

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.11
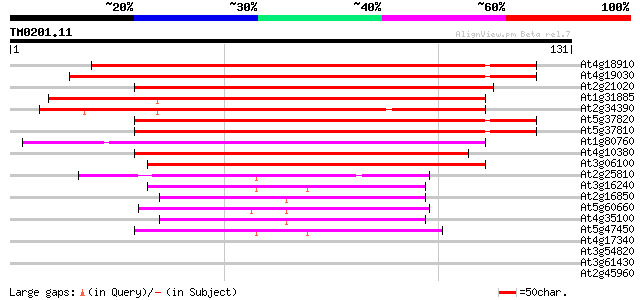
(131 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g18910 major intrinsic protein (MIP)- like 140 2e-34
At4g19030 nodulin-26 like protein 139 3e-34
At2g21020 putative major intrinsic (channel) protein 112 7e-26
At1g31885 major intrinsic protein - like 112 7e-26
At2g34390 putative aquaporin (plasma membrane intrinsic protein) 111 1e-25
At5g37820 Membrane integral protein (MIP) -like 107 1e-24
At5g37810 Membrane integral protein (MIP) -like 107 2e-24
At1g80760 nodulin-like protein 87 3e-18
At4g10380 major intrinsic protein (MIP) - like 76 4e-15
At3g06100 putative major intrinsic protein 68 1e-12
At2g25810 putative aquaporin (tonoplast intrinsic protein) 49 7e-07
At3g16240 delta tonoplast integral protein (delta-TIP) 45 1e-05
At2g16850 putative aquaporin (plasma membrane intrinsic protein) 42 7e-05
At5g60660 mipC protein - like (aquaporin) 41 2e-04
At4g35100 plasma membrane intrinsic protein PIP3 41 2e-04
At5g47450 membrane channel protein-like; aquaporin (tonoplast in... 40 4e-04
At4g17340 membrane channel like protein 38 0.001
At3g54820 aquaporin/MIP - like protein 38 0.001
At3g61430 plasma membrane intrinsic protein 1a 37 0.003
At2g45960 aquaporin (plasma membrane intrinsic protein 1B) 37 0.003
>At4g18910 major intrinsic protein (MIP)- like
Length = 294
Score = 140 bits (352), Expect = 2e-34
Identities = 73/104 (70%), Positives = 82/104 (78%), Gaps = 1/104 (0%)
Query: 20 KKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMV 79
KK DS+ VP LQKL+AEVLGTYFLIFAGCA+V VN +DK VTLPGIAIVWGL VMV
Sbjct: 38 KKQDSLLSISVPFLQKLMAEVLGTYFLIFAGCAAVAVNTQHDKAVTLPGIAIVWGLTVMV 97
Query: 80 LVYSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
LVYS+GHISGAHFNPAVT+A A+ FP+KQV P + V GS
Sbjct: 98 LVYSLGHISGAHFNPAVTIAFASCGRFPLKQV-PAYVISQVIGS 140
Score = 31.2 bits (69), Expect = 0.16
Identities = 23/73 (31%), Positives = 36/73 (48%), Gaps = 5/73 (6%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
LQ V E + T++L+F V +N + L G+A+ G V++ V G +SGA
Sbjct: 175 LQSFVIEFIITFYLMFVISG---VATDNRAIGELAGLAV--GSTVLLNVIIAGPVSGASM 229
Query: 93 NPAVTVAHATTKS 105
NP ++ A S
Sbjct: 230 NPGRSLGPAMVYS 242
>At4g19030 nodulin-26 like protein
Length = 296
Score = 139 bits (351), Expect = 3e-34
Identities = 71/109 (65%), Positives = 82/109 (75%), Gaps = 1/109 (0%)
Query: 15 NNETSKKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWG 74
N KK DS+ VP LQKL+AE LGTYFL+F GCASVVVN+ ND VVTLPGIAIVWG
Sbjct: 36 NPRPLKKQDSLLSVSVPFLQKLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAIVWG 95
Query: 75 LAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
L +MVL+YS+GHISGAH NPAVT+A A+ FP+KQV P + V GS
Sbjct: 96 LTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQV-PAYVISQVIGS 143
>At2g21020 putative major intrinsic (channel) protein
Length = 262
Score = 112 bits (279), Expect = 7e-26
Identities = 50/84 (59%), Positives = 66/84 (78%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
V +QKL+ E +GT+ +IFAGC+++VVN K VTLPGIA+VWGL V V++YSIGH+SG
Sbjct: 50 VSFVQKLIGEFVGTFSMIFAGCSAIVVNETYGKPVTLPGIALVWGLVVTVMIYSIGHVSG 109
Query: 90 AHFNPAVTVAHATTKSFPVKQVFP 113
AHFNPAV++A A++K FP Q P
Sbjct: 110 AHFNPAVSIAFASSKKFPFNQFHP 133
>At1g31885 major intrinsic protein - like
Length = 294
Score = 112 bits (279), Expect = 7e-26
Identities = 55/107 (51%), Positives = 75/107 (69%), Gaps = 5/107 (4%)
Query: 10 VILNVNNETSKKCDSIEEDCVPLL-----QKLVAEVLGTYFLIFAGCASVVVNLNNDKVV 64
V+L++ N S + PL+ QKL+ E +GT+ +IFAGC+++VVN K V
Sbjct: 14 VVLDIENYQSIDDSRSSDLSAPLVSVSFVQKLIGEFVGTFTMIFAGCSAIVVNETYGKPV 73
Query: 65 TLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
TLPGIA+VWGL V V++YSIGH+SGAHFNPAV++A A++K FP QV
Sbjct: 74 TLPGIALVWGLVVTVMIYSIGHVSGAHFNPAVSIAFASSKKFPFNQV 120
Score = 27.3 bits (59), Expect = 2.3
Identities = 20/65 (30%), Positives = 31/65 (46%), Gaps = 5/65 (7%)
Query: 37 VAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAV 96
V E + T+ L+F V+ + DK T I G +++ + G ISGA NPA
Sbjct: 170 VMEFIATFNLMF-----VISAVATDKRATGSFAGIAIGATIVLDILFSGPISGASMNPAR 224
Query: 97 TVAHA 101
++ A
Sbjct: 225 SLGPA 229
>At2g34390 putative aquaporin (plasma membrane intrinsic protein)
Length = 288
Score = 111 bits (277), Expect = 1e-25
Identities = 59/113 (52%), Positives = 81/113 (71%), Gaps = 10/113 (8%)
Query: 8 NEVILNVNN----ETSKKCDSIEEDCVPLL-----QKLVAEVLGTYFLIFAGCASVVVNL 58
N V+LN+ +TS + E PLL QKL+AE++GTY+LIFAGCA++ VN
Sbjct: 13 NVVVLNIKASSLADTSLPSNKHESSSPPLLSVHFLQKLLAELVGTYYLIFAGCAAIAVNA 72
Query: 59 NNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
++ VVTL GIA+VWG+ +MVLVY +GH+S AHFNPAVT+A A+++ FP+ QV
Sbjct: 73 QHNHVVTLVGIAVVWGIVIMVLVYCLGHLS-AHFNPAVTLALASSQRFPLNQV 124
Score = 33.5 bits (75), Expect = 0.032
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 8/100 (8%)
Query: 5 SGSNEVILNVNNETSKKCDSIEEDCVPL---LQKLVAEVLGTYFLIFAGCASVVVNLNND 61
S + ++ ++NN+ K + P LQ V E + T FL+ CA +
Sbjct: 139 SATLRLLFDLNNDVCSKKHDVFLGSSPSGSDLQAFVMEFIITGFLMLVVCAVTTTKRTTE 198
Query: 62 KVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHA 101
+ L G+ I G V + V G +SGA NPA ++ A
Sbjct: 199 E---LEGLII--GATVTLNVIFAGEVSGASMNPARSIGPA 233
>At5g37820 Membrane integral protein (MIP) -like
Length = 283
Score = 107 bits (268), Expect = 1e-24
Identities = 54/94 (57%), Positives = 67/94 (70%), Gaps = 1/94 (1%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
V L QKL+AE++GTYF+IF+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIIFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 90 AHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
AHFNPAVTV A + FP QV PL + + GS
Sbjct: 99 AHFNPAVTVTFAVFRRFPWYQV-PLYIGAQLTGS 131
Score = 36.6 bits (83), Expect = 0.004
Identities = 32/100 (32%), Positives = 51/100 (51%), Gaps = 10/100 (10%)
Query: 34 QKLVAEVLGTYFLIFAGCASVVVNLNNDKVVT--LPGIAIVWGLAVMVLVYSIGHISGAH 91
Q LVAE++ ++ L+F V+ + D T L GIA+ G+ +++ V+ G ISGA
Sbjct: 160 QALVAEIIISFLLMF-----VISGVATDSRATGELAGIAV--GMTIILNVFVAGPISGAS 212
Query: 92 FNPAVTVAHATTKSFPVKQVFPLPLFKHVGGSAKGFHWRF 131
NPA ++ A K ++ + VG A GF + F
Sbjct: 213 MNPARSLGPAIVMG-RYKGIWVYIVGPFVGIFAGGFVYNF 251
>At5g37810 Membrane integral protein (MIP) -like
Length = 283
Score = 107 bits (267), Expect = 2e-24
Identities = 53/94 (56%), Positives = 66/94 (69%), Gaps = 1/94 (1%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
V L QKL+AE++GTYF++F+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIVFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 90 AHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
AHFNPAVTV A + FP QV PL + GS
Sbjct: 99 AHFNPAVTVTFAIFRRFPWHQV-PLYIGAQFAGS 131
Score = 37.7 bits (86), Expect = 0.002
Identities = 26/66 (39%), Positives = 38/66 (57%), Gaps = 5/66 (7%)
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPA 95
LVAE++ ++ L+F V +N V L GIA+ G+ +MV V+ G ISGA NPA
Sbjct: 162 LVAEIIISFLLMFVISG---VATDNRAVGELAGIAV--GMTIMVNVFVAGPISGASMNPA 216
Query: 96 VTVAHA 101
++ A
Sbjct: 217 RSLGPA 222
>At1g80760 nodulin-like protein
Length = 305
Score = 86.7 bits (213), Expect = 3e-18
Identities = 52/108 (48%), Positives = 62/108 (57%), Gaps = 1/108 (0%)
Query: 4 YSGSNEVILNVNNETSKKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV 63
+S NE L C S+ V L +KL AE +GT LIFAG A+ +VN D
Sbjct: 51 FSVDNEWALEDGRLPPVTC-SLPPPNVSLYRKLGAEFVGTLILIFAGTATAIVNQKTDGA 109
Query: 64 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
TL G A GLAVM+++ S GHISGAH NPAVT+A A K FP K V
Sbjct: 110 ETLIGCAASAGLAVMIVILSTGHISGAHLNPAVTIAFAALKHFPWKHV 157
>At4g10380 major intrinsic protein (MIP) - like
Length = 304
Score = 76.3 bits (186), Expect = 4e-15
Identities = 39/78 (50%), Positives = 50/78 (64%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
V L +KL AE +GT+ LIF A +VN D TL G A GLAVM+++ S GHISG
Sbjct: 74 VSLTRKLGAEFVGTFILIFTATAGPIVNQKYDGAETLIGNAACAGLAVMIIILSTGHISG 133
Query: 90 AHFNPAVTVAHATTKSFP 107
AH NP++T+A A + FP
Sbjct: 134 AHLNPSLTIAFAALRHFP 151
>At3g06100 putative major intrinsic protein
Length = 275
Score = 68.2 bits (165), Expect = 1e-12
Identities = 35/79 (44%), Positives = 52/79 (65%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
L+ ++AE++GT+ L+F+ C + + V L A+ GL+V+V+VYSIGHISGAH
Sbjct: 45 LRIVMAELVGTFILMFSVCGVISSTQLSGGHVGLLEYAVTAGLSVVVVVYSIGHISGAHL 104
Query: 93 NPAVTVAHATTKSFPVKQV 111
NP++T+A A FP QV
Sbjct: 105 NPSITIAFAVFGGFPWSQV 123
>At2g25810 putative aquaporin (tonoplast intrinsic protein)
Length = 249
Score = 48.9 bits (115), Expect = 7e-07
Identities = 28/84 (33%), Positives = 46/84 (54%), Gaps = 6/84 (7%)
Query: 17 ETSKKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPGIAIVWG 74
E ++ + DC+ + L+ E + T+ +FAG S + +L + +V L +A+
Sbjct: 5 ELGHHSEAAKPDCI---KALIVEFITTFLFVFAGVGSAMATDSLVGNTLVGLFAVAVAHA 61
Query: 75 LAVMVLVYSIGHISGAHFNPAVTV 98
V V++ S GHISG H NPAVT+
Sbjct: 62 FVVAVMI-SAGHISGGHLNPAVTL 84
>At3g16240 delta tonoplast integral protein (delta-TIP)
Length = 250
Score = 44.7 bits (104), Expect = 1e-05
Identities = 27/70 (38%), Positives = 37/70 (52%), Gaps = 5/70 (7%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSIGHI 87
L+ +AE + T +FAG S + L +D + PG IA+ G A+ V V +I
Sbjct: 18 LRAYLAEFISTLLFVFAGVGSAIAYAKLTSDAALDTPGLVAIAVCHGFALFVAVAIGANI 77
Query: 88 SGAHFNPAVT 97
SG H NPAVT
Sbjct: 78 SGGHVNPAVT 87
>At2g16850 putative aquaporin (plasma membrane intrinsic protein)
Length = 278
Score = 42.4 bits (98), Expect = 7e-05
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
++AE + T ++ A+V+ + N V L GIA +G + VLVY ISG H
Sbjct: 38 IIAEFIATLLFLYVTVATVIGHKNQTGPCGGVGLLGIAWAFGGMIFVLVYCTAGISGGHI 97
Query: 93 NPAVT 97
NPAVT
Sbjct: 98 NPAVT 102
>At5g60660 mipC protein - like (aquaporin)
Length = 291
Score = 40.8 bits (94), Expect = 2e-04
Identities = 26/77 (33%), Positives = 37/77 (47%), Gaps = 9/77 (11%)
Query: 31 PLLQKLVAEVLGTYFLIFAGCASVV-VNLNNDKV--------VTLPGIAIVWGLAVMVLV 81
PL + ++AE + T ++ +V+ D V + GIA +G + VLV
Sbjct: 36 PLYRAVIAEFVATLLFLYVSILTVIGYKAQTDATAGGVDCGGVGILGIAWAFGGMIFVLV 95
Query: 82 YSIGHISGAHFNPAVTV 98
Y ISG H NPAVTV
Sbjct: 96 YCTAGISGGHINPAVTV 112
>At4g35100 plasma membrane intrinsic protein PIP3
Length = 280
Score = 40.8 bits (94), Expect = 2e-04
Identities = 25/65 (38%), Positives = 34/65 (51%), Gaps = 3/65 (4%)
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
L+AE + T ++ A+V+ + V L GIA +G + VLVY ISG H
Sbjct: 40 LIAEFIATLLFLYVTVATVIGHKKQTGPCDGVGLLGIAWAFGGMIFVLVYCTAGISGGHI 99
Query: 93 NPAVT 97
NPAVT
Sbjct: 100 NPAVT 104
>At5g47450 membrane channel protein-like; aquaporin (tonoplast
intrinsic protein)-like
Length = 250
Score = 39.7 bits (91), Expect = 4e-04
Identities = 28/77 (36%), Positives = 38/77 (48%), Gaps = 5/77 (6%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSI 84
V L+ ++E + T +FAG S V L +D + G IAI A+ V V
Sbjct: 15 VSSLKAYLSEFIATLLFVFAGVGSAVAFAKLTSDGALDPAGLVAIAIAHAFALFVGVSIA 74
Query: 85 GHISGAHFNPAVTVAHA 101
+ISG H NPAVT+ A
Sbjct: 75 ANISGGHLNPAVTLGLA 91
>At4g17340 membrane channel like protein
Length = 250
Score = 38.1 bits (87), Expect = 0.001
Identities = 25/77 (32%), Positives = 38/77 (48%), Gaps = 5/77 (6%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSI 84
V L+ ++E + T +FAG S + L +D + G +A+ A+ V V
Sbjct: 15 VASLKAYLSEFIATLLFVFAGVGSALAFAKLTSDAALDPAGLVAVAVAHAFALFVGVSIA 74
Query: 85 GHISGAHFNPAVTVAHA 101
+ISG H NPAVT+ A
Sbjct: 75 ANISGGHLNPAVTLGLA 91
>At3g54820 aquaporin/MIP - like protein
Length = 286
Score = 38.1 bits (87), Expect = 0.001
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 9/71 (12%)
Query: 36 LVAEVLGTYFLIFAGCASVV-------VNLNNDKV--VTLPGIAIVWGLAVMVLVYSIGH 86
L+AE + T ++ +V+ LN D+ V + GIA +G + +LVY
Sbjct: 40 LIAEFIATLLFLYVTIMTVIGYKSQTDPALNPDQCTGVGVLGIAWAFGGMIFILVYCTAG 99
Query: 87 ISGAHFNPAVT 97
ISG H NPAVT
Sbjct: 100 ISGGHINPAVT 110
>At3g61430 plasma membrane intrinsic protein 1a
Length = 286
Score = 37.0 bits (84), Expect = 0.003
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 3/64 (4%)
Query: 37 VAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHFN 93
+AE + T+ ++ +V+ + + V + GIA +G + LVY ISG H N
Sbjct: 55 IAEFIATFLFLYITVLTVMGVKRSPNMCASVGIQGIAWAFGGMIFALVYCTAGISGGHIN 114
Query: 94 PAVT 97
PAVT
Sbjct: 115 PAVT 118
>At2g45960 aquaporin (plasma membrane intrinsic protein 1B)
Length = 286
Score = 37.0 bits (84), Expect = 0.003
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 3/64 (4%)
Query: 37 VAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHFN 93
+AE + T+ ++ +V+ + + V + GIA +G + LVY ISG H N
Sbjct: 55 IAEFIATFLFLYITVLTVMGVKRSPNMCASVGIQGIAWAFGGMIFALVYCTAGISGGHIN 114
Query: 94 PAVT 97
PAVT
Sbjct: 115 PAVT 118
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,846,728
Number of Sequences: 26719
Number of extensions: 102170
Number of successful extensions: 285
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 234
Number of HSP's gapped (non-prelim): 58
length of query: 131
length of database: 11,318,596
effective HSP length: 88
effective length of query: 43
effective length of database: 8,967,324
effective search space: 385594932
effective search space used: 385594932
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0201.11


