

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0192.10
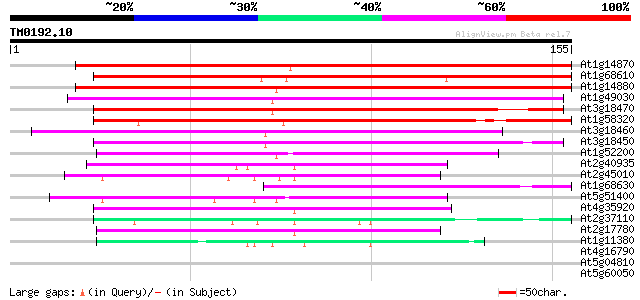
(155 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g14870 unknown protein 166 5e-42
At1g68610 hypothetical protein 157 2e-39
At1g14880 unknown protein 153 3e-38
At1g49030 hypothetical protein; similar to ESTs gb|H76375.1, gb|... 130 4e-31
At3g18470 unknown protein (At3g18480) 127 3e-30
At1g58320 hypothetical protein 124 2e-29
At3g18460 hypothetical protein 120 3e-28
At3g18450 hypothetical protein 111 1e-25
At1g52200 unknown protein 100 4e-22
At2g40935 unknown protein 87 4e-18
At2g45010 unknown protein 65 1e-11
At1g68630 hypothetical protein 61 3e-10
At5g51400 unknown protein 60 6e-10
At4g35920 unknown protein 56 9e-09
At2g37110 unknown protein 55 2e-08
At2g17780 hypothetical protein 50 6e-07
At1g11380 unknown protein 47 4e-06
At4g16790 glycoprotein homolog 34 0.027
At5g04810 membrane-associated salt-inducible protein-like 33 0.080
At5g60050 unknown protein 32 0.18
>At1g14870 unknown protein
Length = 152
Score = 166 bits (420), Expect = 5e-42
Identities = 73/143 (51%), Positives = 96/143 (67%), Gaps = 6/143 (4%)
Query: 19 PQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGV-- 76
P EWSTG DCFSD NCC+T+WCPC+TFG++AEIVD+GSTSCG +G + V
Sbjct: 10 PHAEGEWSTGFCDCFSDCKNCCITFWCPCITFGQVAEIVDRGSTSCGTAGALYALIAVVT 69
Query: 77 ----FYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGN 132
YS YR K+R+QYN+KG++C DCL H C C+L Q+YREL+ +G++M +GW GN
Sbjct: 70 GCACIYSCFYRGKMRAQYNIKGDDCTDCLKHFCCELCSLTQQYRELKHRGYDMSLGWAGN 129
Query: 133 VEQQTRGVAMTSTAPTVEQSMTR 155
VE+Q + AP + MTR
Sbjct: 130 VERQQNQGGVAMGAPVFQGGMTR 152
>At1g68610 hypothetical protein
Length = 160
Score = 157 bits (397), Expect = 2e-39
Identities = 74/141 (52%), Positives = 94/141 (66%), Gaps = 9/141 (6%)
Query: 24 EWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASG------FYFVQHG-- 75
+WST L +C+ D N+CCLT WCPCV FGRIAE+VD+GSTSCG SG F +G
Sbjct: 18 DWSTDLCECWMDINSCCLTCWCPCVAFGRIAEVVDRGSTSCGVSGAMYMIIFMLTGYGGS 77
Query: 76 VFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELE-KQGFNMKIGWHGNVE 134
YS YRTK+R+QYNLK C DC H C CALCQEYR+L+ + ++ IGWHGN+E
Sbjct: 78 SLYSCFYRTKLRAQYNLKERPCCDCCVHFCCEPCALCQEYRQLQHNRDLDLVIGWHGNME 137
Query: 135 QQTRGVAMTSTAPTVEQSMTR 155
+ R A T +AP ++ M+R
Sbjct: 138 RHARLAASTPSAPPLQAPMSR 158
>At1g14880 unknown protein
Length = 151
Score = 153 bits (387), Expect = 3e-38
Identities = 70/143 (48%), Positives = 92/143 (63%), Gaps = 6/143 (4%)
Query: 19 PQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFV------ 72
P EWSTG DCFSD NCC+T CPC+TFG++AEIVD+GS SC A+G ++
Sbjct: 9 PHAQGEWSTGFCDCFSDCRNCCITLCCPCITFGQVAEIVDRGSKSCCAAGALYMLIDLIT 68
Query: 73 QHGVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGN 132
G Y+ Y K+R+QYN+KG+ C DCL H C+ CAL Q+YREL+ +GF+M +GW GN
Sbjct: 69 SCGRMYACFYSGKMRAQYNIKGDGCTDCLKHFCCNLCALTQQYRELKHRGFDMSLGWAGN 128
Query: 133 VEQQTRGVAMTSTAPTVEQSMTR 155
E+Q + AP + MTR
Sbjct: 129 AEKQQNQGGVAMGAPAFQGGMTR 151
>At1g49030 hypothetical protein; similar to ESTs gb|H76375.1,
gb|AI995130.1
Length = 224
Score = 130 bits (326), Expect = 4e-31
Identities = 62/143 (43%), Positives = 85/143 (59%), Gaps = 6/143 (4%)
Query: 17 PPPQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYF----- 71
P P W++GL+DC +D N +T P VTFG+IAE++D+G+TSCG +G +
Sbjct: 80 PVHTQPSNWTSGLFDCMNDGENALITCCFPFVTFGQIAEVIDEGATSCGTAGMLYGLICC 139
Query: 72 -VQHGVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWH 130
Y+ +RTK+RS+Y L D +THCFC CALCQEYREL+ +G + IGW
Sbjct: 140 LFAIPCVYTCTFRTKLRSKYGLPDAPAPDWITHCFCEYCALCQEYRELKNRGLDPSIGWI 199
Query: 131 GNVEQQTRGVAMTSTAPTVEQSM 153
GNV++Q G AP + Q M
Sbjct: 200 GNVQKQRMGQQQEMMAPPMGQRM 222
>At3g18470 unknown protein (At3g18480)
Length = 133
Score = 127 bits (318), Expect = 3e-30
Identities = 60/136 (44%), Positives = 83/136 (60%), Gaps = 14/136 (10%)
Query: 24 EWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYF------VQHGVF 77
+W++GL+ C D CLT +CPCVTFGRIA+I D+G T CG G ++ V
Sbjct: 4 QWTSGLFSCMEDSETACLTCFCPCVTFGRIADISDEGRTGCGRCGVFYGLICCVVGLPCL 63
Query: 78 YSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQQT 137
+S YRTKIRS++ L + DC+TH FC CALCQE+REL+ +G + IGW GN+++
Sbjct: 64 FSCTYRTKIRSKFGLPESPTSDCVTHFFCECCALCQEHRELKTRGLDPSIGWSGNMQR-- 121
Query: 138 RGVAMTSTAPTVEQSM 153
+ AP + Q M
Sbjct: 122 ------TMAPPMSQQM 131
>At1g58320 hypothetical protein
Length = 148
Score = 124 bits (312), Expect = 2e-29
Identities = 65/138 (47%), Positives = 83/138 (60%), Gaps = 11/138 (7%)
Query: 24 EWSTGLYDCFS-DYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQH-----GVF 77
+W+TGLYDC S D + CC T+ CPCV FGRIAEI+DKG TS G +G V G +
Sbjct: 16 QWTTGLYDCLSEDISTCCFTWVCPCVAFGRIAEILDKGETSRGLAGLMVVAMSSIGCGWY 75
Query: 78 YSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQQT 137
Y++ YR K+R QY L C D HCFC CAL QE+REL+ +G + +GW N+E
Sbjct: 76 YASKYRAKLRHQYALPEAPCADGAIHCFCCPCALTQEHRELKHRGLDPSLGW--NIE--- 130
Query: 138 RGVAMTSTAPTVEQSMTR 155
G ++T P V M R
Sbjct: 131 NGGLNSNTPPFVASGMDR 148
>At3g18460 hypothetical protein
Length = 184
Score = 120 bits (301), Expect = 3e-28
Identities = 59/135 (43%), Positives = 78/135 (57%), Gaps = 5/135 (3%)
Query: 7 GSTDYQPTPPPPPQPPVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGA 66
G + Q P Q WS+ L+DC +D N +T PCVT G+IAEIVD+G+T C
Sbjct: 36 GQANIQTGRPVNNQTQNLWSSDLFDCMNDSENAVITCLAPCVTLGQIAEIVDEGATPCAT 95
Query: 67 SGF-----YFVQHGVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQ 121
G +F+ YS +R K+R++Y L D +TH FC CALCQEYREL+ +
Sbjct: 96 GGLLYGMIFFIGVPFVYSCMFRAKMRNKYGLPDAPAPDWITHLFCEHCALCQEYRELKHR 155
Query: 122 GFNMKIGWHGNVEQQ 136
GF+ IGW GNV+ Q
Sbjct: 156 GFDPNIGWAGNVQAQ 170
>At3g18450 hypothetical protein
Length = 184
Score = 111 bits (278), Expect = 1e-25
Identities = 57/135 (42%), Positives = 79/135 (58%), Gaps = 7/135 (5%)
Query: 24 EWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGF-----YFVQHGVFY 78
+WS+ L+DC +D N +T PCVTFG+IAEIVD+G+T C +G +F Y
Sbjct: 51 QWSSQLFDCMNDSENAVITLIAPCVTFGQIAEIVDEGATPCATAGLLYGALFFTGASFVY 110
Query: 79 SADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQQTR 138
S +R +IR ++ L D +TH C ALCQEYREL+ GF+ +GW GNV+Q +
Sbjct: 111 SYMFRARIRKKFGLPDAPAPDWITHLVCMPFALCQEYRELKHHGFDPILGWAGNVQQAQQ 170
Query: 139 GVAMTSTAPTVEQSM 153
M T PT ++ M
Sbjct: 171 QEMM--TPPTGQRMM 183
>At1g52200 unknown protein
Length = 190
Score = 100 bits (248), Expect = 4e-22
Identities = 49/118 (41%), Positives = 70/118 (58%), Gaps = 8/118 (6%)
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFV-------QHGVF 77
WSTGL+DC +D N LT PCVTFG+IAE++D+G +C F ++ H V
Sbjct: 53 WSTGLFDCQADQANAVLTTIVPCVTFGQIAEVMDEGEMTCPLGTFMYLLMMPALCSHWVM 112
Query: 78 YSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQ 135
+ YR K+R ++NL DC +H C C+LCQEYREL+ + + +GW+G + Q
Sbjct: 113 -GSKYREKMRRKFNLVEAPYSDCASHVLCPCCSLCQEYRELKIRNLDPSLGWNGILAQ 169
>At2g40935 unknown protein
Length = 190
Score = 87.0 bits (214), Expect = 4e-18
Identities = 44/121 (36%), Positives = 64/121 (52%), Gaps = 21/121 (17%)
Query: 22 PVEWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGS------TSC----------- 64
P +WS+G+ CF D +CC+ +CPC FG+ AE++ G+ T C
Sbjct: 44 PRQWSSGICACFDDMQSCCVGLFCPCYIFGKNAELLGSGTFAGPCLTHCISWALVNTICC 103
Query: 65 -GASGFYFVQHGVF---YSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEK 120
+G G F Y+ YR +R++YNL+ C D +TH FC CA+CQEYRE+ +
Sbjct: 104 FATNGALLGLPGCFVSCYACGYRKSLRAKYNLQEAPCGDFVTHFFCHLCAICQEYREIRE 163
Query: 121 Q 121
Q
Sbjct: 164 Q 164
>At2g45010 unknown protein
Length = 244
Score = 65.1 bits (157), Expect = 1e-11
Identities = 43/141 (30%), Positives = 64/141 (44%), Gaps = 37/141 (26%)
Query: 16 PPPPQPPVE--WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDK-------------- 59
P +PP + W+TG++ C D +C +CPCV FGR E V +
Sbjct: 51 PETYEPPSDENWTTGIFGCAEDPESCRTGLFCPCVLFGRNIEAVREEIPWTQPCVCHAVC 110
Query: 60 --GSTSCGA-----SGFYFVQ------HGVF--------YSADYRTKIRSQYNLKGNNCL 98
G + A SG+ Q G+F YS +R +++ +Y+LK C
Sbjct: 111 VEGGMALAAVTALFSGYIDPQTTVVICEGLFFAWWMCGIYSGLFRQELQKKYHLKNAPCD 170
Query: 99 DCLTHCFCSRCALCQEYRELE 119
C+ HC CALCQE+RE++
Sbjct: 171 HCMVHCCLHWCALCQEHREMK 191
>At1g68630 hypothetical protein
Length = 93
Score = 60.8 bits (146), Expect = 3e-10
Identities = 32/85 (37%), Positives = 45/85 (52%), Gaps = 3/85 (3%)
Query: 71 FVQHGVFYSADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQGFNMKIGWH 130
F+ Y+ R+++R + L C D L H FC+ CA+CQE REL+ +G + IGW
Sbjct: 12 FIGCSWLYAFPNRSRLREHFALPEEPCRDFLVHLFCTPCAICQESRELKNRGADPSIGWL 71
Query: 131 GNVEQQTRGVAMTSTAPTVEQSMTR 155
NVE+ +R T P V M R
Sbjct: 72 SNVEKWSREKV---TPPIVVPGMIR 93
>At5g51400 unknown protein
Length = 241
Score = 59.7 bits (143), Expect = 6e-10
Identities = 41/147 (27%), Positives = 64/147 (42%), Gaps = 38/147 (25%)
Query: 12 QPTPPPPPQPPVE-WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAE--------------- 55
QP P P E W+TG++ C D N+ L +CP V FGR+ E
Sbjct: 48 QPLPENFEAPADEPWTTGIFGCTEDMNSFWLGLFCPSVLFGRVYETLSDEETSWKKACIC 107
Query: 56 ---IVDKGSTSCGA----------------SGFYFV--QHGVFYSADYRTKIRSQYNLKG 94
+V+ G T+ G FV G+ Y+ + R ++ +Y+L+
Sbjct: 108 HSIVVEGGLTAASMLACVPGIDPHTSLLIWEGLLFVWWMCGI-YTGNVRQTLQRKYHLQN 166
Query: 95 NNCLDCLTHCFCSRCALCQEYRELEKQ 121
C C+ HC CA+CQE+RE++ +
Sbjct: 167 APCDPCMVHCCLHFCAVCQEHREMKNR 193
>At4g35920 unknown protein
Length = 421
Score = 55.8 bits (133), Expect = 9e-09
Identities = 33/104 (31%), Positives = 48/104 (45%), Gaps = 5/104 (4%)
Query: 24 EWSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF-----Y 78
EW T L C S+ + C T++ PC T +IA S + + + + Y
Sbjct: 299 EWHTDLLACCSEPSLCFKTFFFPCGTLAKIATAASNRHISSAEACNELMAYSLILSCCCY 358
Query: 79 SADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELEKQG 122
+ R K+R N+ G D L+H C CAL QE RE+E +G
Sbjct: 359 TCCVRRKLRKTLNITGGFIDDFLSHVMCCCCALVQELREVEIRG 402
>At2g37110 unknown protein
Length = 242
Score = 55.1 bits (131), Expect = 2e-08
Identities = 51/178 (28%), Positives = 72/178 (39%), Gaps = 56/178 (31%)
Query: 24 EWSTGLYDCF--------SDYNNCCLTYWCPCVTFGRIAEIVDKG----STSCGAS-GFY 70
+W++GL+ C SD C L PCV +G AE + S C G Y
Sbjct: 74 QWNSGLFTCLGRNDEFCSSDLEVCLLGSVAPCVLYGTNAERLGSAPGTFSNHCLTYLGLY 133
Query: 71 FVQHGVF--------YSADYRTKIRSQYNLKGN------------NCL------------ 98
FV + +F +S R+ IR ++NL+G+ C+
Sbjct: 134 FVGNSLFGWNCLAPWFSYSSRSAIRRKFNLEGSFEAMNRSCGCCGGCIEDEMQREHLETT 193
Query: 99 -DCLTHCFCSRCALCQEYRELEKQGFNMKIGWHGNVEQQTRGVAMTSTAPTVEQSMTR 155
D +TH C CALCQE REL ++ H Q+ V M P +EQ+M R
Sbjct: 194 CDFVTHVLCHTCALCQEGRELRRKVL------HPGFNAQSTVVVM----PPIEQTMGR 241
>At2g17780 hypothetical protein
Length = 417
Score = 49.7 bits (117), Expect = 6e-07
Identities = 30/100 (30%), Positives = 43/100 (43%), Gaps = 5/100 (5%)
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSCGASGFYFVQHGVF-----YS 79
W L DC S+ C T + PC T +I+ + S + + + Y+
Sbjct: 292 WHADLLDCCSEPCLCLKTLFFPCGTLAKISTVATSRQISSTEVCKNLIVYSLILSCCCYT 351
Query: 80 ADYRTKIRSQYNLKGNNCLDCLTHCFCSRCALCQEYRELE 119
R K+R N+ G D L+H C CAL QE RE+E
Sbjct: 352 CCIRKKLRKTLNITGGCIDDFLSHLMCCCCALVQELREVE 391
>At1g11380 unknown protein
Length = 254
Score = 47.0 bits (110), Expect = 4e-06
Identities = 40/138 (28%), Positives = 56/138 (39%), Gaps = 34/138 (24%)
Query: 25 WSTGLYDCFSDYNNCCLTYWCPCVTFGRIAEIVDKGSTSC---GA------SGFYF---- 71
W + DCF D + C + CPC FG+ + G SC GA +GF F
Sbjct: 86 WEGDVMDCFEDRHLCIESACCPCYRFGK--NMTRTGFGSCFLQGAVHMILIAGFLFNVVA 143
Query: 72 ----VQHGVFYSA------------DYRTKIRSQYNLKGNNCL--DCLTHCFCSRCALCQ 113
+H Y A +R IR ++N++G + D + H C C L Q
Sbjct: 144 FAVTKRHCFLYLAIAFVLLIGSYLGFFRMLIRRKFNIRGTDSFLDDFIHHLVCPFCTLTQ 203
Query: 114 EYRELEKQGFNMKIGWHG 131
E + LE + I WHG
Sbjct: 204 ESKTLEMNNVHDGI-WHG 220
>At4g16790 glycoprotein homolog
Length = 473
Score = 34.3 bits (77), Expect = 0.027
Identities = 12/25 (48%), Positives = 17/25 (68%)
Query: 1 MYKTVAGSTDYQPTPPPPPQPPVEW 25
++KT ++ P PPPPP PPVE+
Sbjct: 317 VHKTKFAGGEFHPPPPPPPPPPVEY 341
Score = 28.9 bits (63), Expect = 1.2
Identities = 8/15 (53%), Positives = 13/15 (86%)
Query: 8 STDYQPTPPPPPQPP 22
++++ P+PPPPP PP
Sbjct: 275 ASEFHPSPPPPPPPP 289
Score = 25.8 bits (55), Expect = 9.8
Identities = 8/11 (72%), Positives = 9/11 (81%)
Query: 13 PTPPPPPQPPV 23
P PPPPP PP+
Sbjct: 282 PPPPPPPPPPL 292
>At5g04810 membrane-associated salt-inducible protein-like
Length = 952
Score = 32.7 bits (73), Expect = 0.080
Identities = 14/26 (53%), Positives = 16/26 (60%)
Query: 8 STDYQPTPPPPPQPPVEWSTGLYDCF 33
S+ P PPPPP PPVE +T D F
Sbjct: 118 SSKLSPPPPPPPPPPVEETTQFRDEF 143
>At5g60050 unknown protein
Length = 499
Score = 31.6 bits (70), Expect = 0.18
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 3/34 (8%)
Query: 8 STDYQPTPPPPPQPPVEWSTG---LYDCFSDYNN 38
ST P PPPPP PP E + L+D SD +N
Sbjct: 34 STASPPLPPPPPPPPNESTLSNPTLFDMMSDEHN 67
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.136 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,161,261
Number of Sequences: 26719
Number of extensions: 184708
Number of successful extensions: 2631
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 87
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1667
Number of HSP's gapped (non-prelim): 554
length of query: 155
length of database: 11,318,596
effective HSP length: 91
effective length of query: 64
effective length of database: 8,887,167
effective search space: 568778688
effective search space used: 568778688
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0192.10


