

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.1
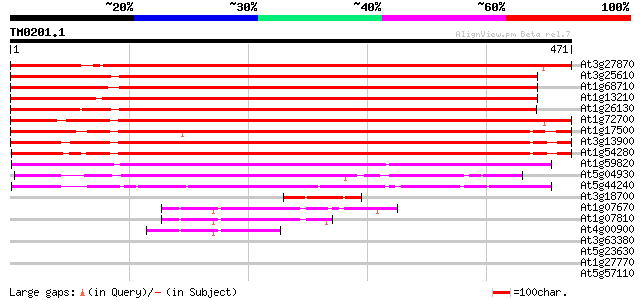
(471 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g27870 P-type ATPase, putative 687 0.0
At3g25610 ATPase II, putative 608 e-174
At1g68710 ATPase, putative 601 e-172
At1g13210 calcium-transporting ATPase like protein 600 e-172
At1g26130 P-type transporting ATPase like protein 576 e-165
At1g72700 putative P-type transporting ATPase 529 e-150
At1g17500 P-type ATPase, putative 520 e-148
At3g13900 putative ATPase 512 e-145
At1g54280 hypothetical protein 503 e-143
At1g59820 hypothetical protein 358 4e-99
At5g04930 ATPase 271 4e-73
At5g44240 ATPase, calcium-transporting 223 2e-58
At3g18700 hypothetical protein 62 7e-10
At1g07670 endoplasmic reticulum-type calcium-transporting ATPase... 48 1e-05
At1g07810 ER-type Ca2+-pump protein 47 2e-05
At4g00900 Ca2+-transporting ATPase - like protein 43 3e-04
At3g63380 Ca2+-transporting ATPase -like protein 40 0.002
At5g23630 cation-transporting ATPase 40 0.003
At1g27770 envelope Ca2+-ATPase 39 0.005
At5g57110 Ca2+-transporting ATPase-like protein (emb|CAB79748.1) 39 0.008
>At3g27870 P-type ATPase, putative
Length = 1174
Score = 687 bits (1773), Expect = 0.0
Identities = 341/476 (71%), Positives = 393/476 (81%), Gaps = 16/476 (3%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KL+QAG+K+WVLTGDK ETA+NIGYACSLLR+ MK+I++TLDS DI +LEKQGDK+A+ K
Sbjct: 707 KLSQAGVKIWVLTGDKTETAINIGYACSLLREGMKQILVTLDSSDIEALEKQGDKEAVAK 766
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSA-FGLIIDGKSLDYSLNKNLEKSFFELAV 119
+ EG+SQ +A + N+ KE S FGL+IDGKSL Y+L+ LEK F ELA+
Sbjct: 767 ---------LREGMSQ--TAAVTDNSAKENSEMFGLVIDGKSLTYALDSKLEKEFLELAI 815
Query: 120 SCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQA 179
C SVICCRSSPKQKA VTRLVK GTG+T L+IGDGANDVGMLQEA IGVGISGAEGMQA
Sbjct: 816 RCNSVICCRSSPKQKALVTRLVKNGTGRTTLAIGDGANDVGMLQEADIGVGISGAEGMQA 875
Query: 180 VMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQP 239
VMASDFAIAQFRFLERLLLVHGHWCYRRI+LMICYFFYKN+AFGFTLFW+EAYASFSG+P
Sbjct: 876 VMASDFAIAQFRFLERLLLVHGHWCYRRITLMICYFFYKNLAFGFTLFWYEAYASFSGKP 935
Query: 240 AYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWML 299
AYNDWYMS YNVFFTSLPVIALGVFDQDVSA+LCLKYP LY EGV++ LFSW RI+GWML
Sbjct: 936 AYNDWYMSCYNVFFTSLPVIALGVFDQDVSARLCLKYPLLYQEGVQNVLFSWERILGWML 995
Query: 300 NGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWI 359
NGVISS+ IFFLT N++ QAFR+DGQVVD+ +LGVTMYS VVWTVNCQMA+SINYFTWI
Sbjct: 996 NGVISSMIIFFLTINTMATQAFRKDGQVVDYSVLGVTMYSSVVWTVNCQMAISINYFTWI 1055
Query: 360 QHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYF 419
QH FIWGSI WY+FL++YG L P S+TA+ VFVE APS +YWL LVV LLPYF
Sbjct: 1056 QHCFIWGSIGVWYLFLVIYGSLPPTFSTTAFQVFVETSAPSPIYWLVLFLVVFSALLPYF 1115
Query: 420 TYRAFQSRFLPMYHDII-QRKRVEGFEV---EISDELPTQVQGKLMHMRERLKQRE 471
TYRAFQ +F PMYHDII +++R E E + ELP QV+ L H+R L +R+
Sbjct: 1116 TYRAFQIKFRPMYHDIIVEQRRTERTETAPNAVLGELPVQVEFTLHHLRANLSRRD 1171
>At3g25610 ATPase II, putative
Length = 1202
Score = 608 bits (1568), Expect = e-174
Identities = 296/443 (66%), Positives = 352/443 (78%), Gaps = 6/443 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+A SLLRQ+MK+I+I L++P I SLEK G KD +
Sbjct: 718 KLAQAGIKIWVLTGDKMETAINIGFASSLLRQEMKQIIINLETPQIKSLEKSGGKDEIEL 777
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
AS ES+ Q+ EG + + ++ SS AF LIIDGKSL Y+L ++K F +LA S
Sbjct: 778 ASRESVVMQLQEGKALLAASGASSE------AFALIIDGKSLTYALEDEIKKMFLDLATS 831
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK GTGKT L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 832 CASVICCRSSPKQKALVTRLVKSGTGKTTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 891
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
M+SD AIAQFR+LERLLLVHGHWCY RI+ MICYFFYKNI FG T+F +EAY SFSGQPA
Sbjct: 892 MSSDIAIAQFRYLERLLLVHGHWCYSRIASMICYFFYKNITFGVTVFLYEAYTSFSGQPA 951
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDW++S +NVFF+SLPVIALGVFDQDVSA+ C K+P LY EGV++ LFSW RIIGWM N
Sbjct: 952 YNDWFLSLFNVFFSSLPVIALGVFDQDVSARFCYKFPLLYQEGVQNILFSWKRIIGWMFN 1011
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
G IS+LAIFFL S+ +Q F DG+ EILG TMY+ VVW VN QMALSI+YFTW+Q
Sbjct: 1012 GFISALAIFFLCKESLKHQLFDPDGKTAGREILGGTMYTCVVWVVNLQMALSISYFTWVQ 1071
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGSI FWY+FL++YG ++P+ S+ AYMVF+EA AP+ YWL T+ V++ L+PYF
Sbjct: 1072 HIVIWGSIAFWYIFLMIYGAMTPSFSTDAYMVFLEALAPAPSYWLTTLFVMIFALIPYFV 1131
Query: 421 YRAFQSRFLPMYHDIIQRKRVEG 443
Y++ Q RF P YH +IQ R EG
Sbjct: 1132 YKSVQMRFFPKYHQMIQWIRYEG 1154
>At1g68710 ATPase, putative
Length = 1200
Score = 601 bits (1549), Expect = e-172
Identities = 298/443 (67%), Positives = 347/443 (78%), Gaps = 8/443 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+ACSLLRQDMK+I+I L++P+I SLEK G+KD + K
Sbjct: 725 KLAQAGIKIWVLTGDKMETAINIGFACSLLRQDMKQIIINLETPEIQSLEKTGEKDVIAK 784
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
AS E++ QI G +Q+K + + AF LIIDGKSL Y+L+ +++ F ELAVS
Sbjct: 785 ASKENVLSQIINGKTQLKYSGGN--------AFALIIDGKSLAYALDDDIKHIFLELAVS 836
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK G GKT L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 837 CASVICCRSSPKQKALVTRLVKSGNGKTTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 896
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
M+SD AIAQFR+LERLLLVHGHWCYRRIS MICYFFYKNI FGFTLF +E Y +FS PA
Sbjct: 897 MSSDIAIAQFRYLERLLLVHGHWCYRRISTMICYFFYKNITFGFTLFLYETYTTFSSTPA 956
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDW++S YNVFF+SLPVIALGVFDQDVSA+ CLK+P LY EGV++ LFSW RI+GWM N
Sbjct: 957 YNDWFLSLYNVFFSSLPVIALGVFDQDVSARYCLKFPLLYQEGVQNVLFSWRRILGWMFN 1016
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
G S++ IFFL +S+ +QAF DG+ EILG TMY+ +VW VN QMAL+I+YFT IQ
Sbjct: 1017 GFYSAVIIFFLCKSSLQSQAFNHDGKTPGREILGGTMYTCIVWVVNLQMALAISYFTLIQ 1076
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IW SI WY F+ VYG L IS+ AY VFVEA APS YWL T+ VVV L+PYF
Sbjct: 1077 HIVIWSSIVVWYFFITVYGELPSRISTGAYKVFVEALAPSLSYWLITLFVVVATLMPYFI 1136
Query: 421 YRAFQSRFLPMYHDIIQRKRVEG 443
Y A Q F PMYH +IQ R EG
Sbjct: 1137 YSALQMSFFPMYHGMIQWLRYEG 1159
>At1g13210 calcium-transporting ATPase like protein
Length = 1203
Score = 600 bits (1547), Expect = e-172
Identities = 288/443 (65%), Positives = 351/443 (79%), Gaps = 4/443 (0%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+ACSLLRQ+MK+I+I L++P I +LEK G+KDA+
Sbjct: 717 KLAQAGIKIWVLTGDKMETAINIGFACSLLRQEMKQIIINLETPHIKALEKAGEKDAIEH 776
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
AS ES+ Q+ EG K+ +S++ AF LIIDGKSL Y+L + +K F +LA
Sbjct: 777 ASRESVVNQMEEG----KALLTASSSASSHEAFALIIDGKSLTYALEDDFKKKFLDLATG 832
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK GTGKT L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 833 CASVICCRSSPKQKALVTRLVKSGTGKTTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 892
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
M+SD AIAQFR+LERLLLVHGHWCY RIS MICYFFYKNI FG T+F +EAY SFS QPA
Sbjct: 893 MSSDIAIAQFRYLERLLLVHGHWCYSRISSMICYFFYKNITFGVTVFLYEAYTSFSAQPA 952
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDW++S +NVFF+SLPVIALGVFDQDVSA+ C K+P LY EGV++ LFSW RIIGWM N
Sbjct: 953 YNDWFLSLFNVFFSSLPVIALGVFDQDVSARYCYKFPLLYQEGVQNLLFSWKRIIGWMFN 1012
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GV ++LAIFFL S+ +Q + +G+ EILG TMY+ VVW VN QMAL+I+YFTW+Q
Sbjct: 1013 GVFTALAIFFLCKESLKHQLYNPNGKTAGREILGGTMYTCVVWVVNLQMALAISYFTWLQ 1072
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGS+ FWY+FL++YG ++P+ S+ AY VF+EA AP+ YWL T+ V+ L+P+F
Sbjct: 1073 HIVIWGSVAFWYIFLMIYGAITPSFSTDAYKVFIEALAPAPSYWLTTLFVMFFALIPFFV 1132
Query: 421 YRAFQSRFLPMYHDIIQRKRVEG 443
+++ Q RF P YH +IQ R EG
Sbjct: 1133 FKSVQMRFFPGYHQMIQWIRYEG 1155
>At1g26130 P-type transporting ATPase like protein
Length = 1184
Score = 576 bits (1485), Expect = e-165
Identities = 280/442 (63%), Positives = 346/442 (77%), Gaps = 7/442 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+ACSLLR+DMK+I+I L++P+I LEK G+KDA+
Sbjct: 720 KLAQAGIKIWVLTGDKMETAINIGFACSLLRRDMKQIIINLETPEIQQLEKSGEKDAIA- 778
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
A E++ QI+ G +Q+K++ ++ AF LIIDGKSL Y+L ++++ F ELA+
Sbjct: 779 ALKENVLHQITSGKAQLKASGGNAK------AFALIIDGKSLAYALEEDMKGIFLELAIG 832
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK G+G+T L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 833 CASVICCRSSPKQKALVTRLVKTGSGQTTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 892
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
M+SD AIAQFR+LERLLLVHGHWCYRRIS MICYFFYKNI FGFTLF +EAY SFS PA
Sbjct: 893 MSSDIAIAQFRYLERLLLVHGHWCYRRISKMICYFFYKNITFGFTLFLYEAYTSFSATPA 952
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDWY+S Y+VFFTSLPVI LG+FDQDVSA CLK+P LY EGV++ LFSW RI+ WM +
Sbjct: 953 YNDWYLSLYSVFFTSLPVICLGIFDQDVSAPFCLKFPVLYQEGVQNLLFSWRRILSWMFH 1012
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
G S++ IFFL S+ +QAF +G+ +ILG TMY+ VVW V+ QM L+I+YFT IQ
Sbjct: 1013 GFCSAIIIFFLCKTSLESQAFNHEGKTAGRDILGGTMYTCVVWVVSLQMVLTISYFTLIQ 1072
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H +WGS+ WY+FL+VYG L +S+ AYMVF+EA AP+ YW+ T+ VV+ ++PYF
Sbjct: 1073 HVVVWGSVVIWYLFLMVYGSLPIRMSTDAYMVFLEALAPAPSYWITTLFVVLSTMMPYFI 1132
Query: 421 YRAFQSRFLPMYHDIIQRKRVE 442
+ A Q RF PM H +Q R E
Sbjct: 1133 FSAIQMRFFPMSHGTVQLLRYE 1154
>At1g72700 putative P-type transporting ATPase
Length = 1228
Score = 529 bits (1363), Expect = e-150
Identities = 260/486 (53%), Positives = 338/486 (69%), Gaps = 28/486 (5%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAG+KLWVLTGDKMETA+NIG+ACSLLRQ M++I IT S+ +G +
Sbjct: 739 KLAQAGLKLWVLTGDKMETAINIGFACSLLRQGMRQICIT-------SMNSEGGSQDSKR 791
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
E+I Q+++ + VK K+ +AF LIIDGK+L Y+L +++ F LAV
Sbjct: 792 VVKENILNQLTKAVQMVKLEKDPH------AAFALIIDGKTLTYALEDDMKYQFLALAVD 845
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCR SPKQKA V RLVK GTGKT L+IGDGANDVGM+QEA IGVGISG EGMQAV
Sbjct: 846 CASVICCRVSPKQKALVVRLVKEGTGKTTLAIGDGANDVGMIQEADIGVGISGVEGMQAV 905
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
MASDF+IAQFRFLERLL+VHGHWCY+RI+ MICYFFYKNIAFG TLF+FEA+ FSGQ
Sbjct: 906 MASDFSIAQFRFLERLLVVHGHWCYKRIAQMICYFFYKNIAFGLTLFYFEAFTGFSGQSV 965
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YND+Y+ +NV TSLPVIALGVF+QDVS+++CL++P LY +G ++ F W RI+GWM N
Sbjct: 966 YNDYYLLLFNVVLTSLPVIALGVFEQDVSSEICLQFPALYQQGTKNLFFDWSRILGWMCN 1025
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GV +SL IFFL I +QAFR +GQ D + +G TM++ ++W N Q+AL++++FTWIQ
Sbjct: 1026 GVYASLVIFFLNIGIIYSQAFRDNGQTADMDAVGTTMFTCIIWAANVQIALTMSHFTWIQ 1085
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGSI WY+F+ +Y + P+ S Y + E AP+ +YW+AT+LV V +LPY
Sbjct: 1086 HVLIWGSIGMWYLFVAIYSMMPPSYSGNIYRILDEILAPAPIYWMATLLVTVAAVLPYVA 1145
Query: 421 YRAFQSRFLPMYHDIIQRKRVEGFEVE---------------ISDELPTQVQGKLMHMRE 465
+ AFQ P+ H IIQ + G ++E +V K+ H+R
Sbjct: 1146 HIAFQRFLNPLDHHIIQEIKYYGRDIEDARLWTRERTKAREKTKIGFTARVDAKIRHLRS 1205
Query: 466 RLKQRE 471
+L +++
Sbjct: 1206 KLNKKQ 1211
>At1g17500 P-type ATPase, putative
Length = 1218
Score = 520 bits (1339), Expect = e-148
Identities = 263/473 (55%), Positives = 335/473 (70%), Gaps = 26/473 (5%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAG+KLWVLTGDKMETA+NIGY+CSLLRQ MK+I IT+ + + S + + KD
Sbjct: 728 KLAQAGLKLWVLTGDKMETAINIGYSCSLLRQGMKQICITVVNSEGASQDAKAVKD---- 783
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
+I QI++ + VK K+ +AF LIIDGK+L Y+L ++ F LAV
Sbjct: 784 ----NILNQITKAVQMVKLEKDPH------AAFALIIDGKTLTYALEDEMKYQFLALAVD 833
Query: 121 CASVICCRSSPKQKARVTRLVKL--GTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQ 178
CASVICCR SPKQKA V L GTGK L+IGDGANDVGM+QEA IGVGISG EGMQ
Sbjct: 834 CASVICCRVSPKQKALVFPLFPYAHGTGKITLAIGDGANDVGMIQEADIGVGISGVEGMQ 893
Query: 179 AVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQ 238
AVMASDF+IAQFRFLERLL+VHGHWCY+RI+ MICYFFYKNIAFG TLF+FEA+ FSGQ
Sbjct: 894 AVMASDFSIAQFRFLERLLVVHGHWCYKRIAQMICYFFYKNIAFGLTLFYFEAFTGFSGQ 953
Query: 239 PAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWM 298
YND+Y+ +NV TSLPVIALGVF+QDVS+++CL++P LY +G ++ F W RI+GWM
Sbjct: 954 SVYNDYYLLLFNVVLTSLPVIALGVFEQDVSSEICLQFPALYQQGKKNLFFDWYRILGWM 1013
Query: 299 LNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTW 358
NGV SSL IFFL I QAFR GQ D + +G TM++ ++W VN Q+AL++++FTW
Sbjct: 1014 GNGVYSSLVIFFLNIGIIYEQAFRVSGQTADMDAVGTTMFTCIIWAVNVQIALTVSHFTW 1073
Query: 359 IQHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPY 418
IQH IWGSI WY+F+ +YG + P++S Y + VE AP+ +YW+AT LV V +LPY
Sbjct: 1074 IQHVLIWGSIGLWYLFVALYGMMPPSLSGNIYRILVEILAPAPIYWIATFLVTVTTVLPY 1133
Query: 419 FTYRAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQGKLMHMRERLKQRE 471
F + +FQ P+ H IIQ ++ ++ ++ D + M RER K RE
Sbjct: 1134 FAHISFQRFLHPLDHHIIQ--EIKYYKRDVED--------RRMWTRERTKARE 1176
>At3g13900 putative ATPase
Length = 1243
Score = 512 bits (1318), Expect = e-145
Identities = 257/471 (54%), Positives = 326/471 (68%), Gaps = 23/471 (4%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAG+K+WVLTGDKMETA+NIGYACSLLRQ MK+I I L + ++G
Sbjct: 746 KLAQAGLKIWVLTGDKMETAINIGYACSLLRQGMKQIYIALRN-------EEGSSQDPEA 798
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
A+ E+I QI +K K+ +AF LIIDGK+L Y+L +++ F LAV
Sbjct: 799 AARENILMQIINASQMIKLEKDPH------AAFALIIDGKTLTYALEDDIKYQFLALAVD 852
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCR SPKQKA VTRL K GTGKT L+IGDGANDVGM+QEA IGVGISG EGMQAV
Sbjct: 853 CASVICCRVSPKQKALVTRLAKEGTGKTTLAIGDGANDVGMIQEADIGVGISGVEGMQAV 912
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
MASDF+IAQFRFLERLL+VHGHWCY+RI+ MICYFFYKNI FG TLF+FEA+ FSGQ
Sbjct: 913 MASDFSIAQFRFLERLLVVHGHWCYKRIAQMICYFFYKNITFGLTLFYFEAFTGFSGQAI 972
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YND Y+ +NV TSLPVIALGVF+QDVS+++CL++P LY +G ++ F W RIIGWM N
Sbjct: 973 YNDSYLLLFNVILTSLPVIALGVFEQDVSSEVCLQFPALYQQGPKNLFFDWYRIIGWMAN 1032
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GV +S+ IF L Q+F GQ D + +G M++ ++W VN Q+AL++++FTWIQ
Sbjct: 1033 GVYASVVIFSLNIGIFHVQSFCSGGQTADMDAMGTAMFTCIIWAVNVQIALTMSHFTWIQ 1092
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGSI WY+FL ++G L P +S + + E AP+ ++WL ++LV+ LPY
Sbjct: 1093 HVLIWGSIVTWYIFLALFGMLPPKVSGNIFHMLSETLAPAPIFWLTSLLVIAATTLPYLA 1152
Query: 421 YRAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQGKLMHMRERLKQRE 471
Y +FQ P+ H IIQ ++ F +++ DE M RER K RE
Sbjct: 1153 YISFQRSLNPLDHHIIQ--EIKHFRIDVQDE--------CMWTRERSKARE 1193
>At1g54280 hypothetical protein
Length = 1240
Score = 503 bits (1295), Expect = e-143
Identities = 255/470 (54%), Positives = 326/470 (69%), Gaps = 23/470 (4%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
LAQAG+K+WVLTGDKMETA+NIGYACSLLRQ MK+I I+L + + E + +A K
Sbjct: 749 LAQAGLKIWVLTGDKMETAINIGYACSLLRQGMKQISISLTNVE----ESSQNSEAAAK- 803
Query: 62 SLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSC 121
ESI QI+ +K K+ +AF LIIDGK+L Y+L +++ F LAV C
Sbjct: 804 --ESILMQITNASQMIKIEKDPH------AAFALIIDGKTLTYALKDDVKYQFLALAVDC 855
Query: 122 ASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVM 181
ASVICCR SPKQKA VTRL K GTGKT L+IGDGANDVGM+QEA IGVGISG EGMQAVM
Sbjct: 856 ASVICCRVSPKQKALVTRLAKEGTGKTTLAIGDGANDVGMIQEADIGVGISGVEGMQAVM 915
Query: 182 ASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAY 241
ASDF+IAQFRFLERLL+VHGHWCY+RI+ MICYFFYKNI FG TLF+FE + FSGQ Y
Sbjct: 916 ASDFSIAQFRFLERLLVVHGHWCYKRIAQMICYFFYKNITFGLTLFYFECFTGFSGQSIY 975
Query: 242 NDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNG 301
ND Y+ +NV TSLPVI+LGVF+QDV + +CL++P LY +G ++ F W RI+GWM NG
Sbjct: 976 NDSYLLLFNVVLTSLPVISLGVFEQDVPSDVCLQFPALYQQGPKNLFFDWYRILGWMGNG 1035
Query: 302 VISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQH 361
V +S+ IF L Q+FR DGQ D +G M++ ++W VN Q+AL++++FTWIQH
Sbjct: 1036 VYASIVIFTLNLGIFHVQSFRSDGQTADMNAMGTAMFTCIIWAVNVQIALTMSHFTWIQH 1095
Query: 362 FFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFTY 421
IWGSI WYVFL +YG L +S + + VE AP+ ++WL ++LV+ LPY +
Sbjct: 1096 VMIWGSIGAWYVFLALYGMLPVKLSGNIFHMLVEILAPAPIFWLTSLLVIAATTLPYLFH 1155
Query: 422 RAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQGKLMHMRERLKQRE 471
++Q P+ H IIQ ++ F +++ DE M RE+ K RE
Sbjct: 1156 ISYQRSVNPLDHHIIQ--EIKHFRIDVEDE--------RMWKREKSKARE 1195
>At1g59820 hypothetical protein
Length = 1213
Score = 358 bits (919), Expect = 4e-99
Identities = 193/456 (42%), Positives = 275/456 (59%), Gaps = 8/456 (1%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
L++AGIK+WVLTGDKMETA+NI YAC+L+ +MK+ VI+ ++ I E++GD+ + +
Sbjct: 697 LSRAGIKIWVLTGDKMETAINIAYACNLINNEMKQFVISSETDAIREAEERGDQVEIARV 756
Query: 62 SLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSC 121
E +K+++ + + + + + + K L+IDGK L Y+L+ +L L+++C
Sbjct: 757 IKEEVKRELKKSLEEAQHSLHTVAGPK----LSLVIDGKCLMYALDPSLRVMLLSLSLNC 812
Query: 122 ASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVM 181
SV+CCR SP QKA+VT LV+ G K LSIGDGANDV M+Q AH+G+GISG EGMQAVM
Sbjct: 813 TSVVCCRVSPLQKAQVTSLVRKGAQKITLSIGDGANDVSMIQAAHVGIGISGMEGMQAVM 872
Query: 182 ASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAY 241
ASDFAIAQFRFL LLLVHG W Y RI ++ YFFYKN+ F T FWF FSGQ Y
Sbjct: 873 ASDFAIAQFRFLTDLLLVHGRWSYLRICKVVMYFFYKNLTFTLTQFWFTFRTGFSGQRFY 932
Query: 242 NDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNG 301
+DW+ S +NV FT+LPVI LG+F++DVSA L +YP LY EG+ ++ F W + W +
Sbjct: 933 DDWFQSLFNVVFTALPVIVLGLFEKDVSASLSKRYPELYREGIRNSFFKWRVVAVWATSA 992
Query: 302 VISSLAIF-FLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
V SL + F+TT+S A G+V + +++ +V VN ++ L N T
Sbjct: 993 VYQSLVCYLFVTTSSF--GAVNSSGKVFGLWDVSTMVFTCLVIAVNVRILLMSNSITRWH 1050
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCV-LLPYF 419
+ + GSI W VF VY + + FV S Y+ T+L+V V LL F
Sbjct: 1051 YITVGGSILAWLVFAFVYCGIMTPHDRNENVYFVIYVLMSTFYFYFTLLLVPIVSLLGDF 1110
Query: 420 TYRAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQ 455
++ + F P + I+Q + +D+L +
Sbjct: 1111 IFQGVERWFFPYDYQIVQEIHRHESDASKADQLEVE 1146
>At5g04930 ATPase
Length = 1158
Score = 271 bits (694), Expect = 4e-73
Identities = 161/431 (37%), Positives = 232/431 (53%), Gaps = 47/431 (10%)
Query: 5 AGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKASLE 64
AGIK+WVLTGDK ETA++IG++ LL ++M++IVI +S L+
Sbjct: 736 AGIKVWVLTGDKQETAISIGFSSRLLTRNMRQIVINSNS-------------------LD 776
Query: 65 SIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSCASV 124
S ++ + E + + S ES N LIIDG SL Y L+ +LE F++A C+++
Sbjct: 777 SCRRSLEEANASIASNDESDNV-------ALIIDGTSLIYVLDNDLEDVLFQVACKCSAI 829
Query: 125 ICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVMASD 184
+CCR +P QKA + LVK T L+IGDGANDV M+Q A +GVGISG EG QAVMASD
Sbjct: 830 LCCRVAPFQKAGIVALVKNRTSDMTLAIGDGANDVSMIQMADVGVGISGQEGRQAVMASD 889
Query: 185 FAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAYNDW 244
FA+ QFRFL LLLVHGHW Y+R+ MI Y FY+N F LFW+ + ++ A +W
Sbjct: 890 FAMGQFRFLVPLLLVHGHWNYQRMGYMILYNFYRNAVFVLILFWYVLFTCYTLTTAITEW 949
Query: 245 YMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLY-----LEGVEDTLFSWPRIIGWML 299
Y+V +T++P I +G+ D+D+ + L +P LY EG TLF + M+
Sbjct: 950 SSVLYSVIYTAIPTIIIGILDKDLGRQTLLDHPQLYGVGQRAEGYSTTLFWYT-----MI 1004
Query: 300 NGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWI 359
+ + S AIFF+ F G +D LG V VN +A+ + + WI
Sbjct: 1005 DTIWQSAAIFFI-------PMFAYWGSTIDTSSLGDLWTIAAVVVVNLHLAMDVIRWNWI 1057
Query: 360 QHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYF 419
H IWGSI + ++V + + Y + + ++W + +VV LLP F
Sbjct: 1058 THAAIWGSIVAACICVIVIDVIP---TLPGYWAIFQV-GKTWMFWFCLLAIVVTSLLPRF 1113
Query: 420 TYRAFQSRFLP 430
+ + P
Sbjct: 1114 AIKFLVEYYRP 1124
>At5g44240 ATPase, calcium-transporting
Length = 1078
Score = 223 bits (568), Expect = 2e-58
Identities = 139/454 (30%), Positives = 234/454 (50%), Gaps = 37/454 (8%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
L +AGI W+LTGDK TA+ I +C+ + + K ++ +D
Sbjct: 596 LRKAGINFWMLTGDKQNTAIQIALSCNFISPEPKGQLLMIDG------------------ 637
Query: 62 SLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSC 121
++ +S + +V + ++ + AF +IDG +L+ +L K+ K F ELA+
Sbjct: 638 ---KTEEDVSRSLERVLLTMRITASEPKDVAF--VIDGWALEIAL-KHHRKDFVELAILS 691
Query: 122 ASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVM 181
+ ICCR +P QKA++ ++K +T L+IGDG NDV M+Q+A IGVGISG EG+QA
Sbjct: 692 RTAICCRVTPSQKAQLVEILKSCDYRT-LAIGDGGNDVRMIQQADIGVGISGREGLQAAR 750
Query: 182 ASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAY 241
A+D++I +FRFL+RL+LVHG + Y R + + Y FYK++ F +F + SG +
Sbjct: 751 AADYSIGRFRFLKRLILVHGRYSYNRTAFLSQYSFYKSLLICFIQIFFSFISGVSGTSLF 810
Query: 242 NDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNG 301
N + YNVF+TS+PV+ + V D+D+S +++P + L + GW
Sbjct: 811 NSVSLMAYNVFYTSVPVL-VSVIDKDLSEASVMQHPQILFYCQAGRLLNPSTFAGWFGRS 869
Query: 302 VISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQH 361
+ ++ +F +T ++ A+ + + E LG+ S +W +A N FT +QH
Sbjct: 870 LFHAIIVFVITIHA---YAYEKS----EMEELGMVALSGCIWLQAFVVAQETNSFTVLQH 922
Query: 362 FFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFTY 421
IWG++ +Y ++ S SS Y + C+ + YW+ L+V + P F
Sbjct: 923 LSIWGNLVGFYAINFLF---SAIPSSGMYTIMFRLCSQPS-YWITMFLIVGAGMGPIFAL 978
Query: 422 RAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQ 455
+ F+ + P +I+Q+ G + + TQ
Sbjct: 979 KYFRYTYRPSKINILQQAERMGGPILTLGNIETQ 1012
>At3g18700 hypothetical protein
Length = 67
Score = 62.0 bits (149), Expect = 7e-10
Identities = 33/65 (50%), Positives = 45/65 (68%), Gaps = 2/65 (3%)
Query: 231 AYASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFS 290
+Y SFS QP N+W++ F N+FF+SL VIAL FDQDVS + K+ FLY +GV++ L
Sbjct: 4 SYTSFSAQPTNNNWFLLF-NIFFSSLLVIALRFFDQDVSDRYSYKFMFLY-QGVQNLLCI 61
Query: 291 WPRII 295
RI+
Sbjct: 62 LKRIV 66
>At1g07670 endoplasmic reticulum-type calcium-transporting ATPase 4
(ECA4)
Length = 1061
Score = 48.1 bits (113), Expect = 1e-05
Identities = 61/207 (29%), Positives = 85/207 (40%), Gaps = 22/207 (10%)
Query: 128 RSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGV--GISGAEGMQAVMASDF 185
R+ PK K + RL+K G+ + GDG ND L+ A IGV GISG E A ASD
Sbjct: 706 RAEPKHKQEIVRLLK-EDGEVVAMTGDGVNDAPALKLADIGVAMGISGTE--VAKEASDL 762
Query: 186 AIAQFRFLERLLLV-HGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQ-PAYND 243
+A F + V G Y + I Y NI ++F A G P
Sbjct: 763 VLADDNFSTIVAAVGEGRSIYNNMKAFIRYMISSNIGEVASIFLTAALGIPEGMIPVQLL 822
Query: 244 WYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNGVI 303
W N+ P ALG D K +K P +D+L + + +M+ G+
Sbjct: 823 W----VNLVTDGPPATALGFNPPD---KDIMKKP---PRRSDDSLITAWILFRYMVIGLY 872
Query: 304 SSLA-----IFFLTTNSIINQAFRRDG 325
+A I + T NS + +DG
Sbjct: 873 VGVATVGVFIIWYTHNSFMGIDLSQDG 899
>At1g07810 ER-type Ca2+-pump protein
Length = 1061
Score = 47.4 bits (111), Expect = 2e-05
Identities = 49/150 (32%), Positives = 63/150 (41%), Gaps = 13/150 (8%)
Query: 128 RSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGV--GISGAEGMQAVMASDF 185
R+ PK K + RL+K G+ + GDG ND L+ A IGV GISG E A ASD
Sbjct: 706 RAEPKHKQEIVRLLK-EDGEVVAMTGDGVNDAPALKLADIGVAMGISGTE--VAKEASDM 762
Query: 186 AIAQFRFLERLLLV-HGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQ-PAYND 243
+A F + V G Y + I Y NI ++F A G P
Sbjct: 763 VLADDNFSTIVAAVGEGRSIYNNMKAFIRYMISSNIGEVASIFLTAALGIPEGMIPVQLL 822
Query: 244 WYMSFYNVFFTSLPVIALGVF--DQDVSAK 271
W N+ P ALG D+D+ K
Sbjct: 823 W----VNLVTDGPPATALGFNPPDKDIMKK 848
>At4g00900 Ca2+-transporting ATPase - like protein
Length = 1054
Score = 43.1 bits (100), Expect = 3e-04
Identities = 33/115 (28%), Positives = 51/115 (43%), Gaps = 6/115 (5%)
Query: 116 ELAVSCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGV--GISG 173
E+ + R+ P+ K + R++K G+ + GDG ND L+ A IG+ GI+G
Sbjct: 690 EILSKSGGKVFSRAEPRHKQEIVRMLK-EMGEIVAMTGDGVNDAPALKLADIGIAMGITG 748
Query: 174 AEGMQAVMASDFAIAQFRFLERLLLV-HGHWCYRRISLMICYFFYKNIAFGFTLF 227
E A ASD +A F + V G Y + I Y N+ ++F
Sbjct: 749 TE--VAKEASDMVLADDNFSTIVSAVAEGRSIYNNMKAFIRYMISSNVGEVISIF 801
>At3g63380 Ca2+-transporting ATPase -like protein
Length = 1033
Score = 40.4 bits (93), Expect = 0.002
Identities = 33/100 (33%), Positives = 45/100 (45%), Gaps = 6/100 (6%)
Query: 125 ICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIG--VGISGAEGMQAVMA 182
+ RSSP K + + ++L G + GDG ND L+EA IG +GI G E A +
Sbjct: 723 VMARSSPSDKLLMVKCLRL-KGHVVAVTGDGTNDAPALKEADIGLSMGIQGTE--VAKES 779
Query: 183 SDFAIAQFRFLE-RLLLVHGHWCYRRISLMICYFFYKNIA 221
SD I F +L G Y I I + N+A
Sbjct: 780 SDIVILDDNFASVATVLKWGRCVYNNIQKFIQFQLTVNVA 819
>At5g23630 cation-transporting ATPase
Length = 1179
Score = 40.0 bits (92), Expect = 0.003
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query: 125 ICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGI 171
+ R +P+QK + K G+ L GDG NDVG L++AH+GV +
Sbjct: 784 VFARVAPQQKELILTTFK-AVGRGTLMCGDGTNDVGALKQAHVGVAL 829
>At1g27770 envelope Ca2+-ATPase
Length = 946
Score = 39.3 bits (90), Expect = 0.005
Identities = 22/53 (41%), Positives = 29/53 (54%), Gaps = 2/53 (3%)
Query: 125 ICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIG--VGISGAE 175
+ RSSP K + RL++ + + GDG ND L EA IG +GISG E
Sbjct: 655 VMARSSPMDKHTLVRLLRTMFQEVVAVTGDGTNDAPALHEADIGLAMGISGTE 707
>At5g57110 Ca2+-transporting ATPase-like protein (emb|CAB79748.1)
Length = 1074
Score = 38.5 bits (88), Expect = 0.008
Identities = 48/180 (26%), Positives = 79/180 (43%), Gaps = 20/180 (11%)
Query: 51 KQGDKDALVKASLESIKKQISEGISQVKSAKESS------NTDKETSAFGLIIDGKSLDY 104
+ G KD++V +K ++ G V++A+ + ++D + S +I+GKS
Sbjct: 685 RPGVKDSVVLCQNAGVKVRMVTG-DNVQTARAIALECGILSSDADLSE-PTLIEGKSFRE 742
Query: 105 SLNKNLEKSFFELAVSCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQE 164
+ +K +S + RSSP K + + ++ G + GDG ND L E
Sbjct: 743 MTDAERDK------ISDKISVMGRSSPNDKLLLVQSLRR-QGHVVAVTGDGTNDAPALHE 795
Query: 165 AHIG--VGISGAEGMQAVMASDFAIAQFRFLERLLLVH-GHWCYRRISLMICYFFYKNIA 221
A IG +GI+G E A +SD I F + +V G Y I I + N+A
Sbjct: 796 ADIGLAMGIAGTE--VAKESSDIIILDDNFASVVKVVRWGRSVYANIQKFIQFQLTVNVA 853
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,932,470
Number of Sequences: 26719
Number of extensions: 413548
Number of successful extensions: 1593
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1517
Number of HSP's gapped (non-prelim): 82
length of query: 471
length of database: 11,318,596
effective HSP length: 103
effective length of query: 368
effective length of database: 8,566,539
effective search space: 3152486352
effective search space used: 3152486352
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0201.1


