREQSSLWIEQKELERKITEAL 189
>At3g25950 hypothetical protein
Length = 251
Score = 29.3 bits (64), Expect = 3.0
Identities = 19/40 (47%), Positives = 22/40 (54%)
Query: 175 VVRNVANMVPPYDQAKFSGSGAAIEYAVLHLKVSNIVVIG 214
VVR +A V YD A F GSGAA +S +VVIG
Sbjct: 184 VVRGLAGPVVLYDMATFYGSGAADGVVPRWAWLSWLVVIG 223
>At3g01120 cystathionine gamma-synthase like protein
Length = 563
Score = 29.3 bits (64), Expect = 3.0
Identities = 25/94 (26%), Positives = 46/94 (48%), Gaps = 13/94 (13%)
Query: 32 ASASSPSSSSSSSFPSLIQDRPVFAAPAPIIPTLDMGKNYEEAIEELQKLLREKNELKAT 91
+SAS+ SS++S++ S PV AAP ++ K+ +E + ++ +REK
Sbjct: 111 SSASAVSSAASAAAASSAAAAPVAAAPPVVL------KSVDEEVVVAEEGIREK------ 158
Query: 92 AAEKVEQITAQLGTTSSDGIPSSEASDRIKTGFL 125
V+ ++ SSDG + A +R+ G +
Sbjct: 159 -IGSVQLTDSKHSFLSSDGSLTVHAGERLGRGIV 191
>At2g40475 unknown protein
Length = 193
Score = 29.3 bits (64), Expect = 3.0
Identities = 20/39 (51%), Positives = 23/39 (58%), Gaps = 3/39 (7%)
Query: 18 KTSLKRVTLRPLVV---ASASSPSSSSSSSFPSLIQDRP 53
KT L R RP AS+SS SS SSSS PS ++ RP
Sbjct: 94 KTLLSRHVSRPSFSWSSASSSSSSSYSSSSPPSKVEHRP 132
>At2g26540 unknown protein
Length = 321
Score = 29.3 bits (64), Expect = 3.0
Identities = 17/33 (51%), Positives = 21/33 (63%), Gaps = 2/33 (6%)
Query: 31 VASASSPSSSSSSSFPSLIQD--RPVFAAPAPI 61
+ S P SSSSS S IQ +PVFA+P+PI
Sbjct: 11 ILSFQPPLSSSSSFHSSHIQSLSKPVFASPSPI 43
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 29.3 bits (64), Expect = 3.0
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 4/59 (6%)
Query: 30 VVASASSPSSSSSSSFPSLIQDRPVFAAPAPIIPTLDMGKNYEEAIEELQKLLREKNEL 88
+V S SS ++SS++++ D P + ++ NY E EELQ LR K +L
Sbjct: 7 LVTSTSSSNTSSTTAYLINASDNPGALISSVVLKE----NNYAEWSEELQNFLRAKQKL 61
>At5g46620 unknown protein
Length = 149
Score = 28.9 bits (63), Expect = 4.0
Identities = 21/68 (30%), Positives = 30/68 (43%), Gaps = 6/68 (8%)
Query: 193 GSGAAIEYAVLHLKVSNIVVIGHSACGGIKGL------LSFPFDGTTSTDFIEDWVSIGL 246
G G ++ AV I+ + GG++GL L DG + +F+ED L
Sbjct: 63 GFGVPVQVAVERSNPGPILQPCAALDGGVQGLRWYSMRLKIDEDGDVADEFLEDQNCKTL 122
Query: 247 PAKAKVKA 254
P K K KA
Sbjct: 123 PRKCKTKA 130
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,308,392
Number of Sequences: 26719
Number of extensions: 316110
Number of successful extensions: 1688
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 1646
Number of HSP's gapped (non-prelim): 46
length of query: 324
length of database: 11,318,596
effective HSP length: 99
effective length of query: 225
effective length of database: 8,673,415
effective search space: 1951518375
effective search space used: 1951518375
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0201.15


