

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0193.6
(491 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
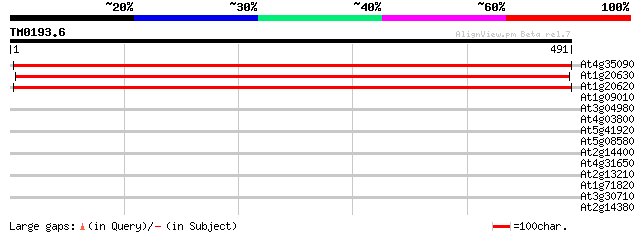
Score E
Sequences producing significant alignments: (bits) Value
At4g35090 catalase 942 0.0
At1g20630 unknown protein 902 0.0
At1g20620 unknown protein 825 0.0
At1g09010 putative Bos beta-mannosidase 35 0.12
At3g04980 hypothetical protein, contains DnaJ motif: prokaryotic... 32 1.0
At4g03800 31 1.4
At5g41920 SCARECROW gene regulator-like protein 30 2.3
At5g08580 Unknown protein (MAH20.14) 30 2.3
At2g14400 putative retroelement pol polyprotein 30 3.0
At4g31650 putative protein 30 4.0
At2g13210 pseudogene 29 5.2
At1g71820 unknown protein (At1g71820) 29 5.2
At3g30710 hypothetical protein 29 6.7
At2g14380 putative retroelement pol polyprotein 29 6.7
>At4g35090 catalase
Length = 492
Score = 942 bits (2434), Expect = 0.0
Identities = 430/488 (88%), Positives = 469/488 (95%)
Query: 4 QHRPSSAFNSPFWTTNSGAPVWNNNNSMTVGVRGPILLEDYHLVEKLANFDRERIPERVV 63
++RP+S++NSPF+TTNSGAPVWNNN+SMTVG RGPILLEDYHLVEKLANFDRERIPERVV
Sbjct: 5 KYRPASSYNSPFFTTNSGAPVWNNNSSMTVGPRGPILLEDYHLVEKLANFDRERIPERVV 64
Query: 64 HARGASAKGFFEVTHDVSHLTCADFLRAPGVQTPIILRFSTVIHERGSPETLRDPRGFAV 123
HARGASAKGFFEVTHD+S+LTCADFLRAPGVQTP+I+RFSTVIHERGSPETLRDPRGFAV
Sbjct: 65 HARGASAKGFFEVTHDISNLTCADFLRAPGVQTPVIVRFSTVIHERGSPETLRDPRGFAV 124
Query: 124 KFYTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHFPESLH 183
KFYTREGNFDLVGNNFPVFF+RDGMKFPDMVHALKPNPKSHIQENWRILDFFSH PESL+
Sbjct: 125 KFYTREGNFDLVGNNFPVFFIRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLN 184
Query: 184 MFTFLFDDVGVPQDYRHMDGFGVNTYTLINKAGKVVYVKFHWKPTCGVKCLLEEEAIKVG 243
MFTFLFDD+G+PQDYRHMDG GVNTY LINKAGK YVKFHWKPTCGVK LLEE+AI+VG
Sbjct: 185 MFTFLFDDIGIPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKPTCGVKSLLEEDAIRVG 244
Query: 244 GSNHSHATQDLYESIAAGNYPEWKLFIQTIDPDHEDRFDFDPLDVTKTWPEDIIPLQPVG 303
G+NHSHATQDLY+SIAAGNYPEWKLFIQ IDP ED+FDFDPLDVTKTWPEDI+PLQPVG
Sbjct: 245 GTNHSHATQDLYDSIAAGNYPEWKLFIQIIDPADEDKFDFDPLDVTKTWPEDILPLQPVG 304
Query: 304 RMVLNKNIDNFFAENEQLAFCPAIVVPGISYSDDKMLQTRIFSYADSQRHRLGPNYLQLP 363
RMVLNKNIDNFFAENEQLAFCPAI+VPGI YSDDK+LQTR+FSYAD+QRHRLGPNYLQLP
Sbjct: 305 RMVLNKNIDNFFAENEQLAFCPAIIVPGIHYSDDKLLQTRVFSYADTQRHRLGPNYLQLP 364
Query: 364 ANAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDPVRHAERYPIPPAICTGSRERCAIEK 423
NAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYD VRHAE+YP PPA+C+G RERC IEK
Sbjct: 365 VNAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDQVRHAEKYPTPPAVCSGKRERCIIEK 424
Query: 424 ENNFKQPGERYRSFAPDRQDRFVRRWVDALSDPRVTHEIRSTWISYWSQADRSLGQKIAS 483
ENNFK+PGERYR+F P+RQ+RF++RW+DALSDPR+THEIRS WISYWSQAD+SLGQK+AS
Sbjct: 425 ENNFKEPGERYRTFTPERQERFIQRWIDALSDPRITHEIRSIWISYWSQADKSLGQKLAS 484
Query: 484 HLNLRPSI 491
LN+RPSI
Sbjct: 485 RLNVRPSI 492
>At1g20630 unknown protein
Length = 492
Score = 902 bits (2332), Expect = 0.0
Identities = 412/485 (84%), Positives = 457/485 (93%)
Query: 6 RPSSAFNSPFWTTNSGAPVWNNNNSMTVGVRGPILLEDYHLVEKLANFDRERIPERVVHA 65
RPSSA +SPF+TTNSGAPVWNNN+S+TVG RGPILLEDYHL+EKLANFDRERIPERVVHA
Sbjct: 7 RPSSAHDSPFFTTNSGAPVWNNNSSLTVGTRGPILLEDYHLLEKLANFDRERIPERVVHA 66
Query: 66 RGASAKGFFEVTHDVSHLTCADFLRAPGVQTPIILRFSTVIHERGSPETLRDPRGFAVKF 125
RGASAKGFFEVTHD++ LT ADFLR PGVQTP+I+RFSTVIHERGSPETLRDPRGFAVKF
Sbjct: 67 RGASAKGFFEVTHDITQLTSADFLRGPGVQTPVIVRFSTVIHERGSPETLRDPRGFAVKF 126
Query: 126 YTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHFPESLHMF 185
YTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSH PESLHMF
Sbjct: 127 YTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLHMF 186
Query: 186 TFLFDDVGVPQDYRHMDGFGVNTYTLINKAGKVVYVKFHWKPTCGVKCLLEEEAIKVGGS 245
+FLFDD+G+PQDYRHM+G GVNTY LINKAGK YVKFHWKPTCG+KCL +EEAI+VGG+
Sbjct: 187 SFLFDDLGIPQDYRHMEGAGVNTYMLINKAGKAHYVKFHWKPTCGIKCLSDEEAIRVGGA 246
Query: 246 NHSHATQDLYESIAAGNYPEWKLFIQTIDPDHEDRFDFDPLDVTKTWPEDIIPLQPVGRM 305
NHSHAT+DLY+SIAAGNYP+W LF+Q +DP HED+FDFDPLDVTK WPEDI+PLQPVGR+
Sbjct: 247 NHSHATKDLYDSIAAGNYPQWNLFVQVMDPAHEDKFDFDPLDVTKIWPEDILPLQPVGRL 306
Query: 306 VLNKNIDNFFAENEQLAFCPAIVVPGISYSDDKMLQTRIFSYADSQRHRLGPNYLQLPAN 365
VLNKNIDNFF ENEQ+AFCPA+VVPGI YSDDK+LQTRIFSYADSQRHRLGPNYLQLP N
Sbjct: 307 VLNKNIDNFFNENEQIAFCPALVVPGIHYSDDKLLQTRIFSYADSQRHRLGPNYLQLPVN 366
Query: 366 APKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDPVRHAERYPIPPAICTGSRERCAIEKEN 425
APKCAHHNNHH+GFMNFMHRDEEVNYFPSR DPVRHAE+YP P +C+G+RE+C I KEN
Sbjct: 367 APKCAHHNNHHDGFMNFMHRDEEVNYFPSRLDPVRHAEKYPTTPIVCSGNREKCFIGKEN 426
Query: 426 NFKQPGERYRSFAPDRQDRFVRRWVDALSDPRVTHEIRSTWISYWSQADRSLGQKIASHL 485
NFKQPGERYRS+ DRQ+RFV+R+V+ALS+PRVTHEIRS WISYWSQAD+SLGQK+A+ L
Sbjct: 427 NFKQPGERYRSWDSDRQERFVKRFVEALSEPRVTHEIRSIWISYWSQADKSLGQKLATRL 486
Query: 486 NLRPS 490
N+RP+
Sbjct: 487 NVRPN 491
>At1g20620 unknown protein
Length = 492
Score = 825 bits (2132), Expect = 0.0
Identities = 376/488 (77%), Positives = 441/488 (90%)
Query: 4 QHRPSSAFNSPFWTTNSGAPVWNNNNSMTVGVRGPILLEDYHLVEKLANFDRERIPERVV 63
++RPSSA+N+PF+TTN GAPV NN +S+T+G RGP+LLEDYHL+EK+ANF RERIPERVV
Sbjct: 5 KYRPSSAYNAPFYTTNGGAPVSNNISSLTIGERGPVLLEDYHLIEKVANFTRERIPERVV 64
Query: 64 HARGASAKGFFEVTHDVSHLTCADFLRAPGVQTPIILRFSTVIHERGSPETLRDPRGFAV 123
HARG SAKGFFEVTHD+S+LTCADFLRAPGVQTP+I+RFSTV+HER SPET+RD RGFAV
Sbjct: 65 HARGISAKGFFEVTHDISNLTCADFLRAPGVQTPVIVRFSTVVHERASPETMRDIRGFAV 124
Query: 124 KFYTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHFPESLH 183
KFYTREGNFDLVGNN PVFF+RDG++FPD+VHALKPNPK++IQE WRILD+ SH PESL
Sbjct: 125 KFYTREGNFDLVGNNTPVFFIRDGIQFPDVVHALKPNPKTNIQEYWRILDYMSHLPESLL 184
Query: 184 MFTFLFDDVGVPQDYRHMDGFGVNTYTLINKAGKVVYVKFHWKPTCGVKCLLEEEAIKVG 243
+ ++FDDVG+PQDYRHM+GFGV+TYTLI K+GKV++VKFHWKPTCG+K L +EEA VG
Sbjct: 185 TWCWMFDDVGIPQDYRHMEGFGVHTYTLIAKSGKVLFVKFHWKPTCGIKNLTDEEAKVVG 244
Query: 244 GSNHSHATQDLYESIAAGNYPEWKLFIQTIDPDHEDRFDFDPLDVTKTWPEDIIPLQPVG 303
G+NHSHAT+DL+++IA+GNYPEWKLFIQT+DP ED+FDFDPLDVTK WPEDI+PLQPVG
Sbjct: 245 GANHSHATKDLHDAIASGNYPEWKLFIQTMDPADEDKFDFDPLDVTKIWPEDILPLQPVG 304
Query: 304 RMVLNKNIDNFFAENEQLAFCPAIVVPGISYSDDKMLQTRIFSYADSQRHRLGPNYLQLP 363
R+VLN+ IDNFF E EQLAF P +VVPGI YSDDK+LQ RIF+Y D+QRHRLGPNYLQLP
Sbjct: 305 RLVLNRTIDNFFNETEQLAFNPGLVVPGIYYSDDKLLQCRIFAYGDTQRHRLGPNYLQLP 364
Query: 364 ANAPKCAHHNNHHEGFMNFMHRDEEVNYFPSRYDPVRHAERYPIPPAICTGSRERCAIEK 423
NAPKCAHHNNHHEGFMNFMHRDEE+NY+PS++DPVR AE+ P P TG R +C I+K
Sbjct: 365 VNAPKCAHHNNHHEGFMNFMHRDEEINYYPSKFDPVRCAEKVPTPTNSYTGIRTKCVIKK 424
Query: 424 ENNFKQPGERYRSFAPDRQDRFVRRWVDALSDPRVTHEIRSTWISYWSQADRSLGQKIAS 483
ENNFKQ G+RYRS+APDRQDRFV+RWV+ LS+PR+THEIR WISYWSQADRSLGQK+AS
Sbjct: 425 ENNFKQAGDRYRSWAPDRQDRFVKRWVEILSEPRLTHEIRGIWISYWSQADRSLGQKLAS 484
Query: 484 HLNLRPSI 491
LN+RPSI
Sbjct: 485 RLNVRPSI 492
>At1g09010 putative Bos beta-mannosidase
Length = 908
Score = 34.7 bits (78), Expect = 0.12
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query: 11 FNSPFWTTNSGAPVWNNNNSMTVGVRGPILLEDYHLVEKLANFDRERIPERVVHARGASA 70
++S WT +G +W N N T G+RG HL+++ A+F R VH + A
Sbjct: 604 WSSQMWTKYTGVLIWKNQNPWT-GLRGQFY---DHLLDQTASFYGCRSAAEPVHVQLNLA 659
Query: 71 KGFFEVTHDVS 81
F EV + S
Sbjct: 660 SYFVEVVNTTS 670
>At3g04980 hypothetical protein, contains DnaJ motif: prokaryotic
heat shock protein motif
Length = 1165
Score = 31.6 bits (70), Expect = 1.0
Identities = 33/131 (25%), Positives = 55/131 (41%), Gaps = 22/131 (16%)
Query: 144 VRDGMKFPDMVHALKPNPKSHIQ----ENWRIL-DFFSHFPESL--HMFTFLFDDVGVPQ 196
V+D F +H L+ N I E W I ++ + SL H F + +D V +
Sbjct: 555 VKDSSIFSGQMHHLRCNNIVSIYPRKGEIWAIFREWEEEWNTSLKKHKFPYKYDFVEIVS 614
Query: 197 DYRHMDGFGVNTYTLINKAGKVVYVKFHWKP---TCGVKCLLEE--------EAIKVGGS 245
D+ ++G GV + K V + FHW+P C ++C ++ A+K+ G
Sbjct: 615 DFHDLNGVGV---AYLGKLKGSVQL-FHWEPQHGICQIQCSPKDMLRFSHKVPAVKMTGK 670
Query: 246 NHSHATQDLYE 256
+ YE
Sbjct: 671 EKESVPPNSYE 681
>At4g03800
Length = 637
Score = 31.2 bits (69), Expect = 1.4
Identities = 22/73 (30%), Positives = 32/73 (43%), Gaps = 7/73 (9%)
Query: 257 SIAAGNYPEWKLFIQTIDPDH-EDRFDFDPLDVTKTWPEDIIPLQPVGRMVLNKNIDNFF 315
SI YP+W + + +DR D D+ K P+D PL + R+V +
Sbjct: 369 SIREVQYPDWVAITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVEST------ 422
Query: 316 AENEQLAFCPAIV 328
A NE L F A +
Sbjct: 423 AGNELLTFMDAFL 435
>At5g41920 SCARECROW gene regulator-like protein
Length = 405
Score = 30.4 bits (67), Expect = 2.3
Identities = 23/86 (26%), Positives = 39/86 (44%), Gaps = 13/86 (15%)
Query: 112 PETLRDPRGFAVKFYTREGN-FDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWR 170
P L +G AV + + +D+ GNN +++ LKPN + +++
Sbjct: 227 PSQLATRQGEAVVVHWMQHRLYDVTGNNLETL---------EILRRLKPNLITVVEQELS 277
Query: 171 ILD---FFSHFPESLHMFTFLFDDVG 193
D F F E+LH ++ LFD +G
Sbjct: 278 YDDGGSFLGRFVEALHYYSALFDALG 303
>At5g08580 Unknown protein (MAH20.14)
Length = 391
Score = 30.4 bits (67), Expect = 2.3
Identities = 22/71 (30%), Positives = 31/71 (42%), Gaps = 9/71 (12%)
Query: 360 LQLPANAPKCAHHNNHHEGFMNFMHR---DEEVNYFPSRYDPVRHAERYPIPPAICTGSR 416
L L + +PK ++HH G N HR N+ P+R+DPV P P + R
Sbjct: 17 LFLVSYSPKKKGDHDHHHGGHNQHHRLKLRSSFNFKPTRHDPV------PFDPLVADMER 70
Query: 417 ERCAIEKENNF 427
R E E +
Sbjct: 71 RREDKEWERQY 81
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 30.0 bits (66), Expect = 3.0
Identities = 22/71 (30%), Positives = 32/71 (44%), Gaps = 7/71 (9%)
Query: 257 SIAAGNYPEWKLFIQTIDPDH-EDRFDFDPLDVTKTWPEDIIPLQPVGRMVLNKNIDNFF 315
SI YP+W + + +DR D D+ K P+D PL + R+V +
Sbjct: 492 SIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVEST------ 545
Query: 316 AENEQLAFCPA 326
A NE L+F A
Sbjct: 546 AGNELLSFMDA 556
>At4g31650 putative protein
Length = 493
Score = 29.6 bits (65), Expect = 4.0
Identities = 22/76 (28%), Positives = 34/76 (43%), Gaps = 8/76 (10%)
Query: 178 FPESLHMFTFLFDDVGVPQDYRHMDGFGVNTY--TLINKAGK------VVYVKFHWKPTC 229
F +S+ + D VGVP+D+ GF + + L+N+ GK + Y
Sbjct: 132 FSQSVTTSNLMRDSVGVPRDFAKRYGFNIGRHEIVLMNEEGKPWESEVISYKSGKVIVAG 191
Query: 230 GVKCLLEEEAIKVGGS 245
G K L E ++VG S
Sbjct: 192 GWKSLCTESKLEVGDS 207
>At2g13210 pseudogene
Length = 152
Score = 29.3 bits (64), Expect = 5.2
Identities = 17/61 (27%), Positives = 27/61 (43%), Gaps = 1/61 (1%)
Query: 257 SIAAGNYPEWKLFIQTIDPDHED-RFDFDPLDVTKTWPEDIIPLQPVGRMVLNKNIDNFF 315
SI YPEW + + R D D+ K P+D P+ +GR+V + + D
Sbjct: 61 SITKAKYPEWLANPVVVKKKNGKWRVCIDFTDLNKACPKDSFPMSHIGRLVESTSGDKLL 120
Query: 316 A 316
+
Sbjct: 121 S 121
>At1g71820 unknown protein (At1g71820)
Length = 752
Score = 29.3 bits (64), Expect = 5.2
Identities = 18/61 (29%), Positives = 26/61 (42%), Gaps = 2/61 (3%)
Query: 41 LEDYHLVEKLANFDRERIPERVVHARGA--SAKGFFEVTHDVSHLTCADFLRAPGVQTPI 98
L Y L +L+N E +P+ + KGF EV + H T PGVQ +
Sbjct: 501 LRCYDLAMELSNSTLEALPQNYAEQVNFEDTCKGFLEVAKEAVHQTVRVIFEDPGVQELL 560
Query: 99 I 99
+
Sbjct: 561 V 561
>At3g30710 hypothetical protein
Length = 523
Score = 28.9 bits (63), Expect = 6.7
Identities = 10/27 (37%), Positives = 16/27 (59%)
Query: 276 DHEDRFDFDPLDVTKTWPEDIIPLQPV 302
DH DFDP +T+TW ++ ++ V
Sbjct: 193 DHHSVSDFDPARITRTWSSNLAKIKEV 219
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 28.9 bits (63), Expect = 6.7
Identities = 20/79 (25%), Positives = 36/79 (45%), Gaps = 6/79 (7%)
Query: 234 LLEEEAIKVGGSNHSHATQDLYESIAAGN-----YPEWKLFIQTIDPDHED-RFDFDPLD 287
L++++ K+G ++ + + AG+ YPEW + ++ R D D
Sbjct: 497 LVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWLANPVVVKKKNDKWRVCIDFTD 556
Query: 288 VTKTWPEDIIPLQPVGRMV 306
+ K P+D PL + RMV
Sbjct: 557 LNKACPKDSFPLPHIDRMV 575
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,646,422
Number of Sequences: 26719
Number of extensions: 595208
Number of successful extensions: 1054
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1047
Number of HSP's gapped (non-prelim): 15
length of query: 491
length of database: 11,318,596
effective HSP length: 103
effective length of query: 388
effective length of database: 8,566,539
effective search space: 3323817132
effective search space used: 3323817132
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0193.6


