

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0193.5
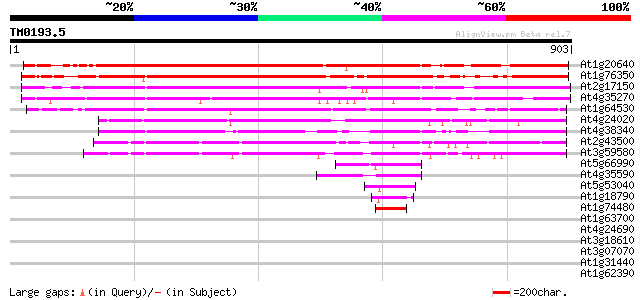
(903 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20640 unknown protein 739 0.0
At1g76350 hypothetical protein 702 0.0
At2g17150 unknown protein 645 0.0
At4g35270 putative protein 624 e-178
At1g64530 unknown protein 449 e-126
At4g24020 unknown protein (At4g24020) 436 e-122
At4g38340 putative protein 419 e-117
At2g43500 hypothetical protein 410 e-114
At3g59580 putative protein 408 e-114
At5g66990 putative protein 52 1e-06
At4g35590 putative protein 52 1e-06
At5g53040 putative protein 52 2e-06
At1g18790 hypothetical protein 49 1e-05
At1g74480 hypothetical protein 44 5e-04
At1g63700 putative protein kinase 36 0.11
At4g24690 unknown protein 34 0.32
At3g18610 unknown protein 34 0.42
At3g07070 putative protein kinase 33 0.55
At1g31440 unknown protein 33 0.55
At1g62390 putative protein 33 0.94
>At1g20640 unknown protein
Length = 844
Score = 739 bits (1907), Expect = 0.0
Identities = 429/887 (48%), Positives = 547/887 (61%), Gaps = 81/887 (9%)
Query: 23 MDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLFDP-SFSWPALETNEPTHVEDQH 81
MD ++M LLLDGCWLE + DGS FL S FDP SF W
Sbjct: 13 MDADFMDGLLLDGCWLETT-DGSEFLNIAPSTSSVSPFDPTSFMW--------------- 56
Query: 82 ESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLG 141
+P +T + S VV Y Q E S+ E + KRWWI P G G
Sbjct: 57 ----SPTQDTSALCTS----GVVSQMYG-QDCVERSSLDEFQWN--KRWWIGPG---GGG 102
Query: 142 PSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYRE 201
S+ E+L++A++ IK + + LIQ+WVPV RG + +L+ PFS D + LA YRE
Sbjct: 103 SSVTERLVQAVEHIKDYTTARGSLIQLWVPVNRGGKRVLTTKEQPFSHDPLCQRLANYRE 162
Query: 202 ISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLA 261
IS + FSAE+D + + GLPGRVF K+PEWTPDVRFF+S+EYPRV HA++ D+ GTLA
Sbjct: 163 ISVNYHFSAEQDDSKALAGLPGRVFLGKLPEWTPDVRFFKSEEYPRVHHAQDCDVRGTLA 222
Query: 262 VPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSY 321
+P+FEQGS+ CLGVIEVVMTT+ + P+LES+C+AL+ VDL S + + K D SY
Sbjct: 223 IPVFEQGSKICLGVIEVVMTTEMVKLRPELESICRALQAVDLRSTELPIPPSLKGCDLSY 282
Query: 322 EAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHSEDNYLHCISPVEQACYVGDP 381
+AALPEI+ +LR AC HKLPLAQTWVSC QQ K GCRH+++NY+HC+S ++ ACYVGDP
Sbjct: 283 KAALPEIRNLLRCACETHKLPLAQTWVSCQQQNKSGCRHNDENYIHCVSTIDDACYVGDP 342
Query: 382 SVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLFGLRAAVA 441
+VR FHEAC EHHLLKGQGVAG+AF+ N P FS+D++ K++YPLSHHA ++GL AVA
Sbjct: 343 TVREFHEACSEHHLLKGQGVAGQAFLTNGPCFSSDVSNYKKSEYPLSHHANMYGLHGAVA 402
Query: 442 IRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDKELERTSS 501
IRLR I++ + D+VLEFFLP +C+D EEQ+ ML +LS I+ RSLR +TDKELE S
Sbjct: 403 IRLRCIHTGSADFVLEFFLPKDCDDLEEQRKMLNALSTIMAHVPRSLRTVTDKELEEESE 462
Query: 502 SVEVMALEDSGFARTVKWSEPQHITSV--ASLEPEEKSSET-----VGGKFSDLREHQED 554
+E + ++ + H S ASLE ++S+ T +G F + +
Sbjct: 463 VIE----REEIVTPKIENASELHGNSPWNASLEEIQRSNNTSNPQNLGLVFDGGDKPNDG 518
Query: 555 SILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKN 614
LK + + + SS G S EK+R KADKTITL+VLRQYF GSLKDAAKN
Sbjct: 519 FGLKRGFDYTMDSNVNESSTFSSGGFSMMAEKKRTKADKTITLDVLRQYFAGSLKDAAKN 578
Query: 615 IGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFP 674
IGVC TTLKR+CRQHGI+RWPSRKIKKVGHSLQK+Q VIDSVQG SG I SFY+NFP
Sbjct: 579 IGVCPTTLKRICRQHGIQRWPSRKIKKVGHSLQKIQRVIDSVQGVSG-PLPIGSFYANFP 637
Query: 675 DLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSM 734
+L S QS+ P S + P P KS SS S S SS CS
Sbjct: 638 NLVS-------------QSQEP-SQQAKTTPPPPPPVQLAKSPVSSYSHSSNSSQCCS-- 681
Query: 735 PEQQPHTSDVACNKDPVVGKDSADV--VLKRIRSEAELKSHSENKAKLFPRSLSQETLGE 792
S+ N S DV LK+ SE EL+S S ++ L SL G
Sbjct: 682 -------SETQLNSGATTDPPSTDVGGALKKTSSEIELQSSSLDETILTLSSLENIPQG- 733
Query: 793 HTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSD 852
T ++ +D R+KV+YG+EK R RM S LL EI +RF++ D
Sbjct: 734 ------------TNLLSSQDDDFLRIKVSYGEEKIRLRMRNSRRLRDLLWEIGKRFSIED 781
Query: 853 MSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS 899
MS++D+KYLD+D EWVLLTCD D+EEC+DVC ++ S TIKL +QASS
Sbjct: 782 MSRYDLKYLDEDNEWVLLTCDEDVEECVDVCRTTPSHTIKLLLQASS 828
>At1g76350 hypothetical protein
Length = 808
Score = 702 bits (1813), Expect = 0.0
Identities = 412/888 (46%), Positives = 539/888 (60%), Gaps = 110/888 (12%)
Query: 20 ETSMDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLFDPS-FSWPALETNEPTHVE 78
+ +MD +M LLL+GCWLE + D S FL SP S FDPS F W T + ++
Sbjct: 9 DPAMDSSFMDGLLLEGCWLETT-DASEFL-NFSPSTSVAPFDPSSFMWSP--TQDTSNSL 64
Query: 79 DQHESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSP 138
Q Q+ P +S E + S +RWWI P+
Sbjct: 65 SQMYGQDCP----------------------ERSSLEDQNQGRDLSTFNRRWWIGPSGHH 102
Query: 139 GLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLAR 198
G S+ME+L++A+ IK F + LIQ+WVPV RG + +L+ PFS D + LA
Sbjct: 103 GF--SVMERLVQAVTHIKDFTSERGSLIQLWVPVDRGGKRVLTTKEQPFSHDPMCQRLAH 160
Query: 199 YREISEGFQFSAEED-----SKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHARE 253
YREISE +QFS E++ S++LV GLPGRVF KVPEWTPDVRFF+++EYPRV+HA++
Sbjct: 161 YREISENYQFSTEQEDSDSSSRDLV-GLPGRVFLGKVPEWTPDVRFFKNEEYPRVQHAQD 219
Query: 254 FDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQN 313
D+ GTLA+P+FEQGS+ CLGVIEVVMTTQ + P LES+C+AL+ VDL S + +
Sbjct: 220 CDVRGTLAIPVFEQGSQICLGVIEVVMTTQMVKLSPDLESICRALQAVDLRSTEIPIPPS 279
Query: 314 AKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHSEDNYLHCISPVE 373
K D SY+AALPEI+ +LR AC HKLPLAQTWVSC +Q K GCRH+++NY+HC+S ++
Sbjct: 280 LKGPDFSYQAALPEIRNLLRCACETHKLPLAQTWVSCLKQSKTGCRHNDENYIHCVSTID 339
Query: 374 QACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARL 433
ACYVGDP+VR FHEAC EHHLLKGQGV G+AF+ N P FS+D++ K++YPLSHHA +
Sbjct: 340 DACYVGDPTVREFHEACSEHHLLKGQGVVGEAFLTNGPCFSSDVSSYKKSEYPLSHHATM 399
Query: 434 FGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITD 493
FGL VAIRLR I++ + D+VLEFFLP NC D EEQ+ ML +LS I+ RSLR +T
Sbjct: 400 FGLHGTVAIRLRCIHTGSVDFVLEFFLPKNCRDIEEQRKMLNALSTIMAHVPRSLRTVTQ 459
Query: 494 KELERTSSSVEVMALEDSGFARTVKWSEPQHITSV-ASLEPEEKSSETVGGKFSDLREHQ 552
KELE S+ +E + V + ++ T V S+ + G +++ E
Sbjct: 460 KELEEEGDSMVSEVIE-----KGVTLPKIENTTEVHQSISTPQNVGLVFDGGTTEMGELG 514
Query: 553 EDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAA 612
+ G E + F S+ G ++ EK+R KA+K ITL+VLRQYF GSLKDAA
Sbjct: 515 SE---YGKGVSVNENNTF----SSASGFNRVTEKKRTKAEKNITLDVLRQYFAGSLKDAA 567
Query: 613 KNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSN 672
K+IGVC TTLKR+CRQHGI+RWPSRKIKKVGHSLQK+Q VIDSV+G SG I SFY++
Sbjct: 568 KSIGVCPTTLKRICRQHGIQRWPSRKIKKVGHSLQKIQRVIDSVEGVSGHHLPIGSFYAS 627
Query: 673 FPDL-ASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSC 731
FP+L ASP S S QS+ LS S KS SSCS S SC
Sbjct: 628 FPNLAASPEAS-----SLQQQSKITTFLS-------YSHSPPAKSPGSSCSHSS----SC 671
Query: 732 SSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLG 791
SS + V+ +D D K +L RS +
Sbjct: 672 SS--------------ETQVIKEDPTD------------------KTRLVSRSFKE---- 695
Query: 792 EHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVS 851
T+T + S ++ ++D RVKV+Y +EK RF+M S + LL EIA+RF++
Sbjct: 696 --TQTTHLS-------PSSQEDDFLRVKVSYEEEKIRFKMRNSHRLKDLLWEIAKRFSIE 746
Query: 852 DMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS 899
D+S++D+KYLD+D EWVLL CD D+EEC+DVC S TIKL +Q SS
Sbjct: 747 DVSRYDLKYLDEDNEWVLLRCDDDVEECVDVCRSFPGQTIKLLLQLSS 794
>At2g17150 unknown protein
Length = 909
Score = 645 bits (1664), Expect = 0.0
Identities = 383/924 (41%), Positives = 538/924 (57%), Gaps = 93/924 (10%)
Query: 20 ETSMDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLFDPSFSWPALETNEPTHVED 79
+T+MD ++M +LL DGCWLE + S +QSP S+ A+ N P
Sbjct: 27 DTAMDLDFMDELLFDGCWLETTDSKSLKQTEQSPSAST----------AMNDNSPFLCFG 76
Query: 80 QHESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPG 139
++ SQ+ N N R Q+E ++E G K WWIAP+ S G
Sbjct: 77 ENPSQD-------------NFSNEETERMFPQAEK---FLLEEAEVG-KSWWIAPSASEG 119
Query: 140 LGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARY 199
S+ E+L++A+ + + +KD L+QIWVP+ + + L+ P + +LA Y
Sbjct: 120 PSSSVKERLLQAISGLNEAVQDKDFLVQIWVPIQQEGKSFLTTWAQPHLFNQEYSSLAEY 179
Query: 200 REISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGT 259
R +SE + F A+E K+ V GLPGRVF K PEWTPDVRFFR DEYPR++ A++ D+ G+
Sbjct: 180 RHVSETYNFPADEGMKDFV-GLPGRVFLQKFPEWTPDVRFFRRDEYPRIKEAQKCDVRGS 238
Query: 260 LAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTS---LKHSSIQNAKA 316
LA+P+FE+GS TCLGV+E+V TTQ++NY +LE +CKALE VDL S L S + +
Sbjct: 239 LALPVFERGSGTCLGVVEIVTTTQKMNYRQELEKMCKALEAVDLRSSSNLNTPSSEFLQV 298
Query: 317 RDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHSEDNYLHCISPVEQAC 376
Y AALPEI++ L + C + PLA +W C +QGK G RHS++N+ C+S ++ AC
Sbjct: 299 YSDFYCAALPEIKDFLATICRSYDFPLALSWAPCARQGKVGSRHSDENFSECVSTIDSAC 358
Query: 377 YVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLFGL 436
V D + F EAC EHHLL+G+G+ GKAF + FF ++ SKT+YPL+HHA++ GL
Sbjct: 359 SVPDEQSKSFWEACSEHHLLQGEGIVGKAFEATKLFFVPEVATFSKTNYPLAHHAKISGL 418
Query: 437 RAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRV-ITDKE 495
AA+A+ L+S S ++VLEFF P C D+E Q+ ML SL + +Q+ RS + I D E
Sbjct: 419 HAALAVPLKS-KSGLVEFVLEFFFPKACLDTEAQQEMLKSLCVTLQQDFRSSNLFIKDLE 477
Query: 496 L--------------------ERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEE 535
L E S E+ E S A +K +E S++ +E
Sbjct: 478 LEVVLPVRETMLFSENLLCGAETVESLTEIQMQESSWIAHMIKANEKGKDVSLSWEYQKE 537
Query: 536 KSSETVGGKFSDLREHQEDSILKGNIECDRE-------------CSPFVE----GNLSSV 578
E G RE+ + + N+ + E P E G + +
Sbjct: 538 DPKELSSG-----RENSQLDPVPNNVPLEAEQLQQASTPGLRVDIGPSTESASTGGGNML 592
Query: 579 GISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRK 638
+ GEK+RAK +KTI LEVLRQYF GSLKDAAK+IGVC TTLKR+CRQHGI RWPSRK
Sbjct: 593 SSRRPGEKKRAKTEKTIGLEVLRQYFAGSLKDAAKSIGVCPTTLKRICRQHGIMRWPSRK 652
Query: 639 IKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNS 698
IKKVGHSL+KLQLV+DSVQGA G S ++DSFY++FP+L SPN+S S +L +E P+
Sbjct: 653 IKKVGHSLKKLQLVMDSVQGAQG-SIQLDSFYTSFPELNSPNMS--SNGPSLKSNEQPSH 709
Query: 699 LSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSAD 758
L+ Q D G ++ E +S SSSCS+ S SS++ N ++ + AD
Sbjct: 710 LNAQTDNGIMAEEN-PRSPSSSCSKSSGSSNNNE--------------NTGNILVAEDAD 754
Query: 759 VVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRV 818
VLKR SEA+L + ++ + K R+ S +T E + S L + + + A +V
Sbjct: 755 AVLKRAHSEAQLHNVNQEETKCLARTQSHKTFKEPLVLDNSSPLTGSSNTSLRARGAIKV 814
Query: 819 KVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEE 878
K T+G+ + RF + SW + L QEIARRFN+ D+S FD+KYLDDD EWVLLTC+ADL E
Sbjct: 815 KATFGEARIRFTLLPSWGFAELKQEIARRFNIDDISWFDLKYLDDDKEWVLLTCEADLVE 874
Query: 879 CIDVCLSSESSTIKLCIQASSGMR 902
CID+ +++ TIK+ + +S ++
Sbjct: 875 CIDIYRLTQTHTIKISLNEASQVK 898
>At4g35270 putative protein
Length = 1031
Score = 624 bits (1608), Expect = e-178
Identities = 380/967 (39%), Positives = 558/967 (57%), Gaps = 112/967 (11%)
Query: 19 NETSMDFEYMSDLLLDGCWLEASADGSNF--LLQQSPPFSSPLFDPS-----FSWPALET 71
++++MD ++M +LL DGCWLE + DG + + Q S+ + D + + + E
Sbjct: 22 SDSAMDMDFMDELLFDGCWLETT-DGKSLKQTMGQQVSDSTTMNDNNNNSYLYGYQYAEN 80
Query: 72 NEPTHVEDQHESQEAPL--GNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKR 129
H+ ++ ++ P + N V + ++ E + E S+G +R
Sbjct: 81 LSQDHISNEETGRKFPPIPPGFLKIEDLSNQVPFDQSAVMSSAQAEKFLLEE--SEGGRR 138
Query: 130 WWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSL 189
+WIAP S G S+ E+L++A++ + + +KD LIQIW+P+ + + L+ + P
Sbjct: 139 YWIAPRTSQGPSSSVKERLVQAIEGLNEEVQDKDFLIQIWLPIQQEGKNFLTTSEQPHFF 198
Query: 190 DSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVE 249
+ +L RYR++S + F A+EDSKE V GLPGRVF K+PEWTPDVRFFRS+EYPR++
Sbjct: 199 NPKYSSLKRYRDVSVAYNFLADEDSKESV-GLPGRVFLKKLPEWTPDVRFFRSEEYPRIK 257
Query: 250 HAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTS---- 305
A + D+ G+LA+P+FE+GS TCLGV+E+V TTQ++NY P+L+++CKALE V+L S
Sbjct: 258 EAEQCDVRGSLALPVFERGSGTCLGVVEIVTTTQKMNYRPELDNICKALESVNLRSSRSL 317
Query: 306 -----------LKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQG 354
L + ++ ++ R+ + L I S C ++ LPLA TW C +QG
Sbjct: 318 NPPSREVCKNGLLNQTLTSSIHRNNEFWL-LNVICPFCFSFCRVYDLPLALTWAPCARQG 376
Query: 355 KDGCRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFS 414
K G RHS++N+ C+S V+ AC V D R F EAC EHHLL+G+G+ GKAF + FF
Sbjct: 377 KVGSRHSDENFSECVSTVDDACIVPDHQSRHFLEACSEHHLLQGEGIVGKAFNATKLFFV 436
Query: 415 TDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNML 474
++T SKT+YPL+HHA++ GL AA+A+ L++ ++S+ ++VLEFF P C D+E Q++ML
Sbjct: 437 PEVTTFSKTNYPLAHHAKISGLHAALAVPLKNKFNSSVEFVLEFFFPKACLDTEAQQDML 496
Query: 475 ISLSIIIQRCCRSLRVITDKELE---------------------RTSSSVEVMALED--- 510
SLS +Q+ RSL + DKELE T ++ + LE+
Sbjct: 497 KSLSATLQQDFRSLNLFIDKELELEVVFPVREEVVFAENPLINAGTGEDMKPLPLEEISQ 556
Query: 511 ---SGFARTVKWSEPQHITSVA----SLEPEEKSSETVG--------GKFSDLREHQE-- 553
S + +K +E S++ EP+E+ T G G + L E ++
Sbjct: 557 EDSSWISHMIKANEKGKGVSLSWEYQKEEPKEEFMLTSGWDNNQIGSGHNNFLSEAEQFQ 616
Query: 554 ---DSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKD 610
+S L+ +++ E + F G + +G + GEKRR K +KTI LEVLRQYF GSLKD
Sbjct: 617 KVTNSGLRIDMDPSFESASFGVGQ-TLLGSRRPGEKRRTKTEKTIGLEVLRQYFAGSLKD 675
Query: 611 AAKNIG--------------VCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSV 656
AAK+IG VC TTLKR+CRQHGI RWPSRKIKKVGHSL+KLQLVIDSV
Sbjct: 676 AAKSIGESDDSFTQFHFHVAVCPTTLKRICRQHGITRWPSRKIKKVGHSLKKLQLVIDSV 735
Query: 657 QGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQ-PDLGPLSPEGATK 715
QG G S ++DSFY++FP+L+SP++SG ++ N +S Q P SP
Sbjct: 736 QGVQG-SIQLDSFYTSFPELSSPHMSGTGTSFKNPNAQTENGVSAQGTAAAPKSPP---- 790
Query: 716 SLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSE 775
SSSCS S SS CS+ Q +T + N + ++A +LKR RSE L + ++
Sbjct: 791 --SSSCSHSSGSSTCCSTGANQSTNTGTTS-NTVTTLMAENASAILKRARSEVRLHTMNQ 847
Query: 776 NKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSW 835
++ K R+LS +T EH E L + + A +VK T+G+ K
Sbjct: 848 DETKSLSRTLSHKTFSEHPLFENPPRLPENSSRKLKAGGASKVKATFGEAK--------- 898
Query: 836 SYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
EIARRFN+ +++ FD+KYLDDD EWVLLTC+ADLEECID+ SS+S TIK+ +
Sbjct: 899 ------HEIARRFNIDNIAPFDLKYLDDDKEWVLLTCEADLEECIDIYRSSQSRTIKISV 952
Query: 896 QASSGMR 902
+S ++
Sbjct: 953 HEASQVK 959
>At1g64530 unknown protein
Length = 841
Score = 449 bits (1155), Expect = e-126
Identities = 307/886 (34%), Positives = 462/886 (51%), Gaps = 86/886 (9%)
Query: 28 MSDLLLDGCW-LEASADGSNFLLQQSPPFSSPLFDP-SFSWPALETNEPTHVEDQHESQE 85
+ DL L G W L+ SNF +SP S P S W ET+ E
Sbjct: 3 LDDLDLSGSWPLDQITFASNF---KSPVIFSSSEQPFSPLWSFSETSGDVG----GELYS 55
Query: 86 APLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLGPSIM 145
A + T+ S + + ++SET T E W I P +P +I
Sbjct: 56 AAVAPTRFTDYSVLLAS-------SESETTTK---ENNQVPSPSWGIMPLENPDSYCAIK 105
Query: 146 EKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEG 205
K+ +AL++ K+ + +L Q+W PV R +L+ + PF L S L +YR +S
Sbjct: 106 AKMTQALRYFKESTGQQHVLAQVWAPVKNRGRYVLTTSGQPFVLGPNSNGLNQYRMVSLT 165
Query: 206 FQFSAE-EDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPI 264
+ FS + E EL GLPGRVFR K+PEWTP+V+++ S E+ R+ HA +++ GTLA+P+
Sbjct: 166 YMFSLDGERDGEL--GLPGRVFRKKLPEWTPNVQYYSSKEFSRLGHALHYNVQGTLALPV 223
Query: 265 FEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSYEAA 324
FE + C+GV+E++MT+ +INY P++E VCKALE V+L + + + + + ++ + A
Sbjct: 224 FEPSRQLCVGVVELIMTSPKINYAPEVEKVCKALEAVNLKTSEILNHETTQICNEGRQNA 283
Query: 325 LPEIQEVLRSACHMHKLPLAQTWVSCFQQG--------KDGCRHSEDNYLH--CISPVEQ 374
L EI E+L C +KLPLAQTWV C + K C + + + C+S +
Sbjct: 284 LAEILEILTVVCETYKLPLAQTWVPCRHRSVLAFGGGFKKSCSSFDGSCMGKVCMSTSDL 343
Query: 375 ACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLF 434
A YV D V F +AC EHHL KGQGVAG+AF F D+T KTDYPL H+AR+F
Sbjct: 344 AVYVVDAHVWGFRDACAEHHLQKGQGVAGRAFQSGNLCFCRDVTRFCKTDYPLVHYARMF 403
Query: 435 GLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDK 494
L + A+ L+S Y+ D+YVLEFFLP D EQ +L SL +++ SL+V+++
Sbjct: 404 KLTSCFAVCLKSTYTGDDEYVLEFFLPPAITDKSEQDCLLGSLLQTMKQHYSSLKVVSET 463
Query: 495 ELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLREHQED 554
EL + S+EV+ + G + EP I A + + K S + E+
Sbjct: 464 ELCENNMSLEVVEASEDGMVYSK--LEPIRIHHPAQISKDYLELNAPEQKVSLNSDFMEN 521
Query: 555 SILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKN 614
+ + +E + P E K E++R K +KTI+LEVL+QYF GSLKDAAK+
Sbjct: 522 NEVDDGVERFQTLDPIPEAK-----TVKKSERKRGKTEKTISLEVLQQYFAGSLKDAAKS 576
Query: 615 IGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFP 674
+GVC TT+KR+CRQHGI RWPSRKI KV SL +L+ VIDSVQGA G+ +P
Sbjct: 577 LGVCPTTMKRICRQHGISRWPSRKINKVNRSLTRLKHVIDSVQGADGSLNLTSLSPRPWP 636
Query: 675 DLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSM 734
P + L ++ P S S +L + E S+ S
Sbjct: 637 HQIPP------IDIQLAKNCPPTSTSPLSNLQDVKIENRDAEDSAGSS------------ 678
Query: 735 PEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHT 794
TS +C +P+ + L +H++ ++ +L
Sbjct: 679 ------TSRASCKVNPI------------CETRFRLPTHNQEPSRQV--ALDDSDSSSKN 718
Query: 795 KTEYQSYLLKTCH-KATPKEDAHR---VKVTYGDEKTRFRM-PKSWSYEHLLQEIARRFN 849
T + ++L TC A+P H+ +K TY ++ RF++ P+S S L Q++A+R
Sbjct: 719 MTNFWAHL--TCQDTASPTILQHKLVSIKATYREDIIRFKISPESVSITELKQQVAKRLK 776
Query: 850 VSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
+ + + F++KYLDDD EWV ++CDADL EC+D +++++T++L +
Sbjct: 777 L-ETAAFELKYLDDDREWVSVSCDADLSECLDTS-AAKANTLRLSV 820
>At4g24020 unknown protein (At4g24020)
Length = 959
Score = 436 bits (1121), Expect = e-122
Identities = 289/827 (34%), Positives = 428/827 (50%), Gaps = 106/827 (12%)
Query: 144 IMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREIS 203
I E++ +AL++ K+ + + +L Q+W PV + R +L+ PF L+ L +YR IS
Sbjct: 147 IKERMTQALRYFKE-STEQHVLAQVWAPVRKNGRDLLTTLGQPFVLNPNGNGLNQYRMIS 205
Query: 204 EGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVP 263
+ FS + +S ++ GLPGRVFR K+PEWTP+V+++ S E+ R++HA +++ GTLA+P
Sbjct: 206 LTYMFSVDSES-DVELGLPGRVFRQKLPEWTPNVQYYSSKEFSRLDHALHYNVRGTLALP 264
Query: 264 IFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSYEA 323
+F ++C+GV+E++MT+++I+Y P+++ VCKALE V+L S + Q + ++S +
Sbjct: 265 VFNPSGQSCIGVVELIMTSEKIHYAPEVDKVCKALEAVNLKSSEILDHQTTQICNESRQN 324
Query: 324 ALPEIQEVLRSACHMHKLPLAQTWVSCFQQG---------KDGCRHSEDNYLH--CISPV 372
AL EI EVL C H LPLAQTWV C Q G K C + + + C+S
Sbjct: 325 ALAEILEVLTVVCETHNLPLAQTWVPC-QHGSVLANGGGLKKNCTSFDGSCMGQICMSTT 383
Query: 373 EQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHAR 432
+ ACYV D V F +AC+EHHL KGQGVAG+AF+ F DIT KT YPL H+A
Sbjct: 384 DMACYVVDAHVWGFRDACLEHHLQKGQGVAGRAFLNGGSCFCRDITKFCKTQYPLVHYAL 443
Query: 433 LFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVIT 492
+F L AI L+S Y+ D Y+LEFFLP + D +EQ +L S+ + ++ +SLRV +
Sbjct: 444 MFKLTTCFAISLQSSYTGDDSYILEFFLPSSITDDQEQDLLLGSILVTMKEHFQSLRVAS 503
Query: 493 DKELERTSSSVE---VMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLR 549
+ + + AL D + E++ FS +
Sbjct: 504 GVDFGEDDDKLSFEIIQALPDKKVHSKI---------------------ESIRVPFSGFK 542
Query: 550 EHQEDSILKGNIECDRECSPFVEGNLSSV-GI---SKTGEKRRAKADKTITLEVLRQYFP 605
+ +++L + N+++V G+ K EK+R K +KTI+L+VL+QYF
Sbjct: 543 SNATETMLIPQPVVQSSDPVNEKINVATVNGVVKEKKKTEKKRGKTEKTISLDVLQQYFT 602
Query: 606 GSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFK 665
GSLKDAAK++GVC TT+KR+CRQHGI RWPSRKIKKV S+ KL+ VI+SVQG G
Sbjct: 603 GSLKDAAKSLGVCPTTMKRICRQHGISRWPSRKIKKVNRSITKLKRVIESVQGTDGGLDL 662
Query: 666 IDSFYSNFP---------DLASPNLSGASLVSALNQSEN-------PNSLSIQPDLGPLS 709
S+ P L SPN S + N S N PN + P+L P +
Sbjct: 663 TSMAVSSIPWTHGQTSAQPLNSPNGSKPPELPNTNNSPNHWSSDHSPNEPNGSPELPPSN 722
Query: 710 PEGATKSLSSSCSQGSLSSH-SCSS-------MPEQQP------------------HTSD 743
G +S + S G+ +SH SC +P Q P H
Sbjct: 723 --GHKRSRTVDESAGTPTSHGSCDGNQLDEPKVPNQDPLFTVGGSPGLLFPPYSRDHDVS 780
Query: 744 VACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLL 803
A P S D + +A S++ L P + + + +
Sbjct: 781 AASFAMPNRLLGSIDHFRGMLIEDA---GSSKDLRNLCPTAAFDDKFQDTNWMNNDNNSN 837
Query: 804 KTCHKATPKEDAHR---------------VKVTYGDEKTRFRMPKSWSYEHLLQEIARRF 848
+ A PKE+A +K +Y D+ RFR+ L E+A+R
Sbjct: 838 NNLY-APPKEEAIANVACEPSGSEMRTVTIKASYKDDIIRFRISSGSGIMELKDEVAKRL 896
Query: 849 NVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
V D FD+KYLDDD EWVL+ CDADL+EC+++ SS + ++L +
Sbjct: 897 KV-DAGTFDIKYLDDDNEWVLIACDADLQECLEIPRSSRTKIVRLLV 942
>At4g38340 putative protein
Length = 767
Score = 419 bits (1076), Expect = e-117
Identities = 276/761 (36%), Positives = 391/761 (51%), Gaps = 127/761 (16%)
Query: 144 IMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLD--SGSENLARYRE 201
+ E++ A+ +++ +++LIQ+WVPV +LS P+S++ S S++LA YR+
Sbjct: 114 LKERVACAMGHLQEVMGERELLIQLWVPVETRSGRVLSTEEQPYSINTFSQSQSLALYRD 173
Query: 202 ISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLA 261
S G+ F+AE S++LV GLPGRVF ++PEWTPDVRFFR +EYPR+ +AR + + TLA
Sbjct: 174 ASAGYSFAAEVGSEQLV-GLPGRVFLRRMPEWTPDVRFFRKEEYPRIGYARRYQVRATLA 232
Query: 262 VPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSI-QNAKARDKS 320
+P+F+ S C+ V+E+V T + + Y QL ++C ALE DL + + S + + K S
Sbjct: 233 LPLFQGTSGNCVAVMEMVTTHRNLEYASQLSTICHALEAFDLRTSQTSIVPASLKVTSSS 292
Query: 321 YEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHSEDNYLHCISPVEQACYVGD 380
++ E+ +L+ C H LPLA TW H + + C+S + A Y D
Sbjct: 293 SSSSRTEVASILQGICSSHGLPLAVTW-----------GHQDSS--SCLSALISASYAAD 339
Query: 381 PSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLFGLRAAV 440
R F AC EHHLL G+G+AG+AF + F+TD+ + SK YPLSH+A++F L AA+
Sbjct: 340 HGSRCFLAACSEHHLLGGEGIAGRAFATKKQCFATDVAIFSKWSYPLSHYAKMFDLHAAL 399
Query: 441 AIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDKELERTS 500
A+ + + + +VLE F P +C D + L S +L+
Sbjct: 400 AVPILTRGNRTVQFVLELFFPRDCLDIQTHSLTLAS------------------QLKLRF 441
Query: 501 SSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLREHQEDSILKGN 560
S + ++D+ A V+ + +T E+ + V FS
Sbjct: 442 QSSPHLMVDDNQIAEEVRDAATPPLTQ------EDPKGKQVSFSFSS------------- 482
Query: 561 IECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTT 620
S KR+ KA+K ITL+ LRQ+F GSLKDAAKNIGVC T
Sbjct: 483 ------------------ASSLENRKRKTKAEKDITLDTLRQHFAGSLKDAAKNIGVCPT 524
Query: 621 TLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFPDLASPN 680
TLKR+CRQ+GI RWPSRKIKKVGHSL+KLQ+V+DSV+G G S + SFYS+FP L S +
Sbjct: 525 TLKRICRQNGISRWPSRKIKKVGHSLRKLQVVMDSVEGVQG-SLHLASFYSSFPQLQSSS 583
Query: 681 LSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPH 740
S + NP P P SP SSS SQ S S +C S EQQ
Sbjct: 584 SSSFPFI-------NPTQTVHVP---PKSPP------SSSGSQSSSGSSTCCSSEEQQ-- 625
Query: 741 TSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTK-TEYQ 799
+ A P+ L+ ++ E +
Sbjct: 626 ------------------------------LGGFQKPALSHPQLLTLSSMHEDQRPVRVT 655
Query: 800 SYLLKTCHKATPK--EDAHRVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDM--SK 855
S L TP+ +D +VK +GD R + L +EIA+RF + D+ S
Sbjct: 656 SSLPPLPSATTPRKAKDGMKVKAMFGDSMLRMSLLPHSRLTDLRREIAKRFGMDDVLRSN 715
Query: 856 FDVKYLDDDLEWVLLTCDADLEECIDVCLSSE-SSTIKLCI 895
F +KYLDDD EWVLLTCDADLEECI V SS TI++ +
Sbjct: 716 FSLKYLDDDQEWVLLTCDADLEECIQVYKSSSLKETIRILV 756
>At2g43500 hypothetical protein
Length = 947
Score = 410 bits (1053), Expect = e-114
Identities = 277/807 (34%), Positives = 428/807 (52%), Gaps = 78/807 (9%)
Query: 135 TCSPGLGPSIMEKLIRALK-WIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGS 193
T L S+ EK+++AL +++ + +L Q+W P+ GD+ +LS + + LD
Sbjct: 154 TIPRSLSHSLDEKMLKALSLFMESSGSGEGILAQVWTPIKTGDQYLLSTCDQAYLLDP-- 211
Query: 194 ENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHARE 253
++YRE+S F F+AE + PGLPGRVF VPEWT +V ++++DEY R++HA +
Sbjct: 212 -RFSQYREVSRRFTFAAEANQCSF-PGLPGRVFISGVPEWTSNVMYYKTDEYLRMKHAID 269
Query: 254 FDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQN 313
++ G++A+PI E +C V+E+V + ++ N+ +++SVC+AL+ V+L + ++I
Sbjct: 270 NEVRGSIAIPILEASGTSCCAVMELVTSKEKPNFDMEMDSVCRALQAVNLRT---AAIPR 326
Query: 314 AKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCF--QQGKDGCRHSEDNYLHCISP 371
+ S AL EIQ+VLR+ CH HKLPLA W+ C Q + + S +N + CI
Sbjct: 327 PQYLSSSQRDALAEIQDVLRTVCHAHKLPLALAWIPCRKDQSIRVSGQKSGENCILCIE- 385
Query: 372 VEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHA 431
E ACYV D + F AC+EH L + +G+ GKAF+ NQPFFS+D+ ++YP+ HA
Sbjct: 386 -ETACYVNDMEMEGFVHACLEHCLREKEGIVGKAFISNQPFFSSDVKAYDISEYPIVQHA 444
Query: 432 RLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVI 491
R +GL AAVAI+LRS Y+ DDY+LE FLPV+ S EQ+ +L SLS +QR CR+LR +
Sbjct: 445 RKYGLNAAVAIKLRSTYTGEDDYILELFLPVSMKGSLEQQLLLDSLSGTMQRICRTLRTV 504
Query: 492 TD-KELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLRE 550
++ ++ + + + S F +T Q I SL+ E S+ ++ FS +
Sbjct: 505 SEVGSTKKEGTKPGFRSSDMSNFPQTTSSENFQTI----SLDSEFNSTRSM---FSGMSS 557
Query: 551 HQEDSIL--KGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSL 608
+E+SI +G +E D V ++T EK+++ +K ++L L+Q+F GSL
Sbjct: 558 DKENSITVSQGTLEQD-------------VSKARTPEKKKSTTEKNVSLSALQQHFSGSL 604
Query: 609 KDAAKNIG-------------VCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDS 655
KDAAK++G C TTLKR+CRQHGI RWPSRKI KV SL+K+Q V+DS
Sbjct: 605 KDAAKSLGGETSAYFQAWVYFFCPTTLKRICRQHGIMRWPSRKINKVNRSLRKIQTVLDS 664
Query: 656 VQGASGASFKIDSFYSNF----PDLASPNLSGASLVSALNQSENPNSLSIQPD------L 705
VQG G K DS F P + + + L S N + S PD
Sbjct: 665 VQGVEG-GLKFDSATGEFIAVRPFIQEID-TQKGLSSLDNDAHARRSQEDMPDDTSFKLQ 722
Query: 706 GPLSPEGATK----SLSSSCSQGSLSSHSCSSMP------EQQPHTSD---VACNKDPVV 752
S + A K + + GS + S P E + S+ CN V
Sbjct: 723 EAKSVDNAIKLEEDTTMNQARPGSFMEVNASGQPWAWMAKESGLNGSEGIKSVCNLSSVE 782
Query: 753 GKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPK 812
D D ++ S E + S + + + S L + T + + H +
Sbjct: 783 ISDGMDPTIRCSGSIVE-PNQSMSCSISDSSNGSGAVLRGSSSTSMEDWNQMRTHNSNSS 841
Query: 813 EDAHR---VKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVL 869
E VK +Y ++ RF+ S L +E+ +RF + D S F +KYLDD+ EWV+
Sbjct: 842 ESGSTTLIVKASYREDTVRFKFEPSVGCPQLYKEVGKRFKLQDGS-FQLKYLDDEEEWVM 900
Query: 870 LTCDADLEECIDVCLSSESSTIKLCIQ 896
L D+DL+EC+++ ++K ++
Sbjct: 901 LVTDSDLQECLEILHGMGKHSVKFLVR 927
>At3g59580 putative protein
Length = 894
Score = 408 bits (1048), Expect = e-114
Identities = 278/833 (33%), Positives = 433/833 (51%), Gaps = 97/833 (11%)
Query: 120 VEGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPI 179
+EG+ KR + L S+ EK+++AL +F+ + +L Q W P+ GD+ +
Sbjct: 82 LEGSYACEKRPLDCTSVPRSLSHSLDEKMLKALSLFMEFS-GEGILAQFWTPIKTGDQYM 140
Query: 180 LSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRF 239
LS + + LDS L+ YRE S F FSAE + PGLPGRVF VPEWT +V +
Sbjct: 141 LSTCDQAYLLDS---RLSGYREASRRFTFSAEANQCSY-PGLPGRVFISGVPEWTSNVMY 196
Query: 240 FRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALE 299
+++ EY R++HA + ++ G++A+P+ E +C V+E+V ++ N+ ++ SVC+AL+
Sbjct: 197 YKTAEYLRMKHALDNEVRGSIAIPVLEASGSSCCAVLELVTCREKPNFDVEMNSVCRALQ 256
Query: 300 VVDLTSLKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSC-FQQGKDG- 357
V+L + S+I + + + AL EI++VLR+ C+ H+LPLA W+ C + +G +
Sbjct: 257 AVNLQT---STIPRRQYLSSNQKEALAEIRDVLRAVCYAHRLPLALAWIPCSYSKGANDE 313
Query: 358 -----CRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPF 412
++S++ L CI E +CYV D + F AC+EH+L +GQG+ GKA + N+P
Sbjct: 314 LVKVYGKNSKECSLLCIE--ETSCYVNDMEMEGFVNACLEHYLREGQGIVGKALISNKPS 371
Query: 413 FSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKN 472
FS+D+ +YPL HAR FGL AAVA +LRS ++ +DY+LEFFLPV+ S EQ+
Sbjct: 372 FSSDVKTFDICEYPLVQHARKFGLNAAVATKLRSTFTGDNDYILEFFLPVSMKGSSEQQL 431
Query: 473 MLISLSIIIQRCCRSLRVITDKE----LERTSSSVEVMALEDSGFARTVKWSEPQHITSV 528
+L SLS +QR CR+L+ ++D E E S SVE+ L PQ SV
Sbjct: 432 LLDSLSGTMQRLCRTLKTVSDAESIDGTEFGSRSVEMTNL-------------PQATVSV 478
Query: 529 ASLEPE--EKSSETVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEK 586
S + + FS++ ++ + + +E S ++ EK
Sbjct: 479 GSFHTTFLDTDVNSTRSTFSNISSNKRNEMAGSQGTLQQEISG-----------ARRLEK 527
Query: 587 RRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSL 646
+++ +K ++L VL+QYF GSLKDAAK++GVC TTLKR+CRQHGI RWPSRKI KV SL
Sbjct: 528 KKSSTEKNVSLNVLQQYFSGSLKDAAKSLGVCPTTLKRICRQHGIMRWPSRKINKVNRSL 587
Query: 647 QKLQLVIDSVQGASGASFKIDSFYSNFPDL--------ASPNLSGASL-VSALNQSENPN 697
+K+Q V+DSVQG G K DS F + +LS A +Q +
Sbjct: 588 RKIQTVLDSVQGVEG-GLKFDSVTGEFVAVGPFIQEFGTQKSLSSHDEDALARSQGDMDE 646
Query: 698 SLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHT-----SDVACNKDPVV 752
+S++P L S +G L ++H ++P T S + + D +
Sbjct: 647 DVSVEP-LEVKSHDGGGVKLEEDVE----TNHQAGPGSLKKPWTWISKQSGLIYSDDTDI 701
Query: 753 G-------KDSADVVLKRIRSEAELKSHSENKA-----------KLFPRSLSQET----- 789
G KD D+ ++R S L N S+S +
Sbjct: 702 GKRSEEVNKDKEDLCVRRCLSSVALAGDGMNTRIERGNGTVEPNHSISSSMSDSSNSSGA 761
Query: 790 --LGEHTKTEYQSYLLKTCHKATPKEDAH---RVKVTYGDEKTRFRM-PKSWSYEHLLQE 843
LG + + Q++ H + + + VK TY ++ RF++ P L +E
Sbjct: 762 VLLGSSSASLEQNWNQIRTHNNSGESGSSSTLTVKATYREDTVRFKLDPYVVGCSQLYRE 821
Query: 844 IARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQ 896
+A+RF + + F +KYLDD+ EWV+L D+DL EC ++ T+K ++
Sbjct: 822 VAKRFKLQE-GAFQLKYLDDEEEWVMLVTDSDLHECFEILNGMRKHTVKFLVR 873
>At5g66990 putative protein
Length = 277
Score = 52.4 bits (124), Expect = 1e-06
Identities = 39/147 (26%), Positives = 71/147 (47%), Gaps = 9/147 (6%)
Query: 525 ITSVASLEPEEKSSETVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTG 584
I+S SL+ G +++ E D I + N D + +++++ KT
Sbjct: 82 ISSTFSLDISSGWWNENNGNYNNQVEPNLDEISRTNTMGDPNMEQILHEDVNTMK-EKTS 140
Query: 585 EKR-----RAKADKTI---TLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPS 636
+KR R + D I + E+++QYF + AAK + + T LK+ CR+ GI RWP
Sbjct: 141 QKRIIMKRRYREDGVINNMSREMMKQYFYMPITKAAKELNIGVTLLKKRCRELGIPRWPH 200
Query: 637 RKIKKVGHSLQKLQLVIDSVQGASGAS 663
RK+ + + L+ ++ + +G + S
Sbjct: 201 RKLTSLNALIANLKDLLGNTKGRTPKS 227
>At4g35590 putative protein
Length = 370
Score = 52.4 bits (124), Expect = 1e-06
Identities = 45/170 (26%), Positives = 76/170 (44%), Gaps = 20/170 (11%)
Query: 495 ELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSE-TVGGKFSDLREHQE 553
E E+ SSS E D R + + + + E EE +E T +F + + Q
Sbjct: 160 EEEQCSSSTEGYYNSDLPKPRKLVLKQDLNCLPDSETESEESVNEKTEHSEFENDKTEQS 219
Query: 554 DSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAK 613
+S K I ++ +P ++ ++LE L +YF ++ +A++
Sbjct: 220 ESDAKTEILKKKKRTP-------------------SRHVAELSLEELSKYFDLTIVEASR 260
Query: 614 NIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGAS 663
N+ V T LK+ CR+ GI RWP RKIK + + LQ + Q + A+
Sbjct: 261 NLKVGLTVLKKKCREFGIPRWPHRKIKSLDCLIHDLQREAEKQQEKNEAA 310
>At5g53040 putative protein
Length = 256
Score = 52.0 bits (123), Expect = 2e-06
Identities = 27/85 (31%), Positives = 47/85 (54%), Gaps = 4/85 (4%)
Query: 572 EGNLSSVGISKTGEKRRAKADKT----ITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCR 627
+G +S+V + K K++ K + + ++Q+F + AAK + V T LK+ CR
Sbjct: 121 DGLISNVKVEKVTVKKKRNLKKKRQDKLEMSEIKQFFDRPIMKAAKELNVGLTVLKKRCR 180
Query: 628 QHGIKRWPSRKIKKVGHSLQKLQLV 652
+ GI RWP RK+K + ++ L+ V
Sbjct: 181 ELGIYRWPHRKLKSLNSLIKNLKNV 205
>At1g18790 hypothetical protein
Length = 269
Score = 49.3 bits (116), Expect = 1e-05
Identities = 29/75 (38%), Positives = 42/75 (55%), Gaps = 12/75 (16%)
Query: 583 TGEKRRAKAD--------KTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRW 634
T +KRR + + KT++ E + YF + AA+ + + T LK+ CR+ GIKRW
Sbjct: 108 TTKKRRCREECFSSCSVSKTLSKETISLYFYMPITQAARELNIGLTLLKKRCRELGIKRW 167
Query: 635 PSRKIKKVGHSLQKL 649
P RK+ SLQKL
Sbjct: 168 PHRKLM----SLQKL 178
>At1g74480 hypothetical protein
Length = 298
Score = 43.5 bits (101), Expect = 5e-04
Identities = 21/51 (41%), Positives = 31/51 (60%)
Query: 589 AKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKI 639
+ A T++ E + +YF + AA + V T LKR CR+ GI+RWP RK+
Sbjct: 129 SSAPTTLSKETVSRYFYMPITQAAIALNVGLTLLKRRCRELGIRRWPHRKL 179
>At1g63700 putative protein kinase
Length = 883
Score = 35.8 bits (81), Expect = 0.11
Identities = 29/100 (29%), Positives = 41/100 (41%), Gaps = 5/100 (5%)
Query: 696 PNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKD 755
P+ + PD P+ AT S+SS S G + S S S P SD V
Sbjct: 123 PHGATSIPDNTGAEPDFATASVSSGSSVGDIPSDSLLS-----PLASDCENGNRTPVNIS 177
Query: 756 SADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTK 795
S D + ++ AE+ NK ++ S + LG H K
Sbjct: 178 SRDQSMHSNKNSAEMFKPVPNKNRILSASPRRRPLGTHVK 217
>At4g24690 unknown protein
Length = 704
Score = 34.3 bits (77), Expect = 0.32
Identities = 23/65 (35%), Positives = 35/65 (53%), Gaps = 6/65 (9%)
Query: 818 VKVTYGDEKTRFRMPKSWSYE------HLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLT 871
VKV+YG RFR+P + + L ++IA FN+S ++ + Y D+D + V L
Sbjct: 10 VKVSYGGVLRRFRVPVKANGQLDLEMAGLKEKIAALFNLSADAELSLTYSDEDGDVVALV 69
Query: 872 CDADL 876
D DL
Sbjct: 70 DDNDL 74
>At3g18610 unknown protein
Length = 636
Score = 33.9 bits (76), Expect = 0.42
Identities = 27/128 (21%), Positives = 51/128 (39%), Gaps = 1/128 (0%)
Query: 707 PLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRS 766
P+ + T + + ++ S S SS E P KDS+
Sbjct: 248 PVVKKKPTTVVKDAKAESSSSEEESSSDDEPTPAKKPTVVKNAKPAAKDSSSSEEDSDEE 307
Query: 767 EAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEK 826
E++ + KAK+ ++ QE+ + + E K K TPK+ V++ ++K
Sbjct: 308 ESDDEKPPTKKAKVSSKTSKQESSSDESSDESDKEESKD-EKVTPKKKDSDVEMVDAEQK 366
Query: 827 TRFRMPKS 834
+ + PK+
Sbjct: 367 SNAKQPKT 374
>At3g07070 putative protein kinase
Length = 414
Score = 33.5 bits (75), Expect = 0.55
Identities = 28/109 (25%), Positives = 47/109 (42%), Gaps = 7/109 (6%)
Query: 671 SNFPDLASPNLSGASLVSALNQSENPNSLSIQPD--LGPLSPEGATK-SLSSSCSQGSLS 727
S FP+LA P+L G ALNQ+ ++ +Q + + PL + T + GS+S
Sbjct: 306 SRFPELADPSLEGVFPEKALNQAVAVAAMCLQEEATVRPLMSDVVTALGFLGTAPDGSIS 365
Query: 728 SHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSEN 776
+ P SD +D V ++ V + + ++HS N
Sbjct: 366 VPHY----DDPPQPSDETSVEDSVAAEERERAVAEAMEWGVASRAHSRN 410
>At1g31440 unknown protein
Length = 439
Score = 33.5 bits (75), Expect = 0.55
Identities = 41/167 (24%), Positives = 77/167 (45%), Gaps = 21/167 (12%)
Query: 685 SLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSH--SCSSMPEQQPHTS 742
+LV A +S + N+L I L L E + + S SL H +S+P+Q+P+++
Sbjct: 231 ALVEA-ERSYHRNALDI---LDKLHSEMIAEEEAIGSSPKSLPLHIEDSASLPQQEPNSN 286
Query: 743 DVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYL 802
K +GK +K R E E+KS+ + K P+ + + E TK+ +Q +
Sbjct: 287 SSGEIKSNPLGK------IKASRRE-EIKSNPQEVTKPSPKDEMKSSPQEETKSNHQKEI 339
Query: 803 LKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFN 849
K++P+E+ +K + G + S + + L ++ F+
Sbjct: 340 -----KSSPQEE---IKKSNGSDDHHNHQLLSQNDSYFLAKVVHPFD 378
>At1g62390 putative protein
Length = 751
Score = 32.7 bits (73), Expect = 0.94
Identities = 48/193 (24%), Positives = 74/193 (37%), Gaps = 29/193 (15%)
Query: 705 LGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSAD-VVLKR 763
LGP P + GS+S + S+ ++P + V K A VVLK
Sbjct: 200 LGPCLPSRNVHKKGVTSPVGSVSLPNASNGKVERPQVVNPVTENGGSVSKGQASRVVLKP 259
Query: 764 IRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHR-VKVTY 822
+ SHS P+ E LG + + K K R +K Y
Sbjct: 260 V-------SHS-------PKGSKVEELGSSS--------VAVVGKVQEKRIRWRPLKFVY 297
Query: 823 GDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADL---EEC 879
+ +MP + ++ L + ++ RF S +KY D+D + V +T A+L E
Sbjct: 298 DHDIRLGQMPVNCRFKELREIVSSRFPSS--KAVLIKYKDNDGDLVTITSTAELKLAESA 355
Query: 880 IDVCLSSESSTIK 892
D L+ E T K
Sbjct: 356 ADCILTKEPDTDK 368
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.131 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,737,896
Number of Sequences: 26719
Number of extensions: 915832
Number of successful extensions: 2855
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 2750
Number of HSP's gapped (non-prelim): 60
length of query: 903
length of database: 11,318,596
effective HSP length: 108
effective length of query: 795
effective length of database: 8,432,944
effective search space: 6704190480
effective search space used: 6704190480
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0193.5


