

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0200.8
(213 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
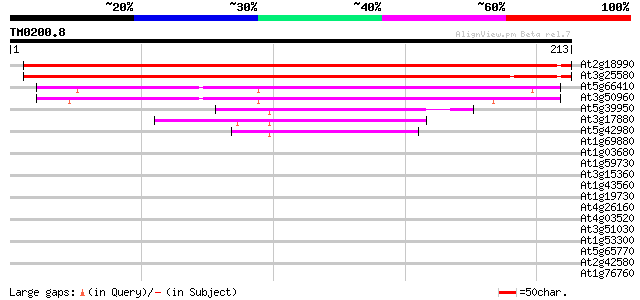
Score E
Sequences producing significant alignments: (bits) Value
At2g18990 putative ATP binding protein 326 6e-90
At3g25580 unknown protein 324 2e-89
At5g66410 unknown protein 130 4e-31
At3g50960 putative protein 128 3e-30
At5g39950 thioredoxin 48 3e-06
At3g17880 thioredoxin, putative 46 2e-05
At5g42980 thioredoxin (clone GIF1) (pir||S58118) 43 1e-04
At1g69880 thioredoxin like protein 39 0.002
At1g03680 putative thioredoxin-m 39 0.003
At1g59730 unknown protein 38 0.005
At3g15360 thioredoxin m4 37 0.006
At1g43560 unknown protein 37 0.008
At1g19730 thioredoxin 37 0.008
At4g26160 thioredoxin like protein 37 0.010
At4g03520 putative M-type thioredoxin 36 0.013
At3g51030 thioredoxin h 36 0.017
At1g53300 unknown protein 34 0.050
At5g65770 nuclear matrix constituent protein 1 (NMCP1)-like 33 0.085
At2g42580 unknown protein 33 0.085
At1g76760 thioredoxin-like protein 33 0.085
>At2g18990 putative ATP binding protein
Length = 211
Score = 326 bits (835), Expect = 6e-90
Identities = 161/208 (77%), Positives = 187/208 (89%), Gaps = 1/208 (0%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
IQE++EKQVLTV +A+EDKIDDEI++L++LD DDLE LRERRL QMKKMAEK+ RWISLG
Sbjct: 5 IQEILEKQVLTVAKAMEDKIDDEIASLEKLDEDDLEVLRERRLKQMKKMAEKKKRWISLG 64
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQHIETRFVKINAEK 125
HG+YSE+ SEKDFFSVVKAS+RVVCHF+RENWPCKV DKH+ +LAKQHIETRFVKI AEK
Sbjct: 65 HGEYSEIHSEKDFFSVVKASERVVCHFYRENWPCKVMDKHMSILAKQHIETRFVKIQAEK 124
Query: 126 SPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGE 185
SPFLAE+LKI VLPTLALIKN KVDDYVVGF+ELGG DDFSTEDLEER+A+AQVI +GE
Sbjct: 125 SPFLAERLKIVVLPTLALIKNTKVDDYVVGFNELGGKDDFSTEDLEERIARAQVIHYDGE 184
Query: 186 SSIHQARSTAQSKRSVRQATRADSSDSE 213
SS + +ST Q +R+VRQ+ R+D SDSE
Sbjct: 185 SSSLKPKSTTQVRRNVRQSARSD-SDSE 211
>At3g25580 unknown protein
Length = 210
Score = 324 bits (831), Expect = 2e-89
Identities = 161/208 (77%), Positives = 188/208 (89%), Gaps = 2/208 (0%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
IQE+IEKQVLTV +A+EDKIDDEI++L++LD DDLE LRERRL QMKKMAEK+ RW+S+G
Sbjct: 5 IQEIIEKQVLTVAKAMEDKIDDEIASLEKLDEDDLEVLRERRLKQMKKMAEKKKRWMSIG 64
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQHIETRFVKINAEK 125
HG+YSE+ SEKDFFSVVK+S+RVVCHF+RENWPCKV DKH+ +LAKQHIETRFVKI AEK
Sbjct: 65 HGEYSEIHSEKDFFSVVKSSERVVCHFYRENWPCKVMDKHMSILAKQHIETRFVKIQAEK 124
Query: 126 SPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGE 185
SPFLAE+LKI VLPTLALIKN KVDDYVVGF+ELGG DDFSTEDLEER+A+AQVI EGE
Sbjct: 125 SPFLAERLKIVVLPTLALIKNTKVDDYVVGFNELGGKDDFSTEDLEERIARAQVIHYEGE 184
Query: 186 SSIHQARSTAQSKRSVRQATRADSSDSE 213
SS+ Q +ST Q +R+VRQ+ R+D SDSE
Sbjct: 185 SSLKQ-KSTTQVRRNVRQSARSD-SDSE 210
>At5g66410 unknown protein
Length = 230
Score = 130 bits (328), Expect = 4e-31
Identities = 73/202 (36%), Positives = 117/202 (57%), Gaps = 4/202 (1%)
Query: 11 EKQVLTVVQAVEDK-IDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHGDY 69
+K+VL +A + +++E+ + +D +LE L R+ +++ EKR + GHG+Y
Sbjct: 27 KKEVLANEKAQGSRPVNEEVDLDELMDDPELEKLHADRIAALRREVEKREAFKRQGHGEY 86
Query: 70 SELPSEKDFFSVVKASDRVVCHFF-RENWPCKVADKHLGVLAKQHIETRFVKINAEKSPF 128
E+ SE DF V S++V+CHF+ +E + CK+ DKHL LA +H++T+F+K++AE +PF
Sbjct: 87 REV-SEGDFLGEVTRSEKVICHFYHKEFYRCKIMDKHLKTLAPRHVDTKFIKMDAENAPF 145
Query: 129 LAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGESSI 188
KL I LP + L D +VGF +LG DDFST LE L K ++ + +
Sbjct: 146 FVTKLAIKTLPCVILFSKGIAMDRLVGFQDLGAKDDFSTTKLENLLVKKGMLSEKRKEED 205
Query: 189 HQARSTAQS-KRSVRQATRADS 209
+ +S +RSVR + DS
Sbjct: 206 EEDYEYQESIRRSVRSSANVDS 227
>At3g50960 putative protein
Length = 225
Score = 128 bits (321), Expect = 3e-30
Identities = 76/215 (35%), Positives = 115/215 (53%), Gaps = 17/215 (7%)
Query: 11 EKQVLTVVQAV-EDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHGDY 69
+K+VL +A + +++E+ + +D +LE L R+ +K+ EKR + GHG+Y
Sbjct: 9 QKEVLANEKAQGSNPVNEEVDLDELMDDPELERLHADRIAALKREVEKRESFKRQGHGEY 68
Query: 70 SELPSEKDFFSVVKASDRVVCHFF-RENWPCKVADKHLGVLAKQHIETRFVKINAEKSPF 128
E+ SE DF V S++V+CHF+ +E + CK+ DKHL LA +H++T+F+K++AE +PF
Sbjct: 69 REV-SEGDFLGEVTRSEKVICHFYHKEFYRCKIMDKHLKTLAPRHVDTKFIKVDAENAPF 127
Query: 129 LAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFL------ 182
KL I LP + L D +VGF +LG DDF+T LE L K FL
Sbjct: 128 FVTKLAIKTLPCVVLFSKGVAMDRLVGFQDLGTKDDFTTNKLENVLLKKVREFLLKECKS 187
Query: 183 --------EGESSIHQARSTAQSKRSVRQATRADS 209
+ E A +RSVR + DS
Sbjct: 188 LTGMLSKKKKEEDDEDAEYQESIRRSVRSSENLDS 222
>At5g39950 thioredoxin
Length = 133
Score = 48.1 bits (113), Expect = 3e-06
Identities = 23/100 (23%), Positives = 57/100 (57%), Gaps = 11/100 (11%)
Query: 79 FSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKIT 136
F+ +K S++++ F +W PC++ + + +A + + FVK++ ++ P +A++ +T
Sbjct: 40 FNEIKESNKLLVVDFSASWCGPCRMIEPAIHAMADKFNDVDFVKLDVDELPDVAKEFNVT 99
Query: 137 VLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAK 176
+PT L+K K + ++G + ++LE++++K
Sbjct: 100 AMPTFVLVKRGKEIERIIGAKK---------DELEKKVSK 130
>At3g17880 thioredoxin, putative
Length = 123
Score = 45.8 bits (107), Expect = 2e-05
Identities = 29/108 (26%), Positives = 52/108 (47%), Gaps = 5/108 (4%)
Query: 56 EKRNRWISLGHGDYSELPSEKDFFSVVKAS---DRVVCHFFRENW--PCKVADKHLGVLA 110
++R +L G+ + S + + KA+ R++ +F W PC+ LA
Sbjct: 2 QEREAQAALNDGEVISIHSTSELEAKTKAAKKASRLLILYFTATWCGPCRYMSPLYSNLA 61
Query: 111 KQHIETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDE 158
QH F+K++ +K+ +A I+ +PT I++ K D VVG D+
Sbjct: 62 TQHSRVVFLKVDIDKANDVAASWNISSVPTFCFIRDGKEVDKVVGADK 109
>At5g42980 thioredoxin (clone GIF1) (pir||S58118)
Length = 118
Score = 42.7 bits (99), Expect = 1e-04
Identities = 20/73 (27%), Positives = 39/73 (53%), Gaps = 2/73 (2%)
Query: 85 SDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLA 142
S +++ F W PC+ LAK+H++ F K++ ++ +AE+ K+ +PT
Sbjct: 26 SKKLIVIDFTATWCPPCRFIAPVFADLAKKHLDVVFFKVDVDELNTVAEEFKVQAMPTFI 85
Query: 143 LIKNAKVDDYVVG 155
+K ++ + VVG
Sbjct: 86 FMKEGEIKETVVG 98
>At1g69880 thioredoxin like protein
Length = 148
Score = 38.9 bits (89), Expect = 0.002
Identities = 24/88 (27%), Positives = 44/88 (49%), Gaps = 4/88 (4%)
Query: 76 KDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKL 133
K + +K +++++ F W PCK + L LA ++ + FVKI+ + + +
Sbjct: 49 KSRLNALKDTNKLLVIEFTAKWCGPCKTLEPKLEELAAKYTDVEFVKIDVDVLMSVWMEF 108
Query: 134 KITVLPTLALIKNAKVDDYVVG--FDEL 159
++ LP + +K + D VVG DEL
Sbjct: 109 NLSTLPAIVFMKRGREVDMVVGVKVDEL 136
>At1g03680 putative thioredoxin-m
Length = 179
Score = 38.5 bits (88), Expect = 0.003
Identities = 22/78 (28%), Positives = 43/78 (54%), Gaps = 4/78 (5%)
Query: 81 VVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHI-ETRFVKINAEKSPFLAEKLKITV 137
V+KA + V F+ W PCK+ D + LA+++ + +F K+N ++SP + +
Sbjct: 88 VLKADEPVFVDFWAP-WCGPCKMIDPIVNELAQKYAGQFKFYKLNTDESPATPGQYGVRS 146
Query: 138 LPTLALIKNAKVDDYVVG 155
+PT+ + N + D ++G
Sbjct: 147 IPTIMIFVNGEKKDTIIG 164
>At1g59730 unknown protein
Length = 129
Score = 37.7 bits (86), Expect = 0.005
Identities = 20/82 (24%), Positives = 39/82 (47%), Gaps = 2/82 (2%)
Query: 76 KDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKL 133
K F +K S++++ F W PCK + + +A ++ E F +++ ++ +A
Sbjct: 33 KSLFDSMKGSNKLLVIDFTAVWCGPCKAMEPRVREIASKYSEAVFARVDVDRLMDVAGTY 92
Query: 134 KITVLPTLALIKNAKVDDYVVG 155
+ LP +K + D VVG
Sbjct: 93 RAITLPAFVFVKRGEEIDRVVG 114
>At3g15360 thioredoxin m4
Length = 193
Score = 37.4 bits (85), Expect = 0.006
Identities = 25/85 (29%), Positives = 43/85 (50%), Gaps = 3/85 (3%)
Query: 74 SEKDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHI-ETRFVKINAEKSPFLA 130
S+ ++ + V SD V F W PC++ + LAK + +F KIN ++SP A
Sbjct: 92 SDSEWQTKVLESDVPVLVEFWAPWCGPCRMIHPIVDQLAKDFAGKFKFYKINTDESPNTA 151
Query: 131 EKLKITVLPTLALIKNAKVDDYVVG 155
+ I +PT+ + K + D ++G
Sbjct: 152 NRYGIRSVPTVIIFKGGEKKDSIIG 176
>At1g43560 unknown protein
Length = 167
Score = 37.0 bits (84), Expect = 0.008
Identities = 27/81 (33%), Positives = 41/81 (50%), Gaps = 3/81 (3%)
Query: 78 FFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIET-RFVKINAEKSPFLAEKLK 134
F +++ SD+ V F W PC++ L +++ + VKI+ EK P LA K +
Sbjct: 68 FDDLLQNSDKPVLVDFYATWCGPCQLMVPILNEVSETLKDIIAVVKIDTEKYPSLANKYQ 127
Query: 135 ITVLPTLALIKNAKVDDYVVG 155
I LPT L K+ K+ D G
Sbjct: 128 IEALPTFILFKDGKLWDRFEG 148
>At1g19730 thioredoxin
Length = 119
Score = 37.0 bits (84), Expect = 0.008
Identities = 24/97 (24%), Positives = 50/97 (50%), Gaps = 12/97 (12%)
Query: 83 KASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETR-FVKINAEKSPFLAEKLKITVLP 139
K S++++ F +W PC++ LAK+ + + F K++ ++ +A++ + +P
Sbjct: 25 KESNKLIVIDFTASWCPPCRMIAPIFNDLAKKFMSSAIFFKVDVDELQSVAKEFGVEAMP 84
Query: 140 TLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAK 176
T IK +V D +VG ++ EDL+ ++ K
Sbjct: 85 TFVFIKAGEVVDKLVGANK---------EDLQAKIVK 112
>At4g26160 thioredoxin like protein
Length = 221
Score = 36.6 bits (83), Expect = 0.010
Identities = 23/80 (28%), Positives = 39/80 (48%), Gaps = 3/80 (3%)
Query: 71 ELPSEKDFFSVVK-ASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSP 127
++ S + F + +K A DR+V F W C+ L AK+H F+K+N +++
Sbjct: 97 DITSAEQFLNALKDAGDRLVIVDFYGTWCGSCRAMFPKLCKTAKEHPNILFLKVNFDENK 156
Query: 128 FLAEKLKITVLPTLALIKNA 147
L + L + VLP + A
Sbjct: 157 SLCKSLNVKVLPYFHFYRGA 176
>At4g03520 putative M-type thioredoxin
Length = 186
Score = 36.2 bits (82), Expect = 0.013
Identities = 22/78 (28%), Positives = 42/78 (53%), Gaps = 4/78 (5%)
Query: 81 VVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHI-ETRFVKINAEKSPFLAEKLKITV 137
V+KA+ VV F+ W PCK+ D + LA+ + + +F K+N ++SP + +
Sbjct: 94 VLKATGPVVVDFWAP-WCGPCKMIDPLVNDLAQHYTGKIKFYKLNTDESPNTPGQYGVRS 152
Query: 138 LPTLALIKNAKVDDYVVG 155
+PT+ + + D ++G
Sbjct: 153 IPTIMIFVGGEKKDTIIG 170
>At3g51030 thioredoxin h
Length = 114
Score = 35.8 bits (81), Expect = 0.017
Identities = 23/74 (31%), Positives = 35/74 (47%), Gaps = 4/74 (5%)
Query: 93 FRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVD 150
F +W PC+ LAK+ F+K++ ++ +A I +PT +K K+
Sbjct: 35 FTASWCGPCRFIAPFFADLAKKLPNVLFLKVDTDELKSVASDWAIQAMPTFMFLKEGKIL 94
Query: 151 DYVVGF--DELGGT 162
D VVG DEL T
Sbjct: 95 DKVVGAKKDELQST 108
>At1g53300 unknown protein
Length = 699
Score = 34.3 bits (77), Expect = 0.050
Identities = 20/81 (24%), Positives = 37/81 (44%), Gaps = 1/81 (1%)
Query: 67 GDYSELPSEKDFFSVVKASDRVVCHFFR-ENWPCKVADKHLGVLAKQHIETRFVKINAEK 125
G+ E+ S + F S + V HF + CK + L ++ F+K++ +K
Sbjct: 595 GEVEEIYSLEQFKSAMNLPGVSVIHFSTASDHQCKQISPFVDSLCTRYPSIHFLKVDIDK 654
Query: 126 SPFLAEKLKITVLPTLALIKN 146
P + + V+PT+ + KN
Sbjct: 655 CPSIGNAENVRVVPTVKIYKN 675
>At5g65770 nuclear matrix constituent protein 1 (NMCP1)-like
Length = 1042
Score = 33.5 bits (75), Expect = 0.085
Identities = 16/41 (39%), Positives = 22/41 (53%)
Query: 23 DKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWIS 63
D I +E AL +D+ESL R M KM E+ + W+S
Sbjct: 595 DNIKEERDALRNQHKNDVESLNREREEFMNKMVEEHSEWLS 635
>At2g42580 unknown protein
Length = 691
Score = 33.5 bits (75), Expect = 0.085
Identities = 33/121 (27%), Positives = 50/121 (41%), Gaps = 7/121 (5%)
Query: 36 DSDDLESLRERRLHQMKKMAEKRNRWISLGHGDYSELPSEKDFFSVVKASDRVVCHFFRE 95
DS+ ESL + M + E + SLG + E S D F A V F+
Sbjct: 559 DSEVAESLERAKTVLMNRSQESK----SLGFNNEVEAVSTLDKFKKSVALPGVSVFHFKS 614
Query: 96 --NWPCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLALIKNA-KVDDY 152
N C+ + L ++ F ++ E+S LA+ I +PT + KN KV +
Sbjct: 615 SSNRQCEEISPFINTLCLRYPLVHFFMVDVEESMALAKAESIRKVPTFKMYKNGDKVKEM 674
Query: 153 V 153
V
Sbjct: 675 V 675
>At1g76760 thioredoxin-like protein
Length = 172
Score = 33.5 bits (75), Expect = 0.085
Identities = 25/81 (30%), Positives = 39/81 (47%), Gaps = 3/81 (3%)
Query: 78 FFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQ-HIETRFVKINAEKSPFLAEKLK 134
F ++ SD+ V + W PC+ L +++ + + VKI+ EK P +A K K
Sbjct: 73 FEDLLVNSDKPVLVDYYATWCGPCQFMVPILNEVSETLKDKIQVVKIDTEKYPSIANKYK 132
Query: 135 ITVLPTLALIKNAKVDDYVVG 155
I LPT L K+ + D G
Sbjct: 133 IEALPTFILFKDGEPCDRFEG 153
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,396,386
Number of Sequences: 26719
Number of extensions: 182852
Number of successful extensions: 655
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 621
Number of HSP's gapped (non-prelim): 46
length of query: 213
length of database: 11,318,596
effective HSP length: 95
effective length of query: 118
effective length of database: 8,780,291
effective search space: 1036074338
effective search space used: 1036074338
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0200.8


