

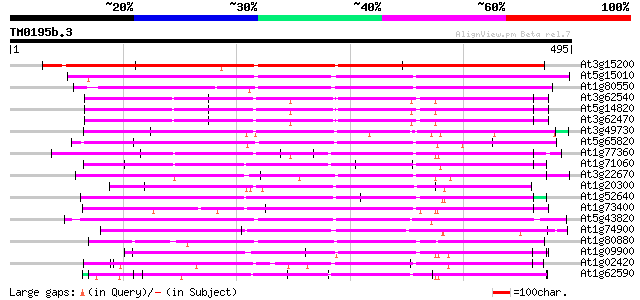
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0195b.3
(495 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g15200 hypothetical protein 526 e-149
At5g15010 putative protein 271 6e-73
At1g80550 unknown protein 225 5e-59
At3g62540 putative protein 218 5e-57
At5g14820 putative protein 218 6e-57
At3g62470 putative protein 218 6e-57
At3g49730 putative protein 207 1e-53
At5g65820 putative protein 193 2e-49
At1g77360 193 2e-49
At1g71060 hypothetical protein 181 6e-46
At3g22670 hypothetical protein 177 1e-44
At1g20300 putative protein 171 7e-43
At1g52640 171 9e-43
At1g73400 unknown protein 166 3e-41
At5g43820 putative protein 164 1e-40
At1g74900 hypothetical protein 160 1e-39
At1g80880 hypothetical protein 154 1e-37
At1g09900 hypothetical protein 150 1e-36
At1g02420 hypothetical protein 150 2e-36
At1g62590 putative membrane-associated salt-inducible protein (A... 148 6e-36
>At3g15200 hypothetical protein
Length = 523
Score = 526 bits (1354), Expect = e-149
Identities = 258/443 (58%), Positives = 329/443 (74%), Gaps = 7/443 (1%)
Query: 30 PPLFTRRFLHSQPEHAAGAGAFVQNLLKFRRDKPTDQVERALDLCGFDLNDDLVLDVLRR 89
P F RF + + +A V N++K R ++++R LD CG DL ++LVL+V+ R
Sbjct: 62 PDKFPNRFNDDKDKQSA---LDVHNIIKHHRGSSPEKIKRILDKCGIDLTEELVLEVVNR 118
Query: 90 HRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNE 149
+RSDWKPA + K + +S + NEILD+LGKM RFEE HQVFDEMS R+G VNE
Sbjct: 119 NRSDWKPAYILSQLVVKQSVHLSSSMLYNEILDVLGKMRRFEEFHQVFDEMSKRDGFVNE 178
Query: 150 DTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHS 209
T+ LL R+AAAHKV+EA+ +F R++FG+D DL AF LLMWLCRYKHVE AETLF S
Sbjct: 179 KTYEVLLNRYAAAHKVDEAVGVFERRKEFGIDDDLVAFHGLLMWLCRYKHVEFAETLFCS 238
Query: 210 KAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKG 269
+ REF DIK N+ILNGWCVLGN HEAKR WKDI+ASKCRPD+ +Y T I ALTKKG
Sbjct: 239 RRREFGC--DIKAMNMILNGWCVLGNVHEAKRFWKDIIASKCRPDVVSYGTMINALTKKG 296
Query: 270 KLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVAT 329
KLG A++L+R MW+ N PDV ICN +IDALCFKKR+PEALEVF+++ E+G +PNV T
Sbjct: 297 KLGKAMELYRAMWDTRRN--PDVKICNNVIDALCFKKRIPEALEVFREISEKGPDPNVVT 354
Query: 330 YNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMER 389
YNSL+KHLCKIRR EKV+ELVE+ME K GSC PN VT+S LL + ++V VLERM +
Sbjct: 355 YNSLLKHLCKIRRTEKVWELVEEMELKGGSCSPNDVTFSYLLKYSQRSKDVDIVLERMAK 414
Query: 390 NGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKD 449
N C ++ D+YNL+ RLY++WD ++ +R+ W EMER+G GPD+R+YTI IHG + GK+ +
Sbjct: 415 NKCEMTSDLYNLMFRLYVQWDKEEKVREIWSEMERSGLGPDQRTYTIRIHGLHTKGKIGE 474
Query: 450 AMRYFREMTSKGMVAEPRTEKLV 472
A+ YF+EM SKGMV EPRTE L+
Sbjct: 475 ALSYFQEMMSKGMVPEPRTEMLL 497
Score = 47.8 bits (112), Expect = 1e-05
Identities = 52/237 (21%), Positives = 89/237 (36%), Gaps = 43/237 (18%)
Query: 112 PTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISM 171
P ++CN ++D L R E +VF E+S + N T+++LL+ + E+ +
Sbjct: 315 PDVKICNNVIDALCFKKRIPEALEVFREISEKGPDPNVVTYNSLLKHLCKIRRTEKVWEL 374
Query: 172 FYTREQFGLDLDLD--AFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNG 229
E G + F LL + R K V+ L + ++ D+ +N++
Sbjct: 375 VEEMELKGGSCSPNDVTFSYLLKYSQRSKDVDIV--LERMAKNKCEMTSDL--YNLMFRL 430
Query: 230 WCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCK 289
+ + + +W ++ S PD TY I L KGK+G
Sbjct: 431 YVQWDKEEKVREIWSEMERSGLGPDQRTYTIRIHGLHTKGKIG----------------- 473
Query: 290 PDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKV 346
EAL FQ+M +G P T L ++ K R +K+
Sbjct: 474 --------------------EALSYFQEMMSKGMVPEPRTEMLLNQNKTKPRVEDKM 510
>At5g15010 putative protein
Length = 532
Score = 271 bits (693), Expect = 6e-73
Identities = 157/448 (35%), Positives = 250/448 (55%), Gaps = 11/448 (2%)
Query: 52 VQNLLKFRRDKPTDQVE--RALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADS 109
V + K +D +D+ E L+ C +++LV+++L R R+DW+ A FF WA K
Sbjct: 57 VGKISKLVKDCGSDRKELRNKLEECDVKPSNELVVEILSRVRNDWETAFTFFVWAGKQQG 116
Query: 110 YAPTSRVCNEILDILGKMSRFEELHQVFDEM-SHREGLVNEDTFSTLLRRFAAAHKVEEA 168
Y + R + ++ ILGKM +F+ + DEM LVN T ++R++ A H V +A
Sbjct: 117 YVRSVREYHSMISILGKMRKFDTAWTLIDEMRKFSPSLVNSQTLLIMIRKYCAVHDVGKA 176
Query: 169 ISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILN 228
I+ F+ ++F L++ +D F++LL LCRYK+V DA L ++ D K++N++LN
Sbjct: 177 INTFHAYKRFKLEMGIDDFQSLLSALCRYKNVSDAGHLIFCNKDKYPF--DAKSFNIVLN 234
Query: 229 GWC-VLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCN 287
GWC V+G+ EA+RVW ++ + D+ +Y++ I +K G L LKLF M E C
Sbjct: 235 GWCNVIGSPREAERVWMEMGNVGVKHDVVSYSSMISCYSKGGSLNKVLKLFDRMKKE-C- 292
Query: 288 CKPDVVICNCIIDALCFKKRVPEALEVFQDMKE-RGCEPNVATYNSLIKHLCKIRRMEKV 346
+PD + N ++ AL V EA + + M+E +G EPNV TYNSLIK LCK R+ E+
Sbjct: 293 IEPDRKVYNAVVHALAKASFVSEARNLMKTMEEEKGIEPNVVTYNSLIKPLCKARKTEEA 352
Query: 347 YELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLY 406
++ ++M K P TY + L+ EEV +L +M + GC + + Y +++R
Sbjct: 353 KQVFDEMLEK--GLFPTIRTYHAFMRILRTGEEVFELLAKMRKMGCEPTVETYIMLIRKL 410
Query: 407 MKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEP 466
+W + D + WDEM+ GPD SY +MIHG + NGK+++A Y++EM KGM
Sbjct: 411 CRWRDFDNVLLLWDEMKEKTVGPDLSSYIVMIHGLFLNGKIEEAYGYYKEMKDKGMRPNE 470
Query: 467 RTEKLVISMNSPLKERTEKQEGKEEETN 494
E ++ S S + ++ + E N
Sbjct: 471 NVEDMIQSWFSGKQYAEQRITDSKGEVN 498
>At1g80550 unknown protein
Length = 448
Score = 225 bits (573), Expect = 5e-59
Identities = 137/427 (32%), Positives = 218/427 (50%), Gaps = 19/427 (4%)
Query: 57 KFRRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRV 116
KFR + DQ + V + L + +DW+ AL FFNW + + T+
Sbjct: 33 KFRSQEEEDQSS---------YDQKTVCEALTCYSNDWQKALEFFNWVERESGFRHTTET 83
Query: 117 CNEILDILGKMSRFEELHQVFDEM-SHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTR 175
N ++DILGK FE + + M + E + N TF + +R+ AH V+EAI +
Sbjct: 84 FNRVIDILGKYFEFEISWALINRMIGNTESVPNHVTFRIVFKRYVTAHLVQEAIDAYDKL 143
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSK---AREFQLHRDIKTWNVILNGWCV 232
+ F L D +F L+ LC +KHV +AE L K F + + K N+IL GW
Sbjct: 144 DDFNLR-DETSFYNLVDALCEHKHVVEAEELCFGKNVIGNGFSVS-NTKIHNLILRGWSK 201
Query: 233 LGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDV 292
LG + K WK + DLF+Y+ ++ + K GK A+KL++ M + K DV
Sbjct: 202 LGWWGKCKEYWKKMDTEGVTKDLFSYSIYMDIMCKSGKPWKAVKLYKEMKSR--RMKLDV 259
Query: 293 VICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVED 352
V N +I A+ + V + VF++M+ERGCEPNVAT+N++IK LC+ RM Y ++++
Sbjct: 260 VAYNTVIRAIGASQGVEFGIRVFREMRERGCEPNVATHNTIIKLLCEDGRMRDAYRMLDE 319
Query: 353 MERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQ 412
M ++ C P+++TY CL + L+ P E+ + RM R+G D Y +++R + +W
Sbjct: 320 MPKR--GCQPDSITYMCLFSRLEKPSEILSLFGRMIRSGVRPKMDTYVMLMRKFERWGFL 377
Query: 413 DGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLV 472
+ W M+ +G PD +Y +I + G + A Y EM +G+ R E +
Sbjct: 378 QPVLYVWKTMKESGDTPDSAAYNAVIDALIQKGMLDMAREYEEEMIERGLSPRRRPELVE 437
Query: 473 ISMNSPL 479
S++ L
Sbjct: 438 KSLDETL 444
>At3g62540 putative protein
Length = 599
Score = 218 bits (556), Expect = 5e-57
Identities = 130/399 (32%), Positives = 214/399 (53%), Gaps = 10/399 (2%)
Query: 67 VERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGK 126
+E LD DL+ DL+++VL R R KPA FF WA++ +A SR N ++ IL K
Sbjct: 148 MEAVLDEMKLDLSHDLIVEVLERFRHARKPAFRFFCWAAERQGFAHASRTYNSMMSILAK 207
Query: 127 MSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDA 186
+FE + V +EM + GL+ +TF+ ++ FAAA + ++A+ +F +++ + ++
Sbjct: 208 TRQFETMVSVLEEMGTK-GLLTMETFTIAMKAFAAAKERKKAVGIFELMKKYKFKIGVET 266
Query: 187 FRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDI 246
LL L R K ++A+ LF F ++ T+ V+LNGWC + N EA R+W D+
Sbjct: 267 INCLLDSLGRAKLGKEAQVLFDKLKERFT--PNMMTYTVLLNGWCRVRNLIEAARIWNDM 324
Query: 247 MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKK 306
+ +PD+ + ++ L + K A+KLF M ++G C P+V +I C +
Sbjct: 325 IDHGLKPDIVAHNVMLEGLLRSMKKSDAIKLFHVMKSKG-PC-PNVRSYTIMIRDFCKQS 382
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
+ A+E F DM + G +P+ A Y LI ++++ VYEL+++M+ K P+ T
Sbjct: 383 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHP--PDGKT 440
Query: 367 YSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEME 423
Y+ L+ + K PE + +M +N S +N++++ Y N + R WDEM
Sbjct: 441 YNALIKLMANQKMPEHGTRIYNKMIQNEIEPSIHTFNMIMKSYFVARNYEMGRAVWDEMI 500
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
+ G PD SYT++I G GK ++A RY EM KGM
Sbjct: 501 KKGICPDDNSYTVLIRGLISEGKSREACRYLEEMLDKGM 539
Score = 65.9 bits (159), Expect = 5e-11
Identities = 71/305 (23%), Positives = 131/305 (42%), Gaps = 15/305 (4%)
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQ-LHRDIKTWNVILNGWCVLG 234
++ LDL D +L R++H F A E Q +T+N +++ +L
Sbjct: 153 DEMKLDLSHDLIVEVLE---RFRHARKPAFRFFCWAAERQGFAHASRTYNSMMS---ILA 206
Query: 235 NAHEAKRVWKDI--MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDV 292
+ + + + M +K + T+ +KA + A+ +F M + K V
Sbjct: 207 KTRQFETMVSVLEEMGTKGLLTMETFTIAMKAFAAAKERKKAVGIFELM--KKYKFKIGV 264
Query: 293 VICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVED 352
NC++D+L K EA +F +KER PN+ TY L+ C++R + + + D
Sbjct: 265 ETINCLLDSLGRAKLGKEAQVLFDKLKER-FTPNMMTYTVLLNGWCRVRNLIEAARIWND 323
Query: 353 M--ERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWD 410
M K + + V LL S+K + + + M+ G + Y +++R + K
Sbjct: 324 MIDHGLKPDIVAHNVMLEGLLRSMKKSDAIK-LFHVMKSKGPCPNVRSYTIMIRDFCKQS 382
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ + + +D+M +G PD YT +I G K+ +EM KG + +T
Sbjct: 383 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYN 442
Query: 471 LVISM 475
+I +
Sbjct: 443 ALIKL 447
>At5g14820 putative protein
Length = 598
Score = 218 bits (555), Expect = 6e-57
Identities = 130/399 (32%), Positives = 214/399 (53%), Gaps = 10/399 (2%)
Query: 67 VERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGK 126
+E LD DL+ DL+++VL R R KPA FF WA++ +A SR N ++ IL K
Sbjct: 147 MEAVLDEMKLDLSHDLIVEVLERFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMSILAK 206
Query: 127 MSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDA 186
+FE + V +EM + GL+ +TF+ ++ FAAA + ++A+ +F +++ + ++
Sbjct: 207 TRQFETMVSVLEEMGTK-GLLTMETFTIAMKAFAAAKERKKAVGIFELMKKYKFKIGVET 265
Query: 187 FRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDI 246
LL L R K ++A+ LF F ++ T+ V+LNGWC + N EA R+W D+
Sbjct: 266 INCLLDSLGRAKLGKEAQVLFDKLKERFT--PNMMTYTVLLNGWCRVRNLIEAARIWNDM 323
Query: 247 MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKK 306
+ +PD+ + ++ L + K A+KLF M ++G C P+V +I C +
Sbjct: 324 IDHGLKPDIVAHNVMLEGLLRSMKKSDAIKLFHVMKSKG-PC-PNVRSYTIMIRDFCKQS 381
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
+ A+E F DM + G +P+ A Y LI ++++ VYEL+++M+ K P+ T
Sbjct: 382 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHP--PDGKT 439
Query: 367 YSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEME 423
Y+ L+ + K PE + +M +N S +N++++ Y N + R WDEM
Sbjct: 440 YNALIKLMANQKMPEHGTRIYNKMIQNEIEPSIHTFNMIMKSYFVARNYEMGRAVWDEMI 499
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
+ G PD SYT++I G GK ++A RY EM KGM
Sbjct: 500 KKGICPDDNSYTVLIRGLISEGKSREACRYLEEMLDKGM 538
Score = 68.9 bits (167), Expect = 6e-12
Identities = 72/305 (23%), Positives = 132/305 (42%), Gaps = 15/305 (4%)
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQ-LHRDIKTWNVILNGWCVLG 234
++ LDL D +L R++H F A E Q D +T+N +++ +L
Sbjct: 152 DEMKLDLSHDLIVEVLE---RFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMS---ILA 205
Query: 235 NAHEAKRVWKDI--MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDV 292
+ + + + M +K + T+ +KA + A+ +F M + K V
Sbjct: 206 KTRQFETMVSVLEEMGTKGLLTMETFTIAMKAFAAAKERKKAVGIFELM--KKYKFKIGV 263
Query: 293 VICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVED 352
NC++D+L K EA +F +KER PN+ TY L+ C++R + + + D
Sbjct: 264 ETINCLLDSLGRAKLGKEAQVLFDKLKER-FTPNMMTYTVLLNGWCRVRNLIEAARIWND 322
Query: 353 M--ERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWD 410
M K + + V LL S+K + + + M+ G + Y +++R + K
Sbjct: 323 MIDHGLKPDIVAHNVMLEGLLRSMKKSDAIK-LFHVMKSKGPCPNVRSYTIMIRDFCKQS 381
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ + + +D+M +G PD YT +I G K+ +EM KG + +T
Sbjct: 382 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYN 441
Query: 471 LVISM 475
+I +
Sbjct: 442 ALIKL 446
>At3g62470 putative protein
Length = 599
Score = 218 bits (555), Expect = 6e-57
Identities = 129/399 (32%), Positives = 214/399 (53%), Gaps = 10/399 (2%)
Query: 67 VERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGK 126
+E LD DL+ DL+++VL R R KPA FF WA++ +A SR N ++ IL K
Sbjct: 148 MEAVLDEMKLDLSHDLIVEVLERFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMSILAK 207
Query: 127 MSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDA 186
+FE + V +EM + GL+ +TF+ ++ FAAA + ++A+ +F +++ + ++
Sbjct: 208 TRQFETMVSVLEEMGTK-GLLTMETFTIAMKAFAAAKERKKAVGIFELMKKYKFKIGVET 266
Query: 187 FRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDI 246
LL L R K ++A+ LF F ++ T+ V+LNGWC + N EA R+W D+
Sbjct: 267 INCLLDSLGRAKLGKEAQVLFDKLKERFT--PNMMTYTVLLNGWCRVRNLIEAARIWNDM 324
Query: 247 MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKK 306
+ +PD+ + ++ L + K A+KLF M ++G C P+V +I C +
Sbjct: 325 IDQGLKPDIVAHNVMLEGLLRSRKKSDAIKLFHVMKSKG-PC-PNVRSYTIMIRDFCKQS 382
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
+ A+E F DM + G +P+ A Y LI ++++ VYEL+++M+ K P+ T
Sbjct: 383 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHP--PDGKT 440
Query: 367 YSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEME 423
Y+ L+ + K PE + +M +N S +N++++ Y N + R W+EM
Sbjct: 441 YNALIKLMANQKMPEHATRIYNKMIQNEIEPSIHTFNMIMKSYFMARNYEMGRAVWEEMI 500
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
+ G PD SYT++I G GK ++A RY EM KGM
Sbjct: 501 KKGICPDDNSYTVLIRGLIGEGKSREACRYLEEMLDKGM 539
Score = 68.2 bits (165), Expect = 1e-11
Identities = 72/305 (23%), Positives = 132/305 (42%), Gaps = 15/305 (4%)
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQ-LHRDIKTWNVILNGWCVLG 234
++ LDL D +L R++H F A E Q D +T+N +++ +L
Sbjct: 153 DEMKLDLSHDLIVEVLE---RFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMS---ILA 206
Query: 235 NAHEAKRVWKDI--MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDV 292
+ + + + M +K + T+ +KA + A+ +F M + K V
Sbjct: 207 KTRQFETMVSVLEEMGTKGLLTMETFTIAMKAFAAAKERKKAVGIFELM--KKYKFKIGV 264
Query: 293 VICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVED 352
NC++D+L K EA +F +KER PN+ TY L+ C++R + + + D
Sbjct: 265 ETINCLLDSLGRAKLGKEAQVLFDKLKER-FTPNMMTYTVLLNGWCRVRNLIEAARIWND 323
Query: 353 M--ERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWD 410
M + K + + V LL S K + + + M+ G + Y +++R + K
Sbjct: 324 MIDQGLKPDIVAHNVMLEGLLRSRKKSDAIK-LFHVMKSKGPCPNVRSYTIMIRDFCKQS 382
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ + + +D+M +G PD YT +I G K+ +EM KG + +T
Sbjct: 383 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYN 442
Query: 471 LVISM 475
+I +
Sbjct: 443 ALIKL 447
>At3g49730 putative protein
Length = 1184
Score = 207 bits (526), Expect = 1e-53
Identities = 134/421 (31%), Positives = 220/421 (51%), Gaps = 11/421 (2%)
Query: 66 QVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILG 125
++E AL+ G DL L++ VL R FF WA+K Y + VC ++ IL
Sbjct: 83 KLELALNESGIDLRPGLIIRVLSRCGDAGNLGYRFFLWATKQPGYFHSYEVCKSMVMILS 142
Query: 126 KMSRFEELHQVFDEMSHREG-LVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDL 184
KM +F + + +EM L+ + F L+RRFA+A+ V++A+ + ++GL+ D
Sbjct: 143 KMRQFGAVWGLIEEMRKTNPELIEPELFVVLMRRFASANMVKKAVEVLDEMPKYGLEPDE 202
Query: 185 DAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWK 244
F LL LC+ V++A +F +F +++ + +L GWC G EAK V
Sbjct: 203 YVFGCLLDALCKNGSVKEASKVFEDMREKFP--PNLRYFTSLLYGWCREGKLMEAKEVLV 260
Query: 245 DIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALC- 303
+ + PD+ + + GK+ A L M G +P+V +I ALC
Sbjct: 261 QMKEAGLEPDIVVFTNLLSGYAHAGKMADAYDLMNDMRKRGF--EPNVNCYTVLIQALCR 318
Query: 304 FKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPN 363
+KR+ EA+ VF +M+ GCE ++ TY +LI CK ++K Y +++DM RKKG MP+
Sbjct: 319 TEKRMDEAMRVFVEMERYGCEADIVTYTALISGFCKWGMIDKGYSVLDDM-RKKG-VMPS 376
Query: 364 AVTYSCLLNSLKGPE---EVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWD 420
VTY ++ + + E E ++E+M+R GC IYN+V+RL K + W+
Sbjct: 377 QVTYMQIMVAHEKKEQFEECLELIEKMKRRGCHPDLLIYNVVIRLACKLGEVKEAVRLWN 436
Query: 421 EMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVISMNSPLK 480
EME NG P ++ IMI+G G + +A +F+EM S+G+ + P+ L +N+ ++
Sbjct: 437 EMEANGLSPGVDTFVIMINGFTSQGFLIEACNHFKEMVSRGIFSAPQYGTLKSLLNNLVR 496
Query: 481 E 481
+
Sbjct: 497 D 497
Score = 70.1 bits (170), Expect = 3e-12
Identities = 86/413 (20%), Positives = 159/413 (37%), Gaps = 64/413 (15%)
Query: 125 GKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHK-VEEAISMFYTREQFGLDLD 183
GKM+ + + + ++M R N + ++ L++ K ++EA+ +F E++G + D
Sbjct: 285 GKMA---DAYDLMNDMRKRGFEPNVNCYTVLIQALCRTEKRMDEAMRVFVEMERYGCEAD 341
Query: 184 LDAFRTLLMWLCRYKHVEDAETLF---------------------HSKAREFQ------- 215
+ + L+ C++ ++ ++ H K +F+
Sbjct: 342 IVTYTALISGFCKWGMIDKGYSVLDDMRKKGVMPSQVTYMQIMVAHEKKEQFEECLELIE 401
Query: 216 ------LHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKG 269
H D+ +NV++ C LG EA R+W ++ A+ P + T+ I T +G
Sbjct: 402 KMKRRGCHPDLLIYNVVIRLACKLGEVKEAVRLWNEMEANGLSPGVDTFVIMINGFTSQG 461
Query: 270 KLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQ--DMKERGCEPNV 327
L A F+ M + G P +++ L ++ A +V+ K CE NV
Sbjct: 462 FLIEACNHFKEMVSRGIFSAPQYGTLKSLLNNLVRDDKLEMAKDVWSCISNKTSSCELNV 521
Query: 328 ATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCL---LNSLKGPEEVPGVL 384
+ + I L +++ DM + MP TY+ L LN L +
Sbjct: 522 SAWTIWIHALYAKGHVKEACSYCLDM--MEMDLMPQPNTYAKLMKGLNKLYNRTIAAEIT 579
Query: 385 ERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNG--CGPDRRSYTIMIHGHY 442
E++ + + ++Y K +D + K + + G G D + Y
Sbjct: 580 EKVVKMASE-----REMSFKMYKKKGEEDLIEKAKPKGNKEGKKKGTDHQRY-------- 626
Query: 443 ENGKMKDAMRYFREMTSKGMVAEPRTEKLVISMNSPL--KERTEKQEGKEEET 493
G++ A R T +A R S +SP K T K EEE+
Sbjct: 627 -KGRVSLARNRLRSETPSSFLARDRLRSKTPS-SSPFSSKRHTPKTSEIEEES 677
>At5g65820 putative protein
Length = 637
Score = 193 bits (490), Expect = 2e-49
Identities = 132/432 (30%), Positives = 216/432 (49%), Gaps = 12/432 (2%)
Query: 55 LLKFRRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTS 114
L KF P ++E AL+ G +L L+ VL R FF WA+K Y +
Sbjct: 90 LRKFHSRVP--KLELALNESGVELRPGLIERVLNRCGDAGNLGYRFFVWAAKQPRYCHSI 147
Query: 115 RVCNEILDILGKMSRFEELHQVFDEM-SHREGLVNEDTFSTLLRRFAAAHKVEEAISMFY 173
V ++ IL KM +F + + +EM L+ + F L++RFA+A V++AI +
Sbjct: 148 EVYKSMVKILSKMRQFGAVWGLIEEMRKENPQLIEPELFVVLVQRFASADMVKKAIEVLD 207
Query: 174 TREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVL 233
+FG + D F LL LC++ V+DA LF F ++ ++ + +L GWC +
Sbjct: 208 EMPKFGFEPDEYVFGCLLDALCKHGSVKDAAKLFEDMRMRFPVN--LRYFTSLLYGWCRV 265
Query: 234 GNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVV 293
G EAK V + + PD+ Y + GK+ A L R M G +P+
Sbjct: 266 GKMMEAKYVLVQMNEAGFEPDIVDYTNLLSGYANAGKMADAYDLLRDMRRRGF--EPNAN 323
Query: 294 ICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDM 353
+I ALC R+ EA++VF +M+ CE +V TY +L+ CK +++K Y +++DM
Sbjct: 324 CYTVLIQALCKVDRMEEAMKVFVEMERYECEADVVTYTALVSGFCKWGKIDKCYIVLDDM 383
Query: 354 ERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMER-NGCSLSDDI--YNLVLRLYMKWD 410
+K MP+ +TY ++ + + E LE ME+ DI YN+V+RL K
Sbjct: 384 IKK--GLMPSELTYMHIMVAHEKKESFEECLELMEKMRQIEYHPDIGIYNVVIRLACKLG 441
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ W+EME NG P ++ IMI+G G + +A +F+EM ++G+ + +
Sbjct: 442 EVKEAVRLWNEMEENGLSPGVDTFVIMINGLASQGCLLEASDHFKEMVTRGLFSVSQYGT 501
Query: 471 LVISMNSPLKER 482
L + +N+ LK++
Sbjct: 502 LKLLLNTVLKDK 513
Score = 70.5 bits (171), Expect = 2e-12
Identities = 74/323 (22%), Positives = 134/323 (40%), Gaps = 16/323 (4%)
Query: 110 YAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAI 169
+ P + ++ L K+ R EE +VF EM E + T++ L+ F K+++
Sbjct: 318 FEPNANCYTVLIQALCKVDRMEEAMKVFVEMERYECEADVVTYTALVSGFCKWGKIDKCY 377
Query: 170 SMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFH--SKAREFQLHRDIKTWNVIL 227
+ + GL + + T + + ++ E E K R+ + H DI +NV++
Sbjct: 378 IVLDDMIKKGL---MPSELTYMHIMVAHEKKESFEECLELMEKMRQIEYHPDIGIYNVVI 434
Query: 228 NGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCN 287
C LG EA R+W ++ + P + T+ I L +G L A F+ M G
Sbjct: 435 RLACKLGEVKEAVRLWNEMEENGLSPGVDTFVIMINGLASQGCLLEASDHFKEMVTRGLF 494
Query: 288 CKPDVVICNCIIDALCFKKRVPEALEVFQDMKERG-CEPNVATYNSLIKHL-CKIRRMEK 345
+++ + K++ A +V+ + +G CE NV ++ I L K E
Sbjct: 495 SVSQYGTLKLLLNTVLKDKKLEMAKDVWSCITSKGACELNVLSWTIWIHALFSKGYEKEA 554
Query: 346 VYELVEDMERKKGSCMPNAVTYSCLLNSLKG--PEEVPGVLERMERNGCSLSDDIYNLVL 403
+E +E MP T++ L+ LK E G + RN + + +
Sbjct: 555 CSYCIEMIEM---DFMPQPDTFAKLMKGLKKLYNREFAGEITEKVRNMAAERE----MSF 607
Query: 404 RLYMKWDNQDGLRKTWDEMERNG 426
++Y + QD K + +R G
Sbjct: 608 KMYKRRGVQDLTEKAKSKQDREG 630
>At1g77360
Length = 481
Score = 193 bits (490), Expect = 2e-49
Identities = 117/427 (27%), Positives = 219/427 (50%), Gaps = 9/427 (2%)
Query: 38 LHSQPEHAAGAGAFVQNLLKFRRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPA 97
L+S E +N+ K P ++ ALD G ++ ++V DVL R R+
Sbjct: 22 LYSSSEQVRDVADVAKNISKVLMSSPQLVLDSALDQSGLRVSQEVVEDVLNRFRNAGLLT 81
Query: 98 LVFFNWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLR 157
FF W+ K Y + R + +++ K+ +++ + + + M ++ ++N +TF ++R
Sbjct: 82 YRFFQWSEKQRHYEHSVRAYHMMIESTAKIRQYKLMWDLINAMRKKK-MLNVETFCIVMR 140
Query: 158 RFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLH 217
++A A KV+EAI F E++ L +L AF LL LC+ K+V A+ +F + F
Sbjct: 141 KYARAQKVDEAIYAFNVMEKYDLPPNLVAFNGLLSALCKSKNVRKAQEVFENMRDRFT-- 198
Query: 218 RDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKL 277
D KT++++L GW N +A+ V+++++ + C PD+ TY+ + L K G++ AL +
Sbjct: 199 PDSKTYSILLEGWGKEPNLPKAREVFREMIDAGCHPDIVTYSIMVDILCKAGRVDEALGI 258
Query: 278 FRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHL 337
R M + CKP I + ++ + R+ EA++ F +M+ G + +VA +NSLI
Sbjct: 259 VRSM--DPSICKPTTFIYSVLVHTYGTENRLEEAVDTFLEMERSGMKADVAVFNSLIGAF 316
Query: 338 CKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL--KGPEEVPGVLERMERNGCSLS 395
CK RM+ VY ++++M+ K PN+ + + +L L +G ++ + R C
Sbjct: 317 CKANRMKNVYRVLKEMKSK--GVTPNSKSCNIILRHLIERGEKDEAFDVFRKMIKVCEPD 374
Query: 396 DDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFR 455
D Y +V++++ + + K W M + G P +++++I+G E + A
Sbjct: 375 ADTYTMVIKMFCEKKEMETADKVWKYMRKKGVFPSMHTFSVLINGLCEERTTQKACVLLE 434
Query: 456 EMTSKGM 462
EM G+
Sbjct: 435 EMIEMGI 441
Score = 84.7 bits (208), Expect = 1e-16
Identities = 69/253 (27%), Positives = 125/253 (49%), Gaps = 13/253 (5%)
Query: 240 KRVWKDI--MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNC 297
K +W I M K ++ T+ ++ + K+ A+ F M E + P++V N
Sbjct: 115 KLMWDLINAMRKKKMLNVETFCIVMRKYARAQKVDEAIYAFNVM--EKYDLPPNLVAFNG 172
Query: 298 IIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKK 357
++ ALC K V +A EVF++M++R P+ TY+ L++ K + K E+ +M
Sbjct: 173 LLSALCKSKNVRKAQEVFENMRDR-FTPDSKTYSILLEGWGKEPNLPKAREVFREMI--D 229
Query: 358 GSCMPNAVTYSCLLNSLKGP---EEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDG 414
C P+ VTYS +++ L +E G++ M+ + C + IY++++ Y + +
Sbjct: 230 AGCHPDIVTYSIMVDILCKAGRVDEALGIVRSMDPSICKPTTFIYSVLVHTYGTENRLEE 289
Query: 415 LRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVIS 474
T+ EMER+G D + +I + +MK+ R +EM SKG+ ++ +++
Sbjct: 290 AVDTFLEMERSGMKADVAVFNSLIGAFCKANRMKNVYRVLKEMKSKGVTPNSKSCNIIL- 348
Query: 475 MNSPLKERTEKQE 487
L ER EK E
Sbjct: 349 --RHLIERGEKDE 359
Score = 43.1 bits (100), Expect = 3e-04
Identities = 31/153 (20%), Positives = 70/153 (45%), Gaps = 2/153 (1%)
Query: 116 VCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTR 175
V N ++ K +R + +++V EM + N + + +LR + +EA +F
Sbjct: 308 VFNSLIGAFCKANRMKNVYRVLKEMKSKGVTPNSKSCNIILRHLIERGEKDEAFDVFRKM 367
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGN 235
+ + D D + ++ C K +E A+ ++ R+ + + T++V++NG C
Sbjct: 368 IKV-CEPDADTYTMVIKMFCEKKEMETADKVW-KYMRKKGVFPSMHTFSVLINGLCEERT 425
Query: 236 AHEAKRVWKDIMASKCRPDLFTYATFIKALTKK 268
+A + ++++ RP T+ + L K+
Sbjct: 426 TQKACVLLEEMIEMGIRPSGVTFGRLRQLLIKE 458
>At1g71060 hypothetical protein
Length = 510
Score = 181 bits (460), Expect = 6e-46
Identities = 116/397 (29%), Positives = 199/397 (49%), Gaps = 6/397 (1%)
Query: 66 QVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILG 125
+VE L+ L+ L+ +VL++ + AL F WA + T+ N +++ LG
Sbjct: 80 KVETLLNEASVKLSPALIEEVLKKLSNAGVLALSVFKWAENQKGFKHTTSNYNALIESLG 139
Query: 126 KMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLD 185
K+ +F+ + + D+M ++ L++++TF+ + RR+A A KV+EAI F+ E+FG ++
Sbjct: 140 KIKQFKLIWSLVDDMKAKK-LLSKETFALISRRYARARKVKEAIGAFHKMEEFGFKMESS 198
Query: 186 AFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKD 245
F +L L + ++V DA+ +F K ++ + DIK++ ++L GW N V ++
Sbjct: 199 DFNRMLDTLSKSRNVGDAQKVF-DKMKKKRFEPDIKSYTILLEGWGQELNLLRVDEVNRE 257
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
+ PD+ Y I A K K A++ F M E NCKP I +I+ L +
Sbjct: 258 MKDEGFEPDVVAYGIIINAHCKAKKYEEAIRFFNEM--EQRNCKPSPHIFCSLINGLGSE 315
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAV 365
K++ +ALE F+ K G TYN+L+ C +RME Y+ V++M K PNA
Sbjct: 316 KKLNDALEFFERSKSSGFPLEAPTYNALVGAYCWSQRMEDAYKTVDEMRLK--GVGPNAR 373
Query: 366 TYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERN 425
TY +L+ L + E + C + Y +++R++ + D K WDEM+
Sbjct: 374 TYDIILHHLIRMQRSKEAYEVYQTMSCEPTVSTYEIMVRMFCNKERLDMAIKIWDEMKGK 433
Query: 426 GCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
G P ++ +I K+ +A YF EM G+
Sbjct: 434 GVLPGMHMFSSLITALCHENKLDEACEYFNEMLDVGI 470
Score = 81.6 bits (200), Expect = 9e-16
Identities = 61/254 (24%), Positives = 118/254 (46%), Gaps = 6/254 (2%)
Query: 102 NWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAA 161
N K + + P I++ K ++EE + F+EM R + F +L+ +
Sbjct: 255 NREMKDEGFEPDVVAYGIIINAHCKAKKYEEAIRFFNEMEQRNCKPSPHIFCSLINGLGS 314
Query: 162 AHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIK 221
K+ +A+ F + G L+ + L+ C + +EDA + R + + +
Sbjct: 315 EKKLNDALEFFERSKSSGFPLEAPTYNALVGAYCWSQRMEDAYKTV-DEMRLKGVGPNAR 373
Query: 222 TWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGM 281
T+++IL+ + + EA V++ + C P + TY ++ K +L A+K++ M
Sbjct: 374 TYDIILHHLIRMQRSKEAYEVYQTM---SCEPTVSTYEIMVRMFCNKERLDMAIKIWDEM 430
Query: 282 WNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIR 341
+G P + + + +I ALC + ++ EA E F +M + G P ++ L + L
Sbjct: 431 KGKGV--LPGMHMFSSLITALCHENKLDEACEYFNEMLDVGIRPPGHMFSRLKQTLLDEG 488
Query: 342 RMEKVYELVEDMER 355
R +KV +LV M+R
Sbjct: 489 RKDKVTDLVVKMDR 502
Score = 56.2 bits (134), Expect = 4e-08
Identities = 40/172 (23%), Positives = 77/172 (44%), Gaps = 7/172 (4%)
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAV 365
++V EA+ F M+E G + + +N ++ L K R + ++ + M++K+ P+
Sbjct: 176 RKVKEAIGAFHKMEEFGFKMESSDFNRMLDTLSKSRNVGDAQKVFDKMKKKRFE--PDIK 233
Query: 366 TYSCLLNSLKGPE----EVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDE 421
+Y+ LL G E V V M+ G Y +++ + K + + ++E
Sbjct: 234 SYTILLEGW-GQELNLLRVDEVNREMKDEGFEPDVVAYGIIINAHCKAKKYEEAIRFFNE 292
Query: 422 MERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVI 473
ME+ C P + +I+G K+ DA+ +F S G E T ++
Sbjct: 293 MEQRNCKPSPHIFCSLINGLGSEKKLNDALEFFERSKSSGFPLEAPTYNALV 344
>At3g22670 hypothetical protein
Length = 562
Score = 177 bits (449), Expect = 1e-44
Identities = 121/425 (28%), Positives = 212/425 (49%), Gaps = 17/425 (4%)
Query: 59 RRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCN 118
++D + V + L C + + LVL VLRR + W A FF WA+ Y + N
Sbjct: 111 KKDTSHEDVVKELSKCDVVVTESLVLQVLRRFSNGWNQAYGFFIWANSQTGYVHSGHTYN 170
Query: 119 EILDILGKMSRFEELHQVFDEMSHRE--GLVNEDTFSTLLRRFAAAHKVEEAISMFYTRE 176
++D+LGK F+ + ++ +EM+ E LV DT S ++RR A + K +A+ F E
Sbjct: 171 AMVDVLGKCRNFDLMWELVNEMNKNEESKLVTLDTMSKVMRRLAKSGKYNKAVDAFLEME 230
Query: 177 Q-FGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQ-LHRDIKTWNVILNGWCVLG 234
+ +G+ D A +L+ L + +E A +F + F + D +T+N++++G+C
Sbjct: 231 KSYGVKTDTIAMNSLMDALVKENSIEHAHEVF---LKLFDTIKPDARTFNILIHGFCKAR 287
Query: 235 NAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVI 294
+A+ + + ++ PD+ TY +F++A K+G ++ M GCN P+VV
Sbjct: 288 KFDDARAMMDLMKVTEFTPDVVTYTSFVEAYCKEGDFRRVNEMLEEMRENGCN--PNVVT 345
Query: 295 CNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDME 354
++ +L K+V EAL V++ MKE GC P+ Y+SLI L K R + E+ EDM
Sbjct: 346 YTIVMHSLGKSKQVAEALGVYEKMKEDGCVPDAKFYSSLIHILSKTGRFKDAAEIFEDMT 405
Query: 355 RKKGSCMPNAVTYSCLLNSL---KGPEEVPGVLERM---ERNGCSLSDDIYNLVLRLYMK 408
+ + + Y+ ++++ E +L+RM E CS + + Y +L++
Sbjct: 406 NQ--GVRRDVLVYNTMISAALHHSRDEMALRLLKRMEDEEGESCSPNVETYAPLLKMCCH 463
Query: 409 WDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRT 468
L M +N D +Y ++I G +GK+++A +F E KGMV T
Sbjct: 464 KKKMKLLGILLHHMVKNDVSIDVSTYILLIRGLCMSGKVEEACLFFEEAVRKGMVPRDST 523
Query: 469 EKLVI 473
K+++
Sbjct: 524 CKMLV 528
Score = 64.3 bits (155), Expect = 1e-10
Identities = 61/278 (21%), Positives = 115/278 (40%), Gaps = 44/278 (15%)
Query: 222 TWNVILNGWCVLGNAHEAKRVWKDIMASKCRPD-----LFTYATFIKALTKKGKLGTALK 276
T+N +++ VLG +W+ + + L T + ++ L K GK A+
Sbjct: 168 TYNAMVD---VLGKCRNFDLMWELVNEMNKNEESKLVTLDTMSKVMRRLAKSGKYNKAVD 224
Query: 277 LFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKH 336
F M + K D + N ++DAL + + A EVF + + +P+ T+N LI
Sbjct: 225 AFLEM-EKSYGVKTDTIAMNSLMDALVKENSIEHAHEVFLKLFDT-IKPDARTFNILIHG 282
Query: 337 LCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSD 396
CK R+ + +++ M K P+ VTY+ + +
Sbjct: 283 FCKARKFDDARAMMDLM--KVTEFTPDVVTYTSFVEA----------------------- 317
Query: 397 DIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFRE 456
Y K + + + +EM NGC P+ +YTI++H ++ ++ +A+ + +
Sbjct: 318 ---------YCKEGDFRRVNEMLEEMRENGCNPNVVTYTIVMHSLGKSKQVAEALGVYEK 368
Query: 457 MTSKGMVAEPRTEKLVISMNSPLKERTEKQEGKEEETN 494
M G V + + +I + S + E E+ TN
Sbjct: 369 MKEDGCVPDAKFYSSLIHILSKTGRFKDAAEIFEDMTN 406
>At1g20300 putative protein
Length = 537
Score = 171 bits (434), Expect = 7e-43
Identities = 104/376 (27%), Positives = 190/376 (49%), Gaps = 10/376 (2%)
Query: 89 RHRSDWKPALVFFNWASKADSYAPTS-RVCNEILDILGKMSRFEELHQVFDEMSHREGLV 147
RH +L FFNWA+ D Y S NE++D+ GK+ +F+ + D M R +
Sbjct: 125 RHGIPLHQSLAFFNWATSRDDYDHKSPHPYNEMIDLSGKVRQFDLAWHLIDLMKSRNVEI 184
Query: 148 NEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLF 207
+ +TF+ L+RR+ A EA+ F E +G D AF ++ L R + +A++ F
Sbjct: 185 SIETFTILIRRYVRAGLASEAVHCFNRMEDYGCVPDKIAFSIVISNLSRKRRASEAQSFF 244
Query: 208 HSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTK 267
S F+ D+ + ++ GWC G EA++V+K++ + P+++TY+ I AL +
Sbjct: 245 DSLKDRFE--PDVIVYTNLVRGWCRAGEISEAEKVFKEMKLAGIEPNVYTYSIVIDALCR 302
Query: 268 KGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNV 327
G++ A +F M + G C P+ + N ++ R + L+V+ MK+ GCEP+
Sbjct: 303 CGQISRAHDVFADMLDSG--CAPNAITFNNLMRVHVKAGRTEKVLQVYNQMKKLGCEPDT 360
Query: 328 ATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPG---VL 384
TYN LI+ C+ +E +++ M +KK C NA T++ + ++ +V G +
Sbjct: 361 ITYNFLIEAHCRDENLENAVKVLNTMIKKK--CEVNASTFNTIFRYIEKKRDVNGAHRMY 418
Query: 385 ERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYEN 444
+M C + YN+++R+++ + D + K EM+ P+ +Y +++
Sbjct: 419 SKMMEAKCEPNTVTYNILMRMFVGSKSTDMVLKMKKEMDDKEVEPNVNTYRLLVTMFCGM 478
Query: 445 GKMKDAMRYFREMTSK 460
G +A + F+EM +
Sbjct: 479 GHWNNAYKLFKEMVEE 494
Score = 70.1 bits (170), Expect = 3e-12
Identities = 61/291 (20%), Positives = 121/291 (40%), Gaps = 40/291 (13%)
Query: 120 ILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFG 179
++ L + R E FD + R + ++ L+R + A ++ EA +F + G
Sbjct: 227 VISNLSRKRRASEAQSFFDSLKDRFE-PDVIVYTNLVRGWCRAGEISEAEKVFKEMKLAG 285
Query: 180 LDLDLDAFRTLLMWLCRYKHVEDAETLF---------------------HSKA----REF 214
++ ++ + ++ LCR + A +F H KA +
Sbjct: 286 IEPNVYTYSIVIDALCRCGQISRAHDVFADMLDSGCAPNAITFNNLMRVHVKAGRTEKVL 345
Query: 215 QLHRDIK---------TWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKAL 265
Q++ +K T+N ++ C N A +V ++ KC + T+ T + +
Sbjct: 346 QVYNQMKKLGCEPDTITYNFLIEAHCRDENLENAVKVLNTMIKKKCEVNASTFNTIFRYI 405
Query: 266 TKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEP 325
KK + A +++ M C+P+ V N ++ K L++ ++M ++ EP
Sbjct: 406 EKKRDVNGAHRMYSKMME--AKCEPNTVTYNILMRMFVGSKSTDMVLKMKKEMDDKEVEP 463
Query: 326 NVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCM-PNAVTYSCLLNSLK 375
NV TY L+ C + Y+L ++M +K C+ P+ Y +L L+
Sbjct: 464 NVNTYRLLVTMFCGMGHWNNAYKLFKEMVEEK--CLTPSLSLYEMVLAQLR 512
>At1g52640
Length = 523
Score = 171 bits (433), Expect = 9e-43
Identities = 120/405 (29%), Positives = 198/405 (48%), Gaps = 10/405 (2%)
Query: 63 PTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILD 122
P D +E L ++ +LV VL+R ++ PA FF WA + +A + + +++
Sbjct: 51 PKDDLEHTLVAYSPRVSSNLVEQVLKRCKNLGFPAHRFFLWARRIPDFAHSLESYHILVE 110
Query: 123 ILGKMSRFEELHQ-VFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLD 181
ILG +F L + + + ++ F + R ++ A+ EA F +FG+
Sbjct: 111 ILGSSKQFALLWDFLIEAREYNYFEISSKVFWIVFRAYSRANLPSEACRAFNRMVEFGIK 170
Query: 182 LDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKR 241
+D LL LC KHV A+ F KA+ F + KT+++++ GW + +A A++
Sbjct: 171 PCVDDLDQLLHSLCDKKHVNHAQEFF-GKAKGFGIVPSAKTYSILVRGWARIRDASGARK 229
Query: 242 VWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDA 301
V+ +++ C DL Y + AL K G + K+F+ M N G KPD I A
Sbjct: 230 VFDEMLERNCVVDLLAYNALLDALCKSGDVDGGYKMFQEMGNLG--LKPDAYSFAIFIHA 287
Query: 302 LCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCM 361
C V A +V MK PNV T+N +IK LCK +++ Y L+++M +K +
Sbjct: 288 YCDAGDVHSAYKVLDRMKRYDLVPNVYTFNHIIKTLCKNEKVDDAYLLLDEMIQKGAN-- 345
Query: 362 PNAVTYSCLLNSLKGPEEV---PGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKT 418
P+ TY+ ++ EV +L RM+R C YN+VL+L ++ D +
Sbjct: 346 PDTWTYNSIMAYHCDHCEVNRATKLLSRMDRTKCLPDRHTYNMVLKLLIRIGRFDRATEI 405
Query: 419 WDEMERNGCGPDRRSYTIMIHGHY-ENGKMKDAMRYFREMTSKGM 462
W+ M P +YT+MIHG + GK+++A RYF M +G+
Sbjct: 406 WEGMSERKFYPTVATYTVMIHGLVRKKGKLEEACRYFEMMIDEGI 450
Score = 55.1 bits (131), Expect = 9e-08
Identities = 45/167 (26%), Positives = 67/167 (39%), Gaps = 5/167 (2%)
Query: 310 EALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSC 369
EA F M E G +P V + L+ LC + + E + K +P+A TYS
Sbjct: 156 EACRAFNRMVEFGIKPCVDDLDQLLHSLCDKKHVNHAQEFFG--KAKGFGIVPSAKTYSI 213
Query: 370 LLNSLKGPEEVPG---VLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNG 426
L+ + G V + M C + YN +L K + DG K + EM G
Sbjct: 214 LVRGWARIRDASGARKVFDEMLERNCVVDLLAYNALLDALCKSGDVDGGYKMFQEMGNLG 273
Query: 427 CGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVI 473
PD S+ I IH + + G + A + M +V T +I
Sbjct: 274 LKPDAYSFAIFIHAYCDAGDVHSAYKVLDRMKRYDLVPNVYTFNHII 320
>At1g73400 unknown protein
Length = 466
Score = 166 bits (420), Expect = 3e-41
Identities = 110/409 (26%), Positives = 202/409 (48%), Gaps = 17/409 (4%)
Query: 65 DQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDIL 124
D +E+ALD DL +V +L+R + + K A FF WA + Y+ NE++DIL
Sbjct: 8 DDMEKALDESSVDLTTPVVCKILQRLQYEEKTAFRFFTWAGHQEHYSHEPIAYNEMIDIL 67
Query: 125 G----KMSRFEELHQVFDEMS-HREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFG 179
K +F + + D M + + +V D +LR++ + + F R++
Sbjct: 68 SSTKYKNKQFRIVIDMLDYMKRNNKTVVLVDVLLEILRKYCERYLTH--VQKFAKRKRIR 125
Query: 180 LDL--DLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAH 237
+ +++AF LL LC+ V++ E L + ++ D T+NV+ GWC + +
Sbjct: 126 VKTQPEINAFNMLLDALCKCGLVKEGEALL--RRMRHRVKPDANTFNVLFFGWCRVRDPK 183
Query: 238 EAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCK-PDVVICN 296
+A ++ ++++ + +P+ FTY I + G + A LF M +G P
Sbjct: 184 KAMKLLEEMIEAGHKPENFTYCAAIDTFCQAGMVDEAADLFDFMITKGSAVSAPTAKTFA 243
Query: 297 CIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERK 356
+I AL + E E+ M GC P+V+TY +I+ +C ++++ Y+ +++M K
Sbjct: 244 LMIVALAKNDKAEECFELIGRMISTGCLPDVSTYKDVIEGMCMAEKVDEAYKFLDEMSNK 303
Query: 357 KGSCMPNAVTYSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQD 413
P+ VTY+C L L + +E + RM + C+ S YN+++ ++ + D+ D
Sbjct: 304 --GYPPDIVTYNCFLRVLCENRKTDEALKLYGRMVESRCAPSVQTYNMLISMFFEMDDPD 361
Query: 414 GLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
G TW EM++ C D +Y MI+G ++ + K+A E+ +KG+
Sbjct: 362 GAFNTWTEMDKRDCVQDVETYCAMINGLFDCHRAKEACFLLEEVVNKGL 410
Score = 68.6 bits (166), Expect = 8e-12
Identities = 63/255 (24%), Positives = 108/255 (41%), Gaps = 9/255 (3%)
Query: 226 ILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEG 285
IL +C H K + + K +P++ + + AL K G + L R M +
Sbjct: 103 ILRKYCERYLTHVQKFAKRKRIRVKTQPEINAFNMLLDALCKCGLVKEGEALLRRMRHR- 161
Query: 286 CNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEK 345
KPD N + C + +A+++ ++M E G +P TY + I C+ +++
Sbjct: 162 --VKPDANTFNVLFFGWCRVRDPKKAMKLLEEMIEAGHKPENFTYCAAIDTFCQAGMVDE 219
Query: 346 VYELVEDMERKKGSCM--PNAVTYSCLLNSLKG---PEEVPGVLERMERNGCSLSDDIYN 400
+L + M K GS + P A T++ ++ +L EE ++ RM GC Y
Sbjct: 220 AADLFDFMITK-GSAVSAPTAKTFALMIVALAKNDKAEECFELIGRMISTGCLPDVSTYK 278
Query: 401 LVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSK 460
V+ + D K DEM G PD +Y + EN K +A++ + M
Sbjct: 279 DVIEGMCMAEKVDEAYKFLDEMSNKGYPPDIVTYNCFLRVLCENRKTDEALKLYGRMVES 338
Query: 461 GMVAEPRTEKLVISM 475
+T ++ISM
Sbjct: 339 RCAPSVQTYNMLISM 353
>At5g43820 putative protein
Length = 680
Score = 164 bits (415), Expect = 1e-40
Identities = 115/446 (25%), Positives = 208/446 (45%), Gaps = 18/446 (4%)
Query: 49 GAFVQNLLKFRRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKAD 108
G F+Q L K ++++L G L+ D+V DVL R + + FF+WA +
Sbjct: 92 GVFLQKL------KGKSAIQKSLSSLGIGLSIDIVADVLNRGNLSGEAMVTFFDWAVREP 145
Query: 109 SYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEA 168
+ IL LG+ F + V M + + + + F H V A
Sbjct: 146 GVTKDVGSYSVILRALGRRKLFSFMMDVLKGMVCEGVNPDLECLTIAMDSFVRVHYVRRA 205
Query: 169 ISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILN 228
I +F E FG+ ++F LL LC HV A+++F++K D ++N++++
Sbjct: 206 IELFEESESFGVKCSTESFNALLRCLCERSHVSAAKSVFNAKKGNIPF--DSCSYNIMIS 263
Query: 229 GWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNC 288
GW LG E ++V K+++ S PD +Y+ I+ L + G++ ++++F + ++G
Sbjct: 264 GWSKLGEVEEMEKVLKEMVESGFGPDCLSYSHLIEGLGRTGRINDSVEIFDNIKHKG--N 321
Query: 289 KPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYE 348
PD + N +I + E++ ++ M + CEPN+ TY+ L+ L K R++ E
Sbjct: 322 VPDANVYNAMICNFISARDFDESMRYYRRMLDEECEPNLETYSKLVSGLIKGRKVSDALE 381
Query: 349 LVEDMERKKGSCMPNAVTYSCL--LNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLY 406
+ E+M +G + S L L S P + ++ + GC +S+ Y L+L+
Sbjct: 382 IFEEM-LSRGVLPTTGLVTSFLKPLCSYGPPHAAMVIYQKSRKAGCRISESAYKLLLKRL 440
Query: 407 MKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEP 466
++ L WDEM+ +G D Y ++ G G +++A+ E KG
Sbjct: 441 SRFGKCGMLLNVWDEMQESGYPSDVEVYEYIVDGLCIIGHLENAVLVMEEAMRKGFC--- 497
Query: 467 RTEKLVISMNSPLKERTEK-QEGKEE 491
+ V S S +++ +K ++GKEE
Sbjct: 498 -PNRFVYSRLSIIRKWQQKFRKGKEE 522
>At1g74900 hypothetical protein
Length = 482
Score = 160 bits (406), Expect = 1e-39
Identities = 112/401 (27%), Positives = 191/401 (46%), Gaps = 13/401 (3%)
Query: 81 DLVLDVLRRHRSDWKPALVFFNWASKAD-SYAPTSRVCNEILDILGKMSRFEELHQVFDE 139
+LV VL+R + AL FF++ Y + + +DI ++ + +
Sbjct: 57 NLVNSVLKRLWNHGPKALQFFHFLDNHHREYVHDASSFDLAIDIAARLHLHPTVWSLIHR 116
Query: 140 MSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKH 199
M + TF+ + R+A+A K ++A+ +F + G DL +F T+L LC+ K
Sbjct: 117 MRSLRIGPSPKTFAIVAERYASAGKPDKAVKLFLNMHEHGCFQDLASFNTILDVLCKSKR 176
Query: 200 VEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYA 259
VE A LF + F + D T+NVILNGWC++ +A V K+++ P+L TY
Sbjct: 177 VEKAYELFRALRGRFSV--DTVTYNVILNGWCLIKRTPKALEVLKEMVERGINPNLTTYN 234
Query: 260 TFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMK 319
T +K + G++ A + F M +C+ DVV ++ + A VF +M
Sbjct: 235 TMLKGFFRAGQIRHAWEFFLEMKKR--DCEIDVVTYTTVVHGFGVAGEIKRARNVFDEMI 292
Query: 320 ERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEE 379
G P+VATYN++I+ LCK +E + E+M R+ PN TY+ L+ L E
Sbjct: 293 REGVLPSVATYNAMIQVLCKKDNVENAVVMFEEMVRR--GYEPNVTTYNVLIRGLFHAGE 350
Query: 380 V---PGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTI 436
+++RME GC + YN+++R Y + + +++M C P+ +Y I
Sbjct: 351 FSRGEELMQRMENEGCEPNFQTYNMMIRYYSECSEVEKALGLFEKMGSGDCLPNLDTYNI 410
Query: 437 MIHGHYENGKMKD---AMRYFREMTSKGMVAEPRTEKLVIS 474
+I G + + +D A + EM +G + T V++
Sbjct: 411 LISGMFVRKRSEDMVVAGKLLLEMVERGFIPRKFTFNRVLN 451
Score = 111 bits (277), Expect = 1e-24
Identities = 77/291 (26%), Positives = 139/291 (47%), Gaps = 10/291 (3%)
Query: 205 TLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKA 264
+L H + R ++ KT+ ++ + G +A +++ ++ C DL ++ T +
Sbjct: 112 SLIH-RMRSLRIGPSPKTFAIVAERYASAGKPDKAVKLFLNMHEHGCFQDLASFNTILDV 170
Query: 265 LTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCE 324
L K ++ A +LFR + D V N I++ C KR P+ALEV ++M ERG
Sbjct: 171 LCKSKRVEKAYELFRALRGR---FSVDTVTYNVILNGWCLIKRTPKALEVLKEMVERGIN 227
Query: 325 PNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVP--- 381
PN+ TYN+++K + ++ +E +E KK C + VTY+ +++ E+
Sbjct: 228 PNLTTYNTMLKGFFRAGQIRHAWEFF--LEMKKRDCEIDVVTYTTVVHGFGVAGEIKRAR 285
Query: 382 GVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGH 441
V + M R G S YN ++++ K DN + ++EM R G P+ +Y ++I G
Sbjct: 286 NVFDEMIREGVLPSVATYNAMIQVLCKKDNVENAVVMFEEMVRRGYEPNVTTYNVLIRGL 345
Query: 442 YENGKMKDAMRYFREMTSKGMVAEPRTEKLVISMNSPLKERTEKQEGKEEE 492
+ G+ + M ++G +T ++I S E EK G E+
Sbjct: 346 FHAGEFSRGEELMQRMENEGCEPNFQTYNMMIRYYSECSE-VEKALGLFEK 395
Score = 35.8 bits (81), Expect = 0.056
Identities = 33/122 (27%), Positives = 55/122 (45%), Gaps = 9/122 (7%)
Query: 125 GKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDL 184
G+ SR EEL Q M + N T++ ++R ++ +VE+A+ +F +L
Sbjct: 349 GEFSRGEELMQ---RMENEGCEPNFQTYNMMIRYYSECSEVEKALGLFEKMGSGDCLPNL 405
Query: 185 DAFRTLLMWLCRYKHVED----AETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAK 240
D + L+ + K ED + L R F + T+N +LNG + GN AK
Sbjct: 406 DTYNILISGMFVRKRSEDMVVAGKLLLEMVERGFIPRKF--TFNRVLNGLLLTGNQAFAK 463
Query: 241 RV 242
+
Sbjct: 464 EI 465
>At1g80880 hypothetical protein
Length = 540
Score = 154 bits (388), Expect = 1e-37
Identities = 110/407 (27%), Positives = 199/407 (48%), Gaps = 17/407 (4%)
Query: 70 ALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGKMSR 129
+L+ FDLN D ++ R +W+ A + F W K + C+ ++ +LG +
Sbjct: 113 SLEDSSFDLNHDSFYSLIWELRDEWRLAFLAFKWGEKRG--CDDQKSCDLMIWVLGNHQK 170
Query: 130 FEELHQVFDEMSHREGLVNEDTFSTL---LRRFAAAHKVEEAISMFYTREQFGLDLDLDA 186
F + +M + V++DT + + R+AAA+ +AI F ++F +A
Sbjct: 171 FNIAWCLIRDMFN----VSKDTRKAMFLMMDRYAAANDTSQAIRTFDIMDKFKHTPYDEA 226
Query: 187 FRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWC-VLGNAHEAKRVWKD 245
F+ LL LCR+ H+E AE + + F + D++ +NVILNGWC + + EAKR+W++
Sbjct: 227 FQGLLCALCRHGHIEKAEEFMLASKKLFPV--DVEGFNVILNGWCNIWTDVTEAKRIWRE 284
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
+ P+ +Y+ I +K G L +L+L+ M G P + + N ++ L +
Sbjct: 285 MGNYCITPNKDSYSHMISCFSKVGNLFDSLRLYDEMKKRGL--APGIEVYNSLVYVLTRE 342
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAV 365
EA+++ + + E G +P+ TYNS+I+ LC+ +++ ++ M + S P
Sbjct: 343 DCFDEAMKLMKKLNEEGLKPDSVTYNSMIRPLCEAGKLDVARNVLATMISENLS--PTVD 400
Query: 366 TYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERN 425
T+ L ++ E+ VL +M+ + +++ + L+L K + K W EM+R
Sbjct: 401 TFHAFLEAVNF-EKTLEVLGQMKISDLGPTEETFLLILGKLFKGKQPENALKIWAEMDRF 459
Query: 426 GCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLV 472
+ Y I G G ++ A + EM SKG V P +KL+
Sbjct: 460 EIVANPALYLATIQGLLSCGWLEKAREIYSEMKSKGFVGNPMLQKLL 506
Score = 33.5 bits (75), Expect = 0.28
Identities = 42/216 (19%), Positives = 82/216 (37%), Gaps = 13/216 (6%)
Query: 270 KLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVAT 329
K A L R M+N + + + + ++D +A+ F M + P
Sbjct: 170 KFNIAWCLIRDMFNVSKDTRKAMFL---MMDRYAAANDTSQAIRTFDIMDKFKHTPYDEA 226
Query: 330 YNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL----KGPEEVPGVLE 385
+ L+ LC+ +EK E M K + ++ +LN E +
Sbjct: 227 FQGLLCALCRHGHIEKAEEF---MLASKKLFPVDVEGFNVILNGWCNIWTDVTEAKRIWR 283
Query: 386 RMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENG 445
M + + D Y+ ++ + K N + +DEM++ G P Y +++
Sbjct: 284 EMGNYCITPNKDSYSHMISCFSKVGNLFDSLRLYDEMKKRGLAPGIEVYNSLVYVLTRED 343
Query: 446 KMKDAMRYFREMTSKGMVAEPRTEKLVISMNSPLKE 481
+AM+ +++ +G+ + T SM PL E
Sbjct: 344 CFDEAMKLMKKLNEEGLKPDSVTYN---SMIRPLCE 376
>At1g09900 hypothetical protein
Length = 598
Score = 150 bits (380), Expect = 1e-36
Identities = 100/363 (27%), Positives = 175/363 (47%), Gaps = 11/363 (3%)
Query: 102 NWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAA 161
+++S S+A N L + + EE + + M + + + +TL+R F
Sbjct: 90 HYSSVNSSFALEDVESNNHLRQMVRTGELEEGFKFLENMVYHGNVPDIIPCTTLIRGFCR 149
Query: 162 AHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIK 221
K +A + E G D+ + ++ C+ + +A ++ + D+
Sbjct: 150 LGKTRKAAKILEILEGSGAVPDVITYNVMISGYCKAGEINNALSVLD----RMSVSPDVV 205
Query: 222 TWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGM 281
T+N IL C G +A V ++ C PD+ TY I+A + +G A+KL M
Sbjct: 206 TYNTILRSLCDSGKLKQAMEVLDRMLQRDCYPDVITYTILIEATCRDSGVGHAMKLLDEM 265
Query: 282 WNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIR 341
+ GC PDVV N +++ +C + R+ EA++ DM GC+PNV T+N +++ +C
Sbjct: 266 RDRGCT--PDVVTYNVLVNGICKEGRLDEAIKFLNDMPSSGCQPNVITHNIILRSMCSTG 323
Query: 342 RMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL--KG-PEEVPGVLERMERNGCSLSDDI 398
R +L+ DM RK S P+ VT++ L+N L KG +LE+M ++GC +
Sbjct: 324 RWMDAEKLLADMLRKGFS--PSVVTFNILINFLCRKGLLGRAIDILEKMPQHGCQPNSLS 381
Query: 399 YNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMT 458
YN +L + K D + + M GC PD +Y M+ ++GK++DA+ +++
Sbjct: 382 YNPLLHGFCKEKKMDRAIEYLERMVSRGCYPDIVTYNTMLTALCKDGKVEDAVEILNQLS 441
Query: 459 SKG 461
SKG
Sbjct: 442 SKG 444
Score = 125 bits (313), Expect = 7e-29
Identities = 95/403 (23%), Positives = 168/403 (41%), Gaps = 39/403 (9%)
Query: 109 SYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEA 168
S +P N IL L + ++ +V D M R+ + T++ L+ V A
Sbjct: 199 SVSPDVVTYNTILRSLCDSGKLKQAMEVLDRMLQRDCYPDVITYTILIEATCRDSGVGHA 258
Query: 169 ISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILN 228
+ + G D+ + L+ +C+ +++A F + ++ T N+IL
Sbjct: 259 MKLLDEMRDRGCTPDVVTYNVLVNGICKEGRLDEA-IKFLNDMPSSGCQPNVITHNIILR 317
Query: 229 GWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCN- 287
C G +A+++ D++ P + T+ I L +KG LG A+ + M GC
Sbjct: 318 SMCSTGRWMDAEKLLADMLRKGFSPSVVTFNILINFLCRKGLLGRAIDILEKMPQHGCQP 377
Query: 288 --------------------------------CKPDVVICNCIIDALCFKKRVPEALEVF 315
C PD+V N ++ ALC +V +A+E+
Sbjct: 378 NSLSYNPLLHGFCKEKKMDRAIEYLERMVSRGCYPDIVTYNTMLTALCKDGKVEDAVEIL 437
Query: 316 QDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLK 375
+ +GC P + TYN++I L K + K +L+++M K P+ +TYS L+ L
Sbjct: 438 NQLSSKGCSPVLITYNTVIDGLAKAGKTGKAIKLLDEMRAK--DLKPDTITYSSLVGGLS 495
Query: 376 GPEEVPGVLE---RMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRR 432
+V ++ ER G + +N ++ K D M GC P+
Sbjct: 496 REGKVDEAIKFFHEFERMGIRPNAVTFNSIMLGLCKSRQTDRAIDFLVFMINRGCKPNET 555
Query: 433 SYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVISM 475
SYTI+I G G K+A+ E+ +KG++ + E++ M
Sbjct: 556 SYTILIEGLAYEGMAKEALELLNELCNKGLMKKSSAEQVAGKM 598
Score = 84.7 bits (208), Expect = 1e-16
Identities = 51/215 (23%), Positives = 99/215 (45%), Gaps = 10/215 (4%)
Query: 262 IKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKER 321
++ + + G+L K M G PD++ C +I C + +A ++ + ++
Sbjct: 109 LRQMVRTGELEEGFKFLENMVYHGN--VPDIIPCTTLIRGFCRLGKTRKAAKILEILEGS 166
Query: 322 GCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGP---E 378
G P+V TYN +I CK + +++ M S P+ VTY+ +L SL +
Sbjct: 167 GAVPDVITYNVMISGYCKAGEINNALSVLDRM-----SVSPDVVTYNTILRSLCDSGKLK 221
Query: 379 EVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMI 438
+ VL+RM + C Y +++ + K DEM GC PD +Y +++
Sbjct: 222 QAMEVLDRMLQRDCYPDVITYTILIEATCRDSGVGHAMKLLDEMRDRGCTPDVVTYNVLV 281
Query: 439 HGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVI 473
+G + G++ +A+++ +M S G T +++
Sbjct: 282 NGICKEGRLDEAIKFLNDMPSSGCQPNVITHNIIL 316
>At1g02420 hypothetical protein
Length = 491
Score = 150 bits (379), Expect = 2e-36
Identities = 110/404 (27%), Positives = 200/404 (49%), Gaps = 17/404 (4%)
Query: 66 QVERALDLCGFDLNDDLVLDVLRRHR-SDWKP--ALVFFNWASKADSYAPTSRVCNEILD 122
+++ +L G L+ DL+ VL+R R S P L F+ +AS + +S + +L
Sbjct: 57 ELKESLSSSGIHLSKDLIDRVLKRVRFSHGNPIQTLEFYRYASAIRGFYHSSFSLDTMLY 116
Query: 123 ILGKMSRFEELHQVFDEMSHRE-GLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLD 181
ILG+ +F+++ ++ E ++ L++ T +L R A V + + F+ ++ D
Sbjct: 117 ILGRNRKFDQIWELLIETKRKDRSLISPRTMQVVLGRVAKLCSVRQTVESFWKFKRLVPD 176
Query: 182 L-DLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAK 240
D F LL LC+ K + DA ++HS +FQ D++T+N++L+GW ++ EA+
Sbjct: 177 FFDTACFNALLRTLCQEKSMTDARNVYHSLKHQFQ--PDLQTFNILLSGW---KSSEEAE 231
Query: 241 RVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIID 300
++++ +PD+ TY + I K ++ A KL M E PDV+ +I
Sbjct: 232 AFFEEMKGKGLKPDVVTYNSLIDVYCKDREIEKAYKLIDKMREE--EETPDVITYTTVIG 289
Query: 301 ALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSC 360
L + +A EV ++MKE GC P+VA YN+ I++ C RR+ +LV++M +K
Sbjct: 290 GLGLIGQPDKAREVLKEMKEYGCYPDVAAYNAAIRNFCIARRLGDADKLVDEMVKK--GL 347
Query: 361 MPNAVTYSCLLNSLKGPEEVPGVLE---RMERNGCSLSDDIYNLVLRLYMKWDNQDGLRK 417
PNA TY+ L ++ E RM N C + +++++ + + D +
Sbjct: 348 SPNATTYNLFFRVLSLANDLGRSWELYVRMLGNECLPNTQSCMFLIKMFKRHEKVDMAMR 407
Query: 418 TWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKG 461
W++M G G +++ + K+++A + EM KG
Sbjct: 408 LWEDMVVKGFGSYSLVSDVLLDLLCDLAKVEEAEKCLLEMVEKG 451
Score = 73.9 bits (180), Expect = 2e-13
Identities = 90/398 (22%), Positives = 164/398 (40%), Gaps = 69/398 (17%)
Query: 90 HRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGL-VN 148
H S+++ ++ F + + +D+ P ++ + +M L E G+ ++
Sbjct: 12 HVSNFRLSVSFLHSVALSDAKVPVEEEGDDAETVF-RMINGSNLQVELKESLSSSGIHLS 70
Query: 149 EDTFSTLLRRFAAAH----------KVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYK 198
+D +L+R +H + AI FY F LD T+L L R +
Sbjct: 71 KDLIDRVLKRVRFSHGNPIQTLEFYRYASAIRGFY-HSSFSLD-------TMLYILGRNR 122
Query: 199 HVEDA-ETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEA-KRVWKDIMASKCRPDLF 256
+ E L +K ++ L +T V+L L + + + WK + PD F
Sbjct: 123 KFDQIWELLIETKRKDRSLISP-RTMQVVLGRVAKLCSVRQTVESFWK---FKRLVPDFF 178
Query: 257 TYATF---IKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALE 313
A F ++ L ++ + A ++ + ++ +PD+ N ++ K EA
Sbjct: 179 DTACFNALLRTLCQEKSMTDARNVYHSLKHQ---FQPDLQTFNILLSGW---KSSEEAEA 232
Query: 314 VFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNS 373
F++MK +G +P+V TYNSLI CK R +EK Y+L++ M ++ + P+ +TY+ ++
Sbjct: 233 FFEEMKGKGLKPDVVTYNSLIDVYCKDREIEKAYKLIDKMREEEET--PDVITYTTVIGG 290
Query: 374 LKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRS 433
L G++ + D R+ EM+ GC PD +
Sbjct: 291 L-------GLIGQ-------------------------PDKAREVLKEMKEYGCYPDVAA 318
Query: 434 YTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKL 471
Y I ++ DA + EM KG+ T L
Sbjct: 319 YNAAIRNFCIARRLGDADKLVDEMVKKGLSPNATTYNL 356
Score = 62.8 bits (151), Expect = 4e-10
Identities = 55/268 (20%), Positives = 120/268 (44%), Gaps = 13/268 (4%)
Query: 92 SDWKP---ALVFFNWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVN 148
S WK A FF K P N ++D+ K E+ +++ D+M E +
Sbjct: 222 SGWKSSEEAEAFFE-EMKGKGLKPDVVTYNSLIDVYCKDREIEKAYKLIDKMREEEETPD 280
Query: 149 EDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFH 208
T++T++ + ++A + +++G D+ A+ + C + + DA+ L
Sbjct: 281 VITYTTVIGGLGLIGQPDKAREVLKEMKEYGCYPDVAAYNAAIRNFCIARRLGDADKLVD 340
Query: 209 SKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWK---DIMASKCRPDLFTYATFIKAL 265
++ L + T+N+ VL A++ R W+ ++ ++C P+ + IK
Sbjct: 341 EMVKK-GLSPNATTYNLFFR---VLSLANDLGRSWELYVRMLGNECLPNTQSCMFLIKMF 396
Query: 266 TKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEP 325
+ K+ A++L+ M +G ++ + ++D LC +V EA + +M E+G P
Sbjct: 397 KRHEKVDMAMRLWEDMVVKGFGSYS--LVSDVLLDLLCDLAKVEEAEKCLLEMVEKGHRP 454
Query: 326 NVATYNSLIKHLCKIRRMEKVYELVEDM 353
+ ++ + + + ++V L++ M
Sbjct: 455 SNVSFKRIKLLMELANKHDEVNNLIQKM 482
>At1g62590 putative membrane-associated salt-inducible protein
(AL021637); similar to EST gb|AA728420
Length = 634
Score = 148 bits (374), Expect = 6e-36
Identities = 111/416 (26%), Positives = 192/416 (45%), Gaps = 18/416 (4%)
Query: 70 ALDLCG-------FDLNDDLVLDVLRRHRSDWK--PALVFFNWASKADSYAPTSRVCNEI 120
++DLCG F ++LR D K A+ F K+ P+ N++
Sbjct: 33 SIDLCGMCYWGRAFSSGSGDYREILRNGLHDMKLDDAIGLFGGMVKSRPL-PSIVEFNKL 91
Query: 121 LDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGL 180
L + KM +F+ + + ++M E + T++ L+ F ++ A+++ + G
Sbjct: 92 LSAIAKMKKFDVVISLGEKMQRLEIVHGLYTYNILINCFCRRSQISLALALLGKMMKLGY 151
Query: 181 DLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAK 240
+ + +LL C K + DA L E D T+ +++G + A EA
Sbjct: 152 EPSIVTLSSLLNGYCHGKRISDAVALVDQMV-EMGYRPDTITFTTLIHGLFLHNKASEAV 210
Query: 241 RVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIID 300
+ ++ C+P+L TY + L K+G AL L M E + DVVI N IID
Sbjct: 211 ALVDRMVQRGCQPNLVTYGVVVNGLCKRGDTDLALNLLNKM--EAAKIEADVVIFNTIID 268
Query: 301 ALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSC 360
+LC + V +AL +F++M+ +G PNV TY+SLI LC R +L+ DM KK
Sbjct: 269 SLCKYRHVDDALNLFKEMETKGIRPNVVTYSSLISCLCSYGRWSDASQLLSDMIEKK--I 326
Query: 361 MPNAVTYSCLLNSLKGP---EEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRK 417
PN VT++ L+++ E + + M + YN ++ + D D ++
Sbjct: 327 NPNLVTFNALIDAFVKEGKFVEAEKLYDDMIKRSIDPDIFTYNSLVNGFCMHDRLDKAKQ 386
Query: 418 TWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVI 473
++ M C PD +Y +I G ++ +++D FREM+ +G+V + T +I
Sbjct: 387 MFEFMVSKDCFPDVVTYNTLIKGFCKSKRVEDGTELFREMSHRGLVGDTVTYTTLI 442
Score = 129 bits (323), Expect = 5e-30
Identities = 92/368 (25%), Positives = 171/368 (46%), Gaps = 8/368 (2%)
Query: 110 YAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAI 169
Y P + ++ L ++ E + D M R N T+ ++ + A+
Sbjct: 186 YRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVVNGLCKRGDTDLAL 245
Query: 170 SMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNG 229
++ E ++ D+ F T++ LC+Y+HV+DA LF + + ++ T++ +++
Sbjct: 246 NLLNKMEAAKIEADVVIFNTIIDSLCKYRHVDDALNLFKEMETK-GIRPNVVTYSSLISC 304
Query: 230 WCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCK 289
C G +A ++ D++ K P+L T+ I A K+GK A KL+ M +
Sbjct: 305 LCSYGRWSDASQLLSDMIEKKINPNLVTFNALIDAFVKEGKFVEAEKLYDDMIKRSID-- 362
Query: 290 PDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYEL 349
PD+ N +++ C R+ +A ++F+ M + C P+V TYN+LIK CK +R+E EL
Sbjct: 363 PDIFTYNSLVNGFCMHDRLDKAKQMFEFMVSKDCFPDVVTYNTLIKGFCKSKRVEDGTEL 422
Query: 350 VEDMERKKGSCMPNAVTYSCLLNSLKGP---EEVPGVLERMERNGCSLSDDIYNLVLRLY 406
+M + + + VTY+ L+ L + V ++M +G Y+++L
Sbjct: 423 FREMSHR--GLVGDTVTYTTLIQGLFHDGDCDNAQKVFKQMVSDGVPPDIMTYSILLDGL 480
Query: 407 MKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEP 466
+ + +D M+++ D YT MI G + GK+ D F ++ KG+
Sbjct: 481 CNNGKLEKALEVFDYMQKSEIKLDIYIYTTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNV 540
Query: 467 RTEKLVIS 474
T +IS
Sbjct: 541 VTYNTMIS 548
Score = 116 bits (291), Expect = 2e-26
Identities = 71/256 (27%), Positives = 132/256 (50%), Gaps = 5/256 (1%)
Query: 118 NEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQ 177
N ++D K +F E +++D+M R + T+++L+ F ++++A MF
Sbjct: 334 NALIDAFVKEGKFVEAEKLYDDMIKRSIDPDIFTYNSLVNGFCMHDRLDKAKQMFEFMVS 393
Query: 178 FGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAH 237
D+ + TL+ C+ K VED LF + L D T+ ++ G G+
Sbjct: 394 KDCFPDVVTYNTLIKGFCKSKRVEDGTELFREMSHR-GLVGDTVTYTTLIQGLFHDGDCD 452
Query: 238 EAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNC 297
A++V+K +++ PD+ TY+ + L GKL AL++F M + K D+ I
Sbjct: 453 NAQKVFKQMVSDGVPPDIMTYSILLDGLCNNGKLEKALEVFDYM--QKSEIKLDIYIYTT 510
Query: 298 IIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKK 357
+I+ +C +V + ++F + +G +PNV TYN++I LC R +++ Y L++ M K+
Sbjct: 511 MIEGMCKAGKVDDGWDLFCSLSLKGVKPNVVTYNTMISGLCSKRLLQEAYALLKKM--KE 568
Query: 358 GSCMPNAVTYSCLLNS 373
+PN+ TY+ L+ +
Sbjct: 569 DGPLPNSGTYNTLIRA 584
Score = 62.0 bits (149), Expect = 7e-10
Identities = 51/232 (21%), Positives = 100/232 (42%), Gaps = 7/232 (3%)
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
++ S+ P + + + A+ K K + L M + + N +I+ C +
Sbjct: 76 MVKSRPLPSIVEFNKLLSAIAKMKKFDVVISLGEKM--QRLEIVHGLYTYNILINCFCRR 133
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAV 365
++ AL + M + G EP++ T +SL+ C +R+ LV+ M P+ +
Sbjct: 134 SQISLALALLGKMMKLGYEPSIVTLSSLLNGYCHGKRISDAVALVDQMVEM--GYRPDTI 191
Query: 366 TYSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEM 422
T++ L++ L E +++RM + GC + Y +V+ K + D ++M
Sbjct: 192 TFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVVNGLCKRGDTDLALNLLNKM 251
Query: 423 ERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVIS 474
E D + +I + + DA+ F+EM +KG+ T +IS
Sbjct: 252 EAAKIEADVVIFNTIIDSLCKYRHVDDALNLFKEMETKGIRPNVVTYSSLIS 303
Score = 51.2 bits (121), Expect = 1e-06
Identities = 60/253 (23%), Positives = 103/253 (39%), Gaps = 42/253 (16%)
Query: 65 DQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDIL 124
D ++R++D F N LV R D A F + D + P N ++
Sbjct: 355 DMIKRSIDPDIFTYNS-LVNGFCMHDRLD--KAKQMFEFMVSKDCF-PDVVTYNTLIKGF 410
Query: 125 GKMSRFEELHQVFDEMSHREGLVNED---------------------------------- 150
K R E+ ++F EMSHR GLV +
Sbjct: 411 CKSKRVEDGTELFREMSHR-GLVGDTVTYTTLIQGLFHDGDCDNAQKVFKQMVSDGVPPD 469
Query: 151 --TFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFH 208
T+S LL K+E+A+ +F ++ + LD+ + T++ +C+ V+D LF
Sbjct: 470 IMTYSILLDGLCNNGKLEKALEVFDYMQKSEIKLDIYIYTTMIEGMCKAGKVDDGWDLFC 529
Query: 209 SKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKK 268
S + + + ++ T+N +++G C EA + K + P+ TY T I+A +
Sbjct: 530 SLSLK-GVKPNVVTYNTMISGLCSKRLLQEAYALLKKMKEDGPLPNSGTYNTLIRAHLRD 588
Query: 269 GKLGTALKLFRGM 281
G + +L R M
Sbjct: 589 GDKAASAELIREM 601
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,866,788
Number of Sequences: 26719
Number of extensions: 537781
Number of successful extensions: 10213
Number of sequences better than 10.0: 493
Number of HSP's better than 10.0 without gapping: 407
Number of HSP's successfully gapped in prelim test: 86
Number of HSP's that attempted gapping in prelim test: 1844
Number of HSP's gapped (non-prelim): 2616
length of query: 495
length of database: 11,318,596
effective HSP length: 103
effective length of query: 392
effective length of database: 8,566,539
effective search space: 3358083288
effective search space used: 3358083288
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0195b.3


