

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0197.6
(158 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
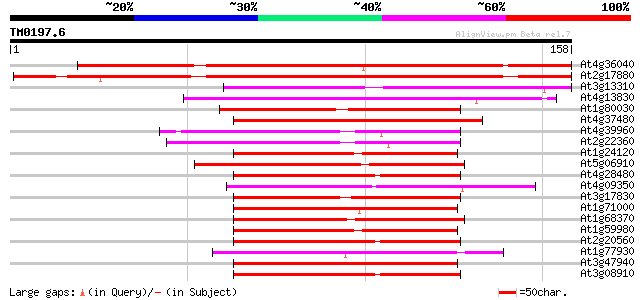
Score E
Sequences producing significant alignments: (bits) Value
At4g36040 DnaJ-like protein 134 3e-32
At2g17880 putative DnaJ protein 134 3e-32
At3g13310 DnaJ protein, putative 84 2e-17
At4g13830 DnaJ-like protein 74 3e-14
At1g80030 unknown protein 64 3e-11
At4g37480 putative protein 64 3e-11
At4g39960 DnaJ like protein 62 1e-10
At2g22360 DnaJ like protein 60 4e-10
At1g24120 DNAJ-like heatshock protein (At1g24120) 60 6e-10
At5g06910 DnaJ homologue (gb|AAB91418.1|) 57 3e-09
At4g28480 heat-shock protein 57 3e-09
At4g09350 putative protein 57 3e-09
At3g17830 DnaJ, putative 57 3e-09
At1g71000 57 5e-09
At1g68370 ARG1 protein (Altered Response to Gravity) 57 5e-09
At1g59980 unknown protein 57 5e-09
At2g20560 putative heat shock protein 56 7e-09
At1g77930 dnaJ like protein 56 7e-09
At3g47940 heat shock protein-like protein 56 9e-09
At3g08910 putative heat shock protein 56 9e-09
>At4g36040 DnaJ-like protein
Length = 161
Score = 134 bits (336), Expect = 3e-32
Identities = 73/140 (52%), Positives = 89/140 (63%), Gaps = 5/140 (3%)
Query: 20 PTGHVRSRPVLAFATATEADYRPTPSLHLSTHGDSGMASCSSSLYEILGIAAAASDQEIK 79
P+ R P L A+ + +P LH + + +SLY++L + A+ Q+IK
Sbjct: 26 PSRTARISPPLVSASCSYTYTEDSPRLHQIPRR---LTTVPASLYDVLEVPLGATSQDIK 82
Query: 80 AAYRRLARVRHPNVAAVDR-KDSSADEFMKIHAAYSTLSDPEKRASYDRSMNRRRRPLTA 138
+AYRRLAR+ HP+VA DR SSADEFMKIHAAY TLSDPEKR+ YDR M RR RPLT
Sbjct: 83 SAYRRLARICHPDVAGTDRTSSSSADEFMKIHAAYCTLSDPEKRSVYDRRMLRRSRPLTV 142
Query: 139 AVTSRFSGYKGRNWETDQCW 158
TS Y GRNWETDQCW
Sbjct: 143 G-TSGLGSYVGRNWETDQCW 161
>At2g17880 putative DnaJ protein
Length = 160
Score = 134 bits (336), Expect = 3e-32
Identities = 80/158 (50%), Positives = 98/158 (61%), Gaps = 12/158 (7%)
Query: 2 LSPSFTTATTFSGQIATSPTGHV-RSRPVLAFATATEADYRPTPSLHLSTHGDSGMASCS 60
LS S +++ +FS +TSP + R P L+ TA+ P L S +
Sbjct: 14 LSSSSSSSPSFS---STSPPSRISRISPSLSTTTASYTCAEDLPRLRQIPQ----RFSAT 66
Query: 61 SSLYEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPE 120
+SLYEIL I ++ QEIK+AYRRLAR+ HP+VA R +SSAD+FMKIHAAY TLSDPE
Sbjct: 67 ASLYEILEIPVGSTSQEIKSAYRRLARICHPDVARNSRDNSSADDFMKIHAAYCTLSDPE 126
Query: 121 KRASYDRSMNRRRRPLTAAVTSRFSGYKGRNWETDQCW 158
KRA YDR R RPLTA + Y GRNWETDQCW
Sbjct: 127 KRAVYDRRTLLRSRPLTAG----YGSYGGRNWETDQCW 160
>At3g13310 DnaJ protein, putative
Length = 157
Score = 84.3 bits (207), Expect = 2e-17
Identities = 47/100 (47%), Positives = 59/100 (59%), Gaps = 7/100 (7%)
Query: 61 SSLYEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPE 120
SSLYE+L + AS EIK AYR LA+V HP+ + D +D FM+IH AY+TL+DP
Sbjct: 63 SSLYELLKVNETASLTEIKTAYRSLAKVYHPDASESDGRD-----FMEIHKAYATLADPT 117
Query: 121 KRASYDRSMNRRRRPLTAAVTSRFSGYKG--RNWETDQCW 158
RA YD ++ RR + A R R WETDQCW
Sbjct: 118 TRAIYDSTLRVPRRRVHAGAMGRSGRVYATTRRWETDQCW 157
>At4g13830 DnaJ-like protein
Length = 197
Score = 73.9 bits (180), Expect = 3e-14
Identities = 41/114 (35%), Positives = 61/114 (52%), Gaps = 10/114 (8%)
Query: 50 THGDSGMASCSSSLYEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKI 109
TH D S S Y++LG+ + + EIK AY++LAR HP+V+ DR + D F+++
Sbjct: 54 THDDPVKQSEDLSFYDLLGVTESVTLPEIKQAYKQLARKYHPDVSPPDRVEEYTDRFIRV 113
Query: 110 HAAYSTLSDPEKRASYDRSMN---------RRRRPLTAAVTSRFSGYKGRNWET 154
AY TLSDP +R YDR ++ RR+ V S +K + W+T
Sbjct: 114 QEAYETLSDPRRRVLYDRDLSMGFSFSFSGRRQNRYDQEVVEEKSEWKAK-WQT 166
>At1g80030 unknown protein
Length = 500
Score = 64.3 bits (155), Expect = 3e-11
Identities = 32/68 (47%), Positives = 48/68 (70%), Gaps = 3/68 (4%)
Query: 60 SSSLYEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDP 119
S Y LG++ +A+++EIKAAYRRLAR HP+ V+++ + ++F +I AAY LSD
Sbjct: 73 SGDYYATLGVSKSANNKEIKAAYRRLARQYHPD---VNKEPGATEKFKEISAAYEVLSDE 129
Query: 120 EKRASYDR 127
+KRA YD+
Sbjct: 130 QKRALYDQ 137
>At4g37480 putative protein
Length = 523
Score = 63.9 bits (154), Expect = 3e-11
Identities = 32/70 (45%), Positives = 47/70 (66%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
Y+IL ++ +S EIKA++RRLA+ HP++ + S++ F++I AAY LSD EKRA
Sbjct: 58 YDILNVSETSSIAEIKASFRRLAKETHPDLIESKKDPSNSRRFVQILAAYEILSDSEKRA 117
Query: 124 SYDRSMNRRR 133
YDR + RR
Sbjct: 118 HYDRYLLSRR 127
>At4g39960 DnaJ like protein
Length = 447
Score = 62.4 bits (150), Expect = 1e-10
Identities = 37/86 (43%), Positives = 51/86 (59%), Gaps = 6/86 (6%)
Query: 43 TPSLHLSTHGDSGMASCSSSLYEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSS 102
+P+ H S G + Y +LG++ A+ EIK+AYR+LAR HP+V KD+
Sbjct: 67 SPNTH-SRRGARFTVRADTDFYSVLGVSKNATKAEIKSAYRKLARSYHPDV----NKDAG 121
Query: 103 A-DEFMKIHAAYSTLSDPEKRASYDR 127
A D+F +I AY LSD EKR+ YDR
Sbjct: 122 AEDKFKEISNAYEILSDDEKRSLYDR 147
>At2g22360 DnaJ like protein
Length = 442
Score = 60.5 bits (145), Expect = 4e-10
Identities = 34/84 (40%), Positives = 48/84 (56%), Gaps = 5/84 (5%)
Query: 45 SLHLSTHGDSGMASCSSSLYEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSAD 104
++H G + Y +LG++ A+ EIK+AYR+LAR HP+V KD A+
Sbjct: 69 NMHPPRRGSRFTVRADADYYSVLGVSKNATKAEIKSAYRKLARNYHPDV----NKDPGAE 124
Query: 105 E-FMKIHAAYSTLSDPEKRASYDR 127
E F +I AY LSD EK++ YDR
Sbjct: 125 EKFKEISNAYEVLSDDEKKSLYDR 148
>At1g24120 DNAJ-like heatshock protein (At1g24120)
Length = 436
Score = 59.7 bits (143), Expect = 6e-10
Identities = 30/63 (47%), Positives = 43/63 (67%), Gaps = 2/63 (3%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
YE+LG+ ++DQEIK+AYR+LA HP+ A D +AD F ++ +Y+ LSDPEKR
Sbjct: 22 YEVLGVLRNSTDQEIKSAYRKLALKYHPDKTAND--PVAADMFKEVTFSYNILSDPEKRR 79
Query: 124 SYD 126
+D
Sbjct: 80 QFD 82
>At5g06910 DnaJ homologue (gb|AAB91418.1|)
Length = 284
Score = 57.4 bits (137), Expect = 3e-09
Identities = 30/76 (39%), Positives = 46/76 (60%), Gaps = 2/76 (2%)
Query: 53 DSGMASCSSSLYEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAA 112
D+ S +SLYE+LG+ A+ QEI+ AY +LA HP+ D++ + D+F ++
Sbjct: 20 DNAGPSSETSLYEVLGVERRATSQEIRKAYHKLALKLHPDKNQDDKE--AKDKFQQLQKV 77
Query: 113 YSTLSDPEKRASYDRS 128
S L D EKRA YD++
Sbjct: 78 ISILGDEEKRAVYDQT 93
>At4g28480 heat-shock protein
Length = 348
Score = 57.4 bits (137), Expect = 3e-09
Identities = 28/64 (43%), Positives = 44/64 (68%), Gaps = 1/64 (1%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
Y++L + +A+D ++K AYR+LA HP+ ++KD+ A +F +I AY LSDP+KRA
Sbjct: 6 YKVLQVDRSANDDDLKKAYRKLAMKWHPDKNPNNKKDAEA-KFKQISEAYDVLSDPQKRA 64
Query: 124 SYDR 127
YD+
Sbjct: 65 VYDQ 68
>At4g09350 putative protein
Length = 249
Score = 57.4 bits (137), Expect = 3e-09
Identities = 34/96 (35%), Positives = 51/96 (52%), Gaps = 10/96 (10%)
Query: 62 SLYEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEK 121
S Y+ LG++ A +EIK+AYRRL++ HP+ ++ K +S ++FMK+ Y+ LSD E
Sbjct: 106 SHYQFLGVSTDADLEEIKSAYRRLSKEYHPDTTSLPLKTAS-EKFMKLREVYNVLSDEET 164
Query: 122 RASYD---------RSMNRRRRPLTAAVTSRFSGYK 148
R YD R + R L F GY+
Sbjct: 165 RRFYDWTLAQEVASRQAEKMRMKLEDPKEQDFRGYE 200
>At3g17830 DnaJ, putative
Length = 517
Score = 57.4 bits (137), Expect = 3e-09
Identities = 29/64 (45%), Positives = 43/64 (66%), Gaps = 3/64 (4%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
Y L + A+ QEIK++YR+LAR HP++ ++ + D+F +I AAY LSD EKR+
Sbjct: 65 YSTLNVNRNATLQEIKSSYRKLARKYHPDM---NKNPGAEDKFKQISAAYEVLSDEEKRS 121
Query: 124 SYDR 127
+YDR
Sbjct: 122 AYDR 125
>At1g71000
Length = 146
Score = 56.6 bits (135), Expect = 5e-09
Identities = 29/65 (44%), Positives = 41/65 (62%), Gaps = 2/65 (3%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVD--RKDSSADEFMKIHAAYSTLSDPEK 121
YEILG+A +S ++I+ AY +LA++ HP+ D R + F +I AYS LSD K
Sbjct: 10 YEILGVAVDSSAEQIRRAYHKLAKIWHPDRWTKDPFRSGEAKRRFQQIQEAYSVLSDERK 69
Query: 122 RASYD 126
R+SYD
Sbjct: 70 RSSYD 74
>At1g68370 ARG1 protein (Altered Response to Gravity)
Length = 410
Score = 56.6 bits (135), Expect = 5e-09
Identities = 29/65 (44%), Positives = 43/65 (65%), Gaps = 2/65 (3%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
YE+L ++ A+DQEIK+AYR+LA HP+ A +++ F ++ +YS LSDPEKR
Sbjct: 19 YEVLCVSKDANDQEIKSAYRKLALKYHPDKNA--NNPDASELFKEVAFSYSILSDPEKRR 76
Query: 124 SYDRS 128
YD +
Sbjct: 77 HYDNA 81
>At1g59980 unknown protein
Length = 414
Score = 56.6 bits (135), Expect = 5e-09
Identities = 30/63 (47%), Positives = 41/63 (64%), Gaps = 2/63 (3%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
YE+LGI + ++DQEIK+AYRR+A HP+ D +A+ F ++ AY LSDPE R
Sbjct: 25 YEVLGIPSNSTDQEIKSAYRRMALRYHPDKNPDD--PVAAEMFKEVTFAYEVLSDPENRR 82
Query: 124 SYD 126
YD
Sbjct: 83 LYD 85
>At2g20560 putative heat shock protein
Length = 337
Score = 56.2 bits (134), Expect = 7e-09
Identities = 28/64 (43%), Positives = 43/64 (66%), Gaps = 1/64 (1%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
Y++L + +ASD ++K AYR+LA HP+ ++KD+ A F +I AY LSDP+K+A
Sbjct: 6 YKVLQVDRSASDDDLKKAYRKLAMKWHPDKNPNNKKDAEA-MFKQISEAYEVLSDPQKKA 64
Query: 124 SYDR 127
YD+
Sbjct: 65 VYDQ 68
>At1g77930 dnaJ like protein
Length = 271
Score = 56.2 bits (134), Expect = 7e-09
Identities = 32/86 (37%), Positives = 49/86 (56%), Gaps = 6/86 (6%)
Query: 58 SCSSSLYEILGIAAAASDQEIKAAYRRLARVRHPNV----AAVDRKDSSADEFMKIHAAY 113
S S Y+ L + A +++IK AYRRLA+ HP+V ++ +++ F+KI AAY
Sbjct: 72 SQEKSPYDTLELDRNAEEEQIKVAYRRLAKFYHPDVYDGKGTLEEGETAEARFIKIQAAY 131
Query: 114 STLSDPEKRASYDRSMNRRRRPLTAA 139
L D EK+ YD M+ R P+ A+
Sbjct: 132 ELLMDSEKKVQYD--MDNRVNPMKAS 155
>At3g47940 heat shock protein-like protein
Length = 350
Score = 55.8 bits (133), Expect = 9e-09
Identities = 26/63 (41%), Positives = 39/63 (61%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
Y IL + A++ ++K AY+RLA + HP+ R+D + +F +I AY LSDP+KR
Sbjct: 6 YNILKVNHNATEDDLKKAYKRLAMIWHPDKNPSTRRDEAEAKFKRISEAYDVLSDPQKRQ 65
Query: 124 SYD 126
YD
Sbjct: 66 IYD 68
>At3g08910 putative heat shock protein
Length = 323
Score = 55.8 bits (133), Expect = 9e-09
Identities = 28/64 (43%), Positives = 42/64 (64%), Gaps = 1/64 (1%)
Query: 64 YEILGIAAAASDQEIKAAYRRLARVRHPNVAAVDRKDSSADEFMKIHAAYSTLSDPEKRA 123
Y++L + A D ++K AYR+LA HP+ ++KD+ A +F +I AY LSDP+KRA
Sbjct: 6 YKVLQVDRNAKDDDLKKAYRKLAMKWHPDKNPNNKKDAEA-KFKQISEAYDVLSDPQKRA 64
Query: 124 SYDR 127
YD+
Sbjct: 65 IYDQ 68
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.125 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,351,526
Number of Sequences: 26719
Number of extensions: 121772
Number of successful extensions: 555
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 405
Number of HSP's gapped (non-prelim): 105
length of query: 158
length of database: 11,318,596
effective HSP length: 91
effective length of query: 67
effective length of database: 8,887,167
effective search space: 595440189
effective search space used: 595440189
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0197.6


