

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0197.2
(366 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
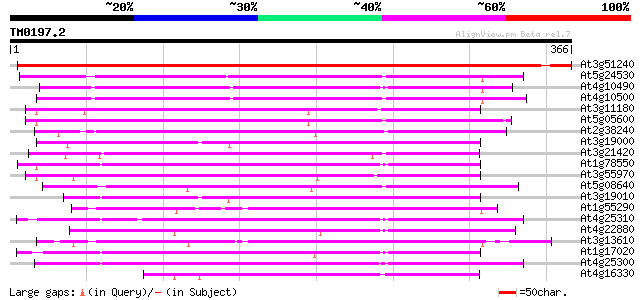
Score E
Sequences producing significant alignments: (bits) Value
At3g51240 flavanone 3-hydroxylase (FH3) 624 e-179
At5g24530 flavanone 3-hydroxylase-like protein 215 3e-56
At4g10490 putative flavanone 3-beta-hydroxylase 208 4e-54
At4g10500 putative Fe(II)/ascorbate oxidase 199 2e-51
At3g11180 putative leucoanthocyanidin dioxygenase 198 3e-51
At5g05600 leucoanthocyanidin dioxygenase-like protein 191 5e-49
At2g38240 putative anthocyanidin synthase 189 3e-48
At3g19000 unknown protein 187 8e-48
At3g21420 unknown protein 185 4e-47
At1g78550 unknown protein 185 4e-47
At3g55970 leucoanthocyanidin dioxygenase -like protein 183 1e-46
At5g08640 flavonol synthase (FLS) (sp|Q96330) 176 2e-44
At3g19010 oxidase like protein 175 3e-44
At1g55290 leucoanthocyanidin dioxygenase 2, putative 175 3e-44
At4g25310 SRG1-like protein 173 2e-43
At4g22880 putative leucoanthocyanidin dioxygenase (LDOX) 170 1e-42
At3g13610 unknown protein 170 1e-42
At1g17020 SRG1-like protein 169 2e-42
At4g25300 SRG1-like protein 166 2e-41
At4g16330 naringenin 3-dioxygenase like protein 165 3e-41
>At3g51240 flavanone 3-hydroxylase (FH3)
Length = 358
Score = 624 bits (1610), Expect = e-179
Identities = 303/361 (83%), Positives = 332/361 (91%), Gaps = 5/361 (1%)
Query: 6 PKTLTTLAQQNTLESSFVRDEDERPKVAYNNFSNEIPVISLAGIDEVDGRRSEICNKIVE 65
P TLT LA ++ L S FVRDEDERPKVAYN FS+EIPVISLAGID+VDG+R EIC +IVE
Sbjct: 3 PGTLTELAGESKLNSKFVRDEDERPKVAYNVFSDEIPVISLAGIDDVDGKRGEICRQIVE 62
Query: 66 ACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGE 125
ACENWGIFQVVDHGVDT LV+ MT LA++FFALPPE+KLRFDM+GGKKGGFIVSSHLQGE
Sbjct: 63 ACENWGIFQVVDHGVDTNLVADMTRLARDFFALPPEDKLRFDMSGGKKGGFIVSSHLQGE 122
Query: 126 SVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLE 185
+VQDWREIVTYFSYP+RNRDYSRWPD P GW VTEEYSE+LM LACKLLEVLSEAMGLE
Sbjct: 123 AVQDWREIVTYFSYPVRNRDYSRWPDKPEGWVKVTEEYSERLMSLACKLLEVLSEAMGLE 182
Query: 186 KEALTKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGK 245
KE+LT ACVDMDQK+VVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGK
Sbjct: 183 KESLTNACVDMDQKIVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGK 242
Query: 246 TWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYP 305
TWITVQPVEGAFVVNLGDHGH+LSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYP
Sbjct: 243 TWITVQPVEGAFVVNLGDHGHFLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYP 302
Query: 306 LKVREGEKSVMEEPITFAEMYRRKMSKDIELARMKKLAKEKKLQDLEKAKLEPKPMNEIF 365
LKVREGEK+++EEPITFAEMY+RKM +D+ELAR+KKLAKE++ K KP+++IF
Sbjct: 303 LKVREGEKAILEEPITFAEMYKRKMGRDLELARLKKLAKEER-----DHKEVDKPVDQIF 357
Query: 366 A 366
A
Sbjct: 358 A 358
>At5g24530 flavanone 3-hydroxylase-like protein
Length = 341
Score = 215 bits (547), Expect = 3e-56
Identities = 117/335 (34%), Positives = 196/335 (57%), Gaps = 14/335 (4%)
Query: 7 KTLTTLAQQNTLESSFVRDEDERPKVAYNNFSNEIPVISLAGIDEVDGRRSEICNKIVEA 66
K ++T + TL ++VR +RP+++ + + P+I L+ D RS + +I +A
Sbjct: 4 KLISTGFRHTTLPENYVRPISDRPRLSEVSQLEDFPLIDLSSTD-----RSFLIQQIHQA 58
Query: 67 CENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLR-FDMTGGKKGGFIVSSHLQGE 125
C +G FQV++HGV+ +++ M ++A+EFF++ EEK++ + K S +++ E
Sbjct: 59 CARFGFFQVINHGVNKQIIDEMVSVAREFFSMSMEEKMKLYSDDPTKTTRLSTSFNVKKE 118
Query: 126 SVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLE 185
V +WR+ + YPI ++ + WP P +K + +YS ++ + K+ E++SE++GLE
Sbjct: 119 EVNNWRDYLRLHCYPI-HKYVNEWPSNPPSFKEIVSKYSREVREVGFKIEELISESLGLE 177
Query: 186 KEALTKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQD-QVGGLQATRDNG 244
K+ + K + Q + VNYYP CP+P+LT GL HTDP +T+LLQD V GLQ D
Sbjct: 178 KDYMKKVLGEQGQHMAVNYYPPCPEPELTYGLPAHTDPNALTILLQDTTVCGLQILIDG- 236
Query: 245 KTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVY 304
W V P AFV+N+GD LSNG +K+ H+AV N+ + RLS+A+F PA A +
Sbjct: 237 -QWFAVNPHPDAFVINIGDQLQALSNGVYKSVWHRAVTNTENPRLSVASFLCPADCAVMS 295
Query: 305 PLK----VREGEKSVMEEPITFAEMYRRKMSKDIE 335
P K + E + + T+AE Y++ S++++
Sbjct: 296 PAKPLWEAEDDETKPVYKDFTYAEYYKKFWSRNLD 330
>At4g10490 putative flavanone 3-beta-hydroxylase
Length = 348
Score = 208 bits (529), Expect = 4e-54
Identities = 119/314 (37%), Positives = 181/314 (56%), Gaps = 10/314 (3%)
Query: 20 SSFVRDEDERPKVAYNNFSNE-IPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDH 78
S++VR +RPK++ S + IP+I L + + R++I N+ AC + G FQ+ +H
Sbjct: 20 SNYVRPVSDRPKMSEVQTSGDSIPLIDLHDLHGPN--RADIINQFAHACSSCGFFQIKNH 77
Query: 79 GVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSS-HLQGESVQDWREIVTYF 137
GV E + M A+EFF E+++ KK + +S ++ E V +WR+ +
Sbjct: 78 GVPEETIKKMMNAAREFFRQSESERVKHYSADTKKTTRLSTSFNVSKEKVSNWRDFLRLH 137
Query: 138 SYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMD 197
YPI + + WP TP ++ VT EY+ + L LLE +SE++GL K+ ++
Sbjct: 138 CYPIEDF-INEWPSTPISFREVTAEYATSVRALVLTLLEAISESLGLAKDRVSNTIGKHG 196
Query: 198 QKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAF 257
Q + +NYYP+CPQP+LT GL H D IT+LLQD+V GLQ +D GK WI V PV F
Sbjct: 197 QHMAINYYPRCPQPELTYGLPGHKDANLITVLLQDEVSGLQVFKD-GK-WIAVNPVPNTF 254
Query: 258 VVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK--VREGEKS- 314
+VNLGD +SN ++K+ H+AVVNS+ R+SI TF P+ DA + P + + E E S
Sbjct: 255 IVNLGDQMQVISNEKYKSVLHRAVVNSDMERISIPTFYCPSEDAVISPAQELINEEEDSP 314
Query: 315 VMEEPITFAEMYRR 328
+ T+AE + +
Sbjct: 315 AIYRNFTYAEYFEK 328
>At4g10500 putative Fe(II)/ascorbate oxidase
Length = 349
Score = 199 bits (507), Expect = 2e-51
Identities = 110/324 (33%), Positives = 179/324 (54%), Gaps = 9/324 (2%)
Query: 18 LESSFVRDEDERPKVAYNNFSNE-IPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVV 76
+ S++VR +RP ++ S + IP+I L + + R+ I ++ AC +G FQ+
Sbjct: 20 IPSNYVRPISDRPNLSEVESSGDSIPLIDLRDLHGPN--RAVIVQQLASACSTYGFFQIK 77
Query: 77 DHGVDTELVSHMTTLAKEFFALPPEEKLR-FDMTGGKKGGFIVSSHLQGESVQDWREIVT 135
+HGV V+ M T+A+EFF P E+++ + K S ++ + V +WR+ +
Sbjct: 78 NHGVPDTTVNKMQTVAREFFHQPESERVKHYSADPTKTTRLSTSFNVGADKVLNWRDFLR 137
Query: 136 YFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVD 195
+PI + WP +P ++ VT EY+ + L +LLE +SE++GLE + ++
Sbjct: 138 LHCFPIEDF-IEEWPSSPISFREVTAEYATSVRALVLRLLEAISESLGLESDHISNILGK 196
Query: 196 MDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEG 255
Q + NYYP CP+P+LT GL H DP IT+LLQDQV GLQ +D+ W+ V P+
Sbjct: 197 HAQHMAFNYYPPCPEPELTYGLPGHKDPTVITVLLQDQVSGLQVFKDD--KWVAVSPIPN 254
Query: 256 AFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK--VREGEK 313
F+VN+GD +SN ++K+ H+AVVN+ + RLSI TF P+ DA + P V E +
Sbjct: 255 TFIVNIGDQMQVISNDKYKSVLHRAVVNTENERLSIPTFYFPSTDAVIGPAHELVNEQDS 314
Query: 314 SVMEEPITFAEMYRRKMSKDIELA 337
+ F E + + ++ + A
Sbjct: 315 LAIYRTYPFVEYWDKFWNRSLATA 338
>At3g11180 putative leucoanthocyanidin dioxygenase
Length = 400
Score = 198 bits (504), Expect = 3e-51
Identities = 107/304 (35%), Positives = 172/304 (56%), Gaps = 9/304 (2%)
Query: 11 TLAQQN--TLESSFVRDEDERPKVAYNNFSNEIPVISLA--GIDEVDGRRSEICNKIVEA 66
+LA+ N +L +++ +RP+ + E+ I++ +D + + +I EA
Sbjct: 58 SLAESNLTSLPDRYIKPPSQRPQTTIIDHQPEVADINIPIIDLDSLFSGNEDDKKRISEA 117
Query: 67 CENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGES 126
C WG FQV++HGV EL+ K FF LP E K + + G+ ++ +
Sbjct: 118 CREWGFFQVINHGVKPELMDAARETWKSFFNLPVEAKEVYSNSPRTYEGYGSRLGVEKGA 177
Query: 127 VQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEK 186
+ DW + P+ +D+++WP P+ + + +EY ++L+ L +L+ +LS +GL
Sbjct: 178 ILDWNDYYYLHFLPLALKDFNKWPSLPSNIREMNDEYGKELVKLGGRLMTILSSNLGLRA 237
Query: 187 EALTKAC--VDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLL-QDQVGGLQATRDN 243
E L +A D+ + VNYYPKCPQP+L LGL H+DPG +T+LL DQV GLQ +
Sbjct: 238 EQLQEAFGGEDVGACLRVNYYPKCPQPELALGLSPHSDPGGMTILLPDDQVVGLQVR--H 295
Query: 244 GKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATV 303
G TWITV P+ AF+VN+GD LSN ++K+ +H+ +VNS R+S+A F NP D +
Sbjct: 296 GDTWITVNPLRHAFIVNIGDQIQILSNSKYKSVEHRVIVNSEKERVSLAFFYNPKSDIPI 355
Query: 304 YPLK 307
P++
Sbjct: 356 QPMQ 359
>At5g05600 leucoanthocyanidin dioxygenase-like protein
Length = 371
Score = 191 bits (485), Expect = 5e-49
Identities = 114/325 (35%), Positives = 182/325 (55%), Gaps = 11/325 (3%)
Query: 11 TLAQQN--TLESSFVRDEDERPKVAYNN-FSNEIPVISLAGIDEVDGRRSE-ICNKIVEA 66
+LA+ N +L +++ RP + + IP+I L G+ +G + I +I EA
Sbjct: 29 SLAESNLSSLPDRYIKPASLRPTTTEDAPTATNIPIIDLEGLFSEEGLSDDVIMARISEA 88
Query: 67 CENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGES 126
C WG FQVV+HGV EL+ +EFF +P K + + G+ ++ +
Sbjct: 89 CRGWGFFQVVNHGVKPELMDAARENWREFFHMPVNAKETYSNSPRTYEGYGSRLGVEKGA 148
Query: 127 VQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEK 186
DW + P +D+++WP P + V +EY E+L+ L+ +++ VLS +GL++
Sbjct: 149 SLDWSDYYFLHLLPHHLKDFNKWPSFPPTIREVIDEYGEELVKLSGRIMRVLSTNLGLKE 208
Query: 187 EALTKAC--VDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLL-QDQVGGLQATRDN 243
+ +A ++ + VNYYPKCP+P+L LGL H+DPG +T+LL DQV GLQ +D+
Sbjct: 209 DKFQEAFGGENIGACLRVNYYPKCPRPELALGLSPHSDPGGMTILLPDDQVFGLQVRKDD 268
Query: 244 GKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATV 303
TWITV+P AF+VN+GD LSN +K+ +H+ +VNS+ R+S+A F NP D +
Sbjct: 269 --TWITVKPHPHAFIVNIGDQIQILSNSTYKSVEHRVIVNSDKERVSLAFFYNPKSDIPI 326
Query: 304 YPL-KVREGEKSVMEEPITFAEMYR 327
PL ++ + P+TF + YR
Sbjct: 327 QPLQELVSTHNPPLYPPMTF-DQYR 350
>At2g38240 putative anthocyanidin synthase
Length = 353
Score = 189 bits (479), Expect = 3e-48
Identities = 113/315 (35%), Positives = 175/315 (54%), Gaps = 13/315 (4%)
Query: 17 TLESSFVRDEDERP--KVAYNNFSNEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQ 74
T+ + +V+ +RP ++ EIPV+ + ++V G+ E + ACE WG FQ
Sbjct: 22 TVPNRYVKPAHQRPVFNTTQSDAGIEIPVLDM---NDVWGK-PEGLRLVRSACEEWGFFQ 77
Query: 75 VVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIV 134
+V+HGV L+ + +EFF LP EEK ++ + G+ + ++ DW +
Sbjct: 78 MVNHGVTHSLMERVRGAWREFFELPLEEKRKYANSPDTYEGYGSRLGVVKDAKLDWSDYF 137
Query: 135 TYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACV 194
P R+ S+WP P + + E+Y E++ L +L E LSE++GL+ L +A
Sbjct: 138 FLNYLPSSIRNPSKWPSQPPKIRELIEKYGEEVRKLCERLTETLSESLGLKPNKLMQALG 197
Query: 195 DMDQ---KVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQ-VGGLQATRDNGKTWITV 250
D+ + N+YPKCPQP LTLGL H+DPG IT+LL D+ V GLQ R +G W+T+
Sbjct: 198 GGDKVGASLRTNFYPKCPQPQLTLGLSSHSDPGGITILLPDEKVAGLQVRRGDG--WVTI 255
Query: 251 QPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPL-KVR 309
+ V A +VN+GD LSNG +K+ +HQ +VNS R+S+A F NP D V P+ ++
Sbjct: 256 KSVPNALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERVSLAFFYNPRSDIPVGPIEELV 315
Query: 310 EGEKSVMEEPITFAE 324
+ + +PI F E
Sbjct: 316 TANRPALYKPIRFDE 330
>At3g19000 unknown protein
Length = 352
Score = 187 bits (475), Expect = 8e-48
Identities = 101/308 (32%), Positives = 169/308 (54%), Gaps = 20/308 (6%)
Query: 18 LESSFVRDEDERPKVAYNN-----FSNEIPVISLAGIDEVDGRRSEICNKIVEACENWGI 72
L+ +F++ + RP N FS+EIP I L+ +++ ++ I +I EAC+ WG
Sbjct: 4 LDEAFIQAPEHRPNTHLTNSGDFIFSDEIPTIDLSSLEDTHHDKTAIAKEIAEACKRWGF 63
Query: 73 FQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGESVQDWRE 132
FQV++HG+ + L + A EFF L EEK + G+ H + +V+DW+E
Sbjct: 64 FQVINHGLPSALRHRVEKTAAEFFNLTTEEKRKVKRDEVNPMGYHDEEHTK--NVRDWKE 121
Query: 133 IVTYFSYPIR-------------NRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLS 179
I +F + ++WP P+ ++ V +EY+ ++ LA +LLE++S
Sbjct: 122 IFDFFLQDSTIVPASPEPEDTELRKLTNQWPQNPSHFREVCQEYAREVEKLAFRLLELVS 181
Query: 180 EAMGLEKEALTKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQA 239
++GL + LT + + N+YP CP P+L LG+ RH D G +T+L QD VGGLQ
Sbjct: 182 ISLGLPGDRLTGFFNEQTSFLRFNHYPPCPNPELALGVGRHKDGGALTVLAQDSVGGLQV 241
Query: 240 TRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAP 299
+R + WI V+P+ A ++N+G+ +N + +A+H+ VVN++ R SI F P+
Sbjct: 242 SRRSDGQWIPVKPISDALIINMGNCIQVWTNDEYWSAEHRVVVNTSKERFSIPFFFFPSH 301
Query: 300 DATVYPLK 307
+A + PL+
Sbjct: 302 EANIEPLE 309
>At3g21420 unknown protein
Length = 364
Score = 185 bits (469), Expect = 4e-47
Identities = 104/302 (34%), Positives = 170/302 (55%), Gaps = 11/302 (3%)
Query: 13 AQQNTLESSFVRDEDERPKVAYN----NFSNEIPVISLAGIDEVDGRRS--EICNKIVEA 66
++ N + F+R+E ER V + + ++IPVI L+ + + D EI K+ +A
Sbjct: 23 SKPNKVPERFIREEYERGVVVSSLKTHHLHHQIPVIDLSKLSKPDNDDFFFEIL-KLSQA 81
Query: 67 CENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGES 126
CE+WG FQV++HG++ E+V + +A EFF +P EEK ++ M G G+ + +
Sbjct: 82 CEDWGFFQVINHGIEVEVVEDIEEVASEFFDMPLEEKKKYPMEPGTVQGYGQAFIFSEDQ 141
Query: 127 VQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEK 186
DW + +P + R+ WP PA + E YS+++ L +LL+ ++ ++GL++
Sbjct: 142 KLDWCNMFALGVHPPQIRNPKLWPSKPARFSESLEGYSKEIRELCKRLLKYIAISLGLKE 201
Query: 187 EALTKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVG--GLQATRDNG 244
E + + Q V +NYYP C PDL LGL H+D +T+L Q + GLQ +DN
Sbjct: 202 ERFEEMFGEAVQAVRMNYYPPCSSPDLVLGLSPHSDGSALTVLQQSKNSCVGLQILKDN- 260
Query: 245 KTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVY 304
TW+ V+P+ A V+N+GD LSNG++K+ +H+AV N RL+I TF P + +
Sbjct: 261 -TWVPVKPLPNALVINIGDTIEVLSNGKYKSVEHRAVTNREKERLTIVTFYAPNYEVEIE 319
Query: 305 PL 306
P+
Sbjct: 320 PM 321
>At1g78550 unknown protein
Length = 356
Score = 185 bits (469), Expect = 4e-47
Identities = 103/306 (33%), Positives = 175/306 (56%), Gaps = 7/306 (2%)
Query: 6 PKTLTTLAQQN--TLESSFVRDEDERPKVAYNN-FSNEIPVISLAGIDEVDGRRSEICNK 62
P L + ++N T+ +VR + E+ ++ ++ S+EIPVI + + V SE+ K
Sbjct: 15 PFVLEIVKEKNFTTIPPRYVRVDQEKTEILNDSSLSSEIPVIDMTRLCSVSAMDSEL-KK 73
Query: 63 IVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHL 122
+ AC++WG FQ+V+HG+D+ + + T +EFF LP +EK + G+ GF + +
Sbjct: 74 LDFACQDWGFFQLVNHGIDSSFLEKLETEVQEFFNLPMKEKQKLWQRSGEFEGFGQVNIV 133
Query: 123 QGESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAM 182
DW ++ + PIR+R + P ++ E YS ++ +A L ++ +
Sbjct: 134 SENQKLDWGDMFILTTEPIRSRKSHLFSKLPPPFRETLETYSSEVKSIAKILFAKMASVL 193
Query: 183 GLEKEALTKACVDMDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQ-DQVGGLQATR 241
++ E + D+ Q + +NYYP CPQPD +GL +H+D +T+LLQ +QV GLQ +
Sbjct: 194 EIKHEEMEDLFDDVWQSIKINYYPPCPQPDQVMGLTQHSDAAGLTILLQVNQVEGLQIKK 253
Query: 242 DNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDA 301
D GK W+ V+P+ A VVN+G+ ++NGR+++ +H+AVVNS RLS+A F +P +
Sbjct: 254 D-GK-WVVVKPLRDALVVNVGEILEIITNGRYRSIEHRAVVNSEKERLSVAMFHSPGKET 311
Query: 302 TVYPLK 307
+ P K
Sbjct: 312 IIRPAK 317
>At3g55970 leucoanthocyanidin dioxygenase -like protein
Length = 363
Score = 183 bits (464), Expect = 1e-46
Identities = 104/308 (33%), Positives = 173/308 (55%), Gaps = 12/308 (3%)
Query: 11 TLAQQN--TLESSFVRDEDERPKVAYNNFSNE----IPVISLAGIDEVD-GRRSEICNKI 63
+L++ N + + +V+ +RP + +N N IP+I L + D +++ ++I
Sbjct: 16 SLSESNLGAIPNRYVKPLSQRPNITPHNKHNPQTTTIPIIDLGRLYTDDLTLQAKTLDEI 75
Query: 64 VEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQ 123
+AC G FQVV+HG+ +L+ +EFF LP E K + G+ ++
Sbjct: 76 SKACRELGFFQVVNHGMSPQLMDQAKATWREFFNLPMELKNMHANSPKTYEGYGSRLGVE 135
Query: 124 GESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMG 183
++ DW + P +DY++WP P + + E+Y ++++ L L+++LS+ +G
Sbjct: 136 KGAILDWSDYYYLHYQPSSLKDYTKWPSLPLHCREILEDYCKEMVKLCENLMKILSKNLG 195
Query: 184 LEKEALTKACVDMDQK---VVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQD-QVGGLQA 239
L+++ L A ++ + VNYYPKCPQP+LTLG+ H+DPG +T+LL D QV LQ
Sbjct: 196 LQEDRLQNAFGGKEESGGCLRVNYYPKCPQPELTLGISPHSDPGGLTILLPDEQVASLQ- 254
Query: 240 TRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAP 299
R + WITV+P AF+VN+GD LSN +K+ +H+ +VN + RLS+A F NP
Sbjct: 255 VRGSDDAWITVEPAPHAFIVNMGDQIQMLSNSIYKSVEHRVIVNPENERLSLAFFYNPKG 314
Query: 300 DATVYPLK 307
+ + PLK
Sbjct: 315 NVPIEPLK 322
>At5g08640 flavonol synthase (FLS) (sp|Q96330)
Length = 336
Score = 176 bits (445), Expect = 2e-44
Identities = 105/317 (33%), Positives = 170/317 (53%), Gaps = 13/317 (4%)
Query: 22 FVRDEDERPKVA-YNNFSNEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGV 80
F+R E E+P + + + IPV+ L+ DE RR+ +V+A E WG+FQVV+HG+
Sbjct: 23 FIRSEKEQPAITTFRGPTPAIPVVDLSDPDEESVRRA-----VVKASEEWGLFQVVNHGI 77
Query: 81 DTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKG--GFIVSSHLQGESVQDWREIVTYFS 138
TEL+ + + ++FF LP EK K G+ E + W + + +
Sbjct: 78 PTELIRRLQDVGRKFFELPSSEKESVAKPEDSKDIEGYGTKLQKDPEGKKAWVDHLFHRI 137
Query: 139 YPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVD--M 196
+P +Y WP P ++ V EEY+ + L+ LL +LS+ +GL+++AL +
Sbjct: 138 WPPSCVNYRFWPKNPPEYREVNEEYAVHVKKLSETLLGILSDGLGLKRDALKEGLGGEMA 197
Query: 197 DQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGA 256
+ + +NYYP CP+PDL LG+ HTD ITLL+ ++V GLQ +D+ W + + A
Sbjct: 198 EYMMKINYYPPCPRPDLALGVPAHTDLSGITLLVPNEVPGLQVFKDD--HWFDAEYIPSA 255
Query: 257 FVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLKVREGEKSVM 316
+V++GD LSNGR+KN H+ V+ +R+S F P + V PL G+ +
Sbjct: 256 VIVHIGDQILRLSNGRYKNVLHRTTVDKEKTRMSWPVFLEPPREKIVGPLPELTGDDNPP 315
Query: 317 E-EPITFAEMYRRKMSK 332
+ +P F + RK++K
Sbjct: 316 KFKPFAFKDYSYRKLNK 332
>At3g19010 oxidase like protein
Length = 349
Score = 175 bits (444), Expect = 3e-44
Identities = 100/285 (35%), Positives = 155/285 (54%), Gaps = 16/285 (5%)
Query: 36 NFSNEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEF 95
N EIPVI L+ +D+ + ++ I ++I +ACE WG FQV++HGV ++ + K F
Sbjct: 23 NQPEEIPVIDLSRLDDPEDVQNVI-SEIGDACEKWGFFQVLNHGVPSDARQRVEKTVKMF 81
Query: 96 FALPPEEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIVT-YFSYPI------------R 142
F LP EEK++ G+ H + +V+DW+E+ YF P+
Sbjct: 82 FDLPMEEKIKVKRDDVNPVGYHDGEHTK--NVKDWKEVFDIYFKDPMVIPSTTDPEDEGL 139
Query: 143 NRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVVV 202
Y++WP +P+ ++ E Y+ LA KLLE++S ++GL KE + +
Sbjct: 140 RLVYNKWPQSPSDFREACEVYARHAEKLAFKLLELISLSLGLPKERFHDYFKEQMSFFRI 199
Query: 203 NYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLG 262
N YP CP+PDL LG+ H D I+LL QD VGGLQ +R + W ++PV A V+N+G
Sbjct: 200 NRYPPCPRPDLALGVGHHKDADVISLLAQDDVGGLQVSRRSDGVWFPIRPVPNALVINIG 259
Query: 263 DHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK 307
+ +N ++ +A+H+ VVN+ R SI F P+ D V PL+
Sbjct: 260 NCMEIWTNDKYWSAEHRVVVNTTRERYSIPFFLLPSHDVEVKPLE 304
>At1g55290 leucoanthocyanidin dioxygenase 2, putative
Length = 361
Score = 175 bits (444), Expect = 3e-44
Identities = 96/286 (33%), Positives = 161/286 (55%), Gaps = 20/286 (6%)
Query: 41 IPVISLAGIDEVDGRRSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPP 100
IPVI ++ +DE + + +A E WG FQV++HGV E++ +M T FF LP
Sbjct: 62 IPVIDISNLDE-----KSVSKAVCDAAEEWGFFQVINHGVSMEVLENMKTATHRFFGLPV 116
Query: 101 EEKLRFD----MTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSR-WPDTPAG 155
EEK +F ++ + G S H E +W++ ++ F + + S+ WPD+
Sbjct: 117 EEKRKFSREKSLSTNVRFGTSFSPH--AEKALEWKDYLSLFF--VSEAEASQLWPDS--- 169
Query: 156 WKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQ-KVVVNYYPKCPQPDLT 214
++ T EY + L KLL L E + +++ TK M ++ +NYYP CP P+LT
Sbjct: 170 CRSETLEYMNETKPLVKKLLRFLGENLNVKELDKTKESFFMGSTRINLNYYPICPNPELT 229
Query: 215 LGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFK 274
+G+ RH+D ++T+LLQD++GGL W+ V P+ G+ V+N+GD +SNGR+K
Sbjct: 230 VGVGRHSDVSSLTILLQDEIGGLHVRSLTTGRWVHVPPISGSLVINIGDAMQIMSNGRYK 289
Query: 275 NADHQAVVNSNSSRLSIATFQNPAPDATVYPL--KVREGEKSVMEE 318
+ +H+ + N + +R+S+ F +P P++ + PL + GEK V ++
Sbjct: 290 SVEHRVLANGSYNRISVPIFVSPKPESVIGPLLEVIENGEKPVYKD 335
>At4g25310 SRG1-like protein
Length = 353
Score = 173 bits (438), Expect = 2e-43
Identities = 106/334 (31%), Positives = 183/334 (54%), Gaps = 14/334 (4%)
Query: 5 KPKTLTTLAQQNTLESSFVRDEDERPKVAYNNFSNEIPVISLAGIDEVDGRRSEICNKIV 64
K K +TT+ L +VR + E+ + A ++ N+IP+I ++ + SEI +K+
Sbjct: 21 KEKVITTV-----LPPRYVRSDQEKGEAAIDSGENQIPIIDMSLLSSSTSMDSEI-DKLD 74
Query: 65 EACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQG 124
AC+ WG FQ+V+HG+D + + ++FF LP EEK + G GF +
Sbjct: 75 FACKEWGFFQLVNHGMDLD---KFKSDIQDFFNLPMEEKKKLWQQPGDIEGFGQAFVFSE 131
Query: 125 ESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGL 184
E DW ++ P+ R +P P ++ + YS +L +A L L+ A+ +
Sbjct: 132 EQKLDWADVFFLTMQPVPLRKPHLFPKLPLPFRDTLDTYSAELKSIAKVLFAKLASALKI 191
Query: 185 EKEALTKACVD-MDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQ-DQVGGLQATRD 242
+ E + K D + Q++ +NYYP CP+PD +GL H+D +T+LLQ ++V GLQ +D
Sbjct: 192 KPEEMEKLFDDELGQRIRMNYYPPCPEPDKAIGLTPHSDATGLTILLQVNEVEGLQIKKD 251
Query: 243 NGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDAT 302
GK W++V+P+ A VVN+GD ++NG +++ +H+ VVNS RLS+A+F N
Sbjct: 252 -GK-WVSVKPLPNALVVNVGDILEIITNGTYRSIEHRGVVNSEKERLSVASFHNTGFGKE 309
Query: 303 VYPLK-VREGEKSVMEEPITFAEMYRRKMSKDIE 335
+ P++ + E K + + +T E + S++++
Sbjct: 310 IGPMRSLVERHKGALFKTLTTEEYFHGLFSRELD 343
>At4g22880 putative leucoanthocyanidin dioxygenase (LDOX)
Length = 356
Score = 170 bits (430), Expect = 1e-42
Identities = 100/299 (33%), Positives = 162/299 (53%), Gaps = 10/299 (3%)
Query: 40 EIPVISLAGIDEVDGRRSEIC-NKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFAL 98
++P I L I+ D + E C ++ +A +WG+ +++HG+ +L+ + +EFF+L
Sbjct: 46 QVPTIDLKNIESDDEKIRENCIEELKKASLDWGVMHLINHGIPADLMERVKKAGEEFFSL 105
Query: 99 PPEEKLRF--DMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSRWPDTPAGW 156
EEK ++ D GK G+ +W + + +YP RD S WP TP+ +
Sbjct: 106 SVEEKEKYANDQATGKIQGYGSKLANNASGQLEWEDYFFHLAYPEEKRDLSIWPKTPSDY 165
Query: 157 KAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVV---VNYYPKCPQPDL 213
T EY++ L LA K+ + LS +GLE + L K +++ ++ +NYYPKCPQP+L
Sbjct: 166 IEATSEYAKCLRLLATKVFKALSVGLGLEPDRLEKEVGGLEELLLQMKINYYPKCPQPEL 225
Query: 214 TLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRF 273
LG++ HTD +T +L + V GLQ + GK W+T + V + V+++GD LSNG++
Sbjct: 226 ALGVEAHTDVSALTFILHNMVPGLQLFYE-GK-WVTAKCVPDSIVMHIGDTLEILSNGKY 283
Query: 274 KNADHQAVVNSNSSRLSIATFQNPAPDATVY-PL-KVREGEKSVMEEPITFAEMYRRKM 330
K+ H+ +VN R+S A F P D V PL ++ E P TFA+ K+
Sbjct: 284 KSILHRGLVNKEKVRISWAVFCEPPKDKIVLKPLPEMVSVESPAKFPPRTFAQHIEHKL 342
>At3g13610 unknown protein
Length = 361
Score = 170 bits (430), Expect = 1e-42
Identities = 112/345 (32%), Positives = 172/345 (49%), Gaps = 31/345 (8%)
Query: 18 LESSFVRDEDERPKVAYNNFSNE----IPVISLAGIDEVDGRRSEICNKIVEACENWGIF 73
L +++ +ER N F NE IPVI ++ DE + + +A E WG F
Sbjct: 38 LPEQYIQPLEER---LINKFVNETDEAIPVIDMSNPDE-----DRVAEAVCDAAEKWGFF 89
Query: 74 QVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGG--FIVSSHLQGESVQDWR 131
QV++HGV E++ + +FF LP EEK +F F S E +W+
Sbjct: 90 QVINHGVPLEVLDDVKAATHKFFNLPVEEKRKFTKENSLSTTVRFGTSFSPLAEQALEWK 149
Query: 132 EIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTK 191
+ ++ F + WPD + T EY K + +LLE L + + +++ TK
Sbjct: 150 DYLSLFFVSEAEAEQF-WPDI---CRNETLEYINKSKKMVRRLLEYLGKNLNVKELDETK 205
Query: 192 ACVDMDQ-KVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITV 250
+ M +V +NYYP CP PDLT+G+ RH+D ++T+LLQDQ+GGL W+ V
Sbjct: 206 ESLFMGSIRVNLNYYPICPNPDLTVGVGRHSDVSSLTILLQDQIGGLHVRSLASGNWVHV 265
Query: 251 QPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK--V 308
PV G+FV+N+GD +SNG +K+ +H+ + N ++R+S+ F NP P++ + PL +
Sbjct: 266 PPVAGSFVINIGDAMQIMSNGLYKSVEHRVLANGYNNRISVPIFVNPKPESVIGPLPEVI 325
Query: 309 REGEKSVMEEPITFAEMYRRKMSKDIELARMKKLAKEKKLQDLEK 353
G EEPI YR + D +K KK D K
Sbjct: 326 ANG-----EEPI-----YRDVLYSDYVKYFFRKAHDGKKTVDYAK 360
>At1g17020 SRG1-like protein
Length = 358
Score = 169 bits (428), Expect = 2e-42
Identities = 103/307 (33%), Positives = 171/307 (55%), Gaps = 13/307 (4%)
Query: 5 KPKTLTTLAQQNTLESSFVRDEDERPKVAYN-NFSNEIPVISLAGIDEVDGRRSEICNKI 63
K KT+TT+ + +VR + ++ +V + + EIP+I + + SE+ K+
Sbjct: 22 KEKTITTVPPR------YVRSDQDKTEVDDDFDVKIEIPIIDMKRLCSSTTMDSEV-EKL 74
Query: 64 VEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQ 123
AC+ WG FQ+V+HG+D+ + + + ++FF LP EEK +F + GF + +
Sbjct: 75 DFACKEWGFFQLVNHGIDSSFLDKVKSEIQDFFNLPMEEKKKFWQRPDEIEGFGQAFVVS 134
Query: 124 GESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMG 183
+ DW ++ + P+ R +P P ++ E YS ++ +A L+ ++ A+
Sbjct: 135 EDQKLDWADLFFHTVQPVELRKPHLFPKLPLPFRDTLEMYSSEVQSVAKILIAKMARALE 194
Query: 184 LEKEALTKACVDMD--QKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQ-DQVGGLQAT 240
++ E L K D+D Q + +NYYP CPQPD +GL H+D +T+L+Q + V GLQ
Sbjct: 195 IKPEELEKLFDDVDSVQSMRMNYYPPCPQPDQVIGLTPHSDSVGLTVLMQVNDVEGLQIK 254
Query: 241 RDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPD 300
+D GK W+ V+P+ AF+VN+GD ++NG +++ +H+ VVNS RLSIATF N
Sbjct: 255 KD-GK-WVPVKPLPNAFIVNIGDVLEIITNGTYRSIEHRGVVNSEKERLSIATFHNVGMY 312
Query: 301 ATVYPLK 307
V P K
Sbjct: 313 KEVGPAK 319
>At4g25300 SRG1-like protein
Length = 356
Score = 166 bits (420), Expect = 2e-41
Identities = 98/323 (30%), Positives = 181/323 (55%), Gaps = 7/323 (2%)
Query: 17 TLESSFVRDEDERPKVAYNN-FSNEIPVISLAGIDEVDGRRSEICNKIVEACENWGIFQV 75
T+ +VR + + ++A ++ N+IP+I ++ + SEI +K+ AC+ WG FQ+
Sbjct: 27 TVPPRYVRSDQDVAEIAVDSGLRNQIPIIDMSLLCSSTSMDSEI-DKLDSACKEWGFFQL 85
Query: 76 VDHGVDTELVSHMTTLAKEFFALPPEEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIVT 135
V+HG+++ ++ + + ++FF LP EEK + GF + E DW ++
Sbjct: 86 VNHGMESSFLNKVKSEVQDFFNLPMEEKKNLWQQPDEIEGFGQVFVVSEEQKLDWADMFF 145
Query: 136 YFSYPIRNRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVD 195
P+R R +P P ++ + YS ++ +A LL ++ A+ ++ E + K D
Sbjct: 146 LTMQPVRLRKPHLFPKLPLPFRDTLDMYSAEVKSIAKILLGKIAVALKIKPEEMDKLFDD 205
Query: 196 -MDQKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQ-DQVGGLQATRDNGKTWITVQPV 253
+ Q++ +NYYP+CP+PD +GL H+D +T+LLQ ++V GLQ + N K W++V+P+
Sbjct: 206 ELGQRIRLNYYPRCPEPDKVIGLTPHSDSTGLTILLQANEVEGLQIKK-NAK-WVSVKPL 263
Query: 254 EGAFVVNLGDHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPLK-VREGE 312
A VVN+GD ++NG +++ +H+ VVNS RLS+A F N + P++ + E
Sbjct: 264 PNALVVNVGDILEIITNGTYRSIEHRGVVNSEKERLSVAAFHNIGLGKEIGPMRSLVERH 323
Query: 313 KSVMEEPITFAEMYRRKMSKDIE 335
K+ + +T E + S++++
Sbjct: 324 KAAFFKSVTTEEYFNGLFSRELD 346
>At4g16330 naringenin 3-dioxygenase like protein
Length = 258
Score = 165 bits (418), Expect = 3e-41
Identities = 89/224 (39%), Positives = 136/224 (59%), Gaps = 7/224 (3%)
Query: 88 MTTLAKEFFA-LPPEEKLRF--DMTGGKKGGFIVSSHL--QGESVQDWREIVTYFSYPIR 142
M +L FF P EEKLR+ D T G+ L + + V DWR+ + ++P
Sbjct: 1 MRSLGLSFFQDSPMEEKLRYACDSTSAASEGYGSRMLLGAKDDVVLDWRDYFDHHTFPSS 60
Query: 143 NRDYSRWPDTPAGWKAVTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKACVDMDQKVVV 202
R+ S WP P+ ++ V EY +++ LA LL ++SE++GL ++ +A ++ Q + V
Sbjct: 61 RRNPSHWPIHPSDYRQVVGEYGDEMKKLAQMLLGLISESLGLPCSSIEEAVGEIYQNITV 120
Query: 203 NYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLG 262
YP CPQPDLTLGL+ H+D G ITLL+QD V GLQ +D W+TV P+ A ++ +
Sbjct: 121 TNYPPCPQPDLTLGLQSHSDFGAITLLIQDDVEGLQLYKD--AQWLTVPPISDAILILIA 178
Query: 263 DHGHYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDATVYPL 306
D ++NGR+K+A H+AV N+N +RLS+ATF +P+ A + P+
Sbjct: 179 DQTEIITNGRYKSAQHRAVTNANRARLSVATFHDPSKTARIAPV 222
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,422,899
Number of Sequences: 26719
Number of extensions: 360072
Number of successful extensions: 1405
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 102
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1071
Number of HSP's gapped (non-prelim): 124
length of query: 366
length of database: 11,318,596
effective HSP length: 101
effective length of query: 265
effective length of database: 8,619,977
effective search space: 2284293905
effective search space used: 2284293905
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0197.2


