

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0191.7
(680 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
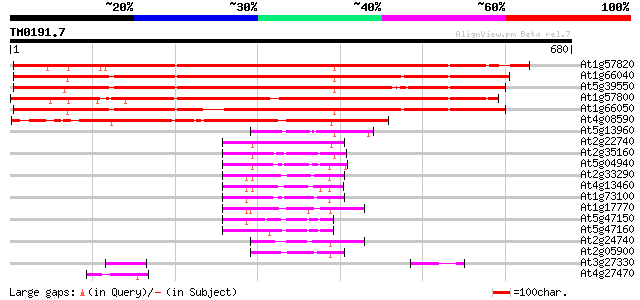
Score E
Sequences producing significant alignments: (bits) Value
At1g57820 putative transcription factor protein (At1g57820) 825 0.0
At1g66040 hypothetical protein 758 0.0
At5g39550 zinc finger -like protein 756 0.0
At1g57800 hypothetical protein 704 0.0
At1g66050 hypothetical protein 700 0.0
At4g08590 unknown protein with zinc finger motive 479 e-135
At5g13960 unknown protein (At5g13960) 83 4e-16
At2g22740 similar to mammalian MHC III region protein G9a 79 8e-15
At2g35160 similar to mammalian MHC III region protein G9a 74 4e-13
At5g04940 SET-domain protein-like 71 2e-12
At2g33290 similar to mammalian MHC III region protein G9a 69 1e-11
At4g13460 putative protein 67 4e-11
At1g73100 putative protein 64 2e-10
At1g17770 putative protein 61 2e-09
At5g47150 putative protein 53 6e-07
At5g47160 putative protein 52 1e-06
At2g24740 pseudogene 51 2e-06
At2g05900 hypothetical protein 47 4e-05
At3g27330 hypothetical protein 46 8e-05
At4g27470 putative RING zinc finger protein 45 1e-04
>At1g57820 putative transcription factor protein (At1g57820)
Length = 645
Score = 825 bits (2130), Expect = 0.0
Identities = 400/650 (61%), Positives = 482/650 (73%), Gaps = 43/650 (6%)
Query: 5 LPCDADGTCMLCKLKPPPSETLQCHTCITPWHLPCLPTPP---ASFVNWDCPDCSSSISD 61
LPCD DG CM CK PPP E+L C TC+TPWH+ CL +PP AS + W CPDCS I
Sbjct: 7 LPCDGDGVCMRCKSNPPPEESLTCGTCVTPWHVSCLSSPPKTLASTLQWHCPDCSGEIDP 66
Query: 62 FHAPAPAP---SAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGPSSGE-------KPP 111
A SAG+DLV+AIRAI++D SL+ ++KAK +Q L+ G E K
Sbjct: 67 LPVSGGATGFESAGSDLVAAIRAIEADESLSTEEKAKMRQRLLSGKGVEEDDEEEKRKKK 126
Query: 112 AAG-----GILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRTCANCRR 166
G +L ++ CSFCMQLP+RPVT PCGHN CLKCFEKW+GQGKRTC CR
Sbjct: 127 GKGKNPNLDVLSALGDNLMCSFCMQLPERPVTKPCGHNACLKCFEKWMGQGKRTCGKCRS 186
Query: 167 DIPAKIASQPRINSQLAIAIRLAKLAKSEGKVSGEPKIYHFMRNQDRPDKAFTTDRAKKT 226
IP K+A PRINS L AIRLAK++KS + K++HF+ NQDRPDKAFTT+RAKKT
Sbjct: 187 IIPEKMAKNPRINSSLVAAIRLAKVSKSAAATTS--KVFHFISNQDRPDKAFTTERAKKT 244
Query: 227 GKANACSGKIFVTVPPDHFGPIPAENDPTRNQGVLVGETWEDRMECRQWGAHLPHVAGIA 286
GKANA SGKI+VT+PPDHFGPIPAENDP RNQG+LVGE+WEDR+ECRQWGAH PHVAGIA
Sbjct: 245 GKANAASGKIYVTIPPDHFGPIPAENDPVRNQGLLVGESWEDRLECRQWGAHFPHVAGIA 304
Query: 287 GQSTYGAQSVALSGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALR 346
GQSTYGAQSVALSGGY DDEDHGEWFLYTGSGGRDLSGNKRTNK QSFDQKFE N AL+
Sbjct: 305 GQSTYGAQSVALSGGYKDDEDHGEWFLYTGSGGRDLSGNKRTNKEQSFDQKFEKSNAALK 364
Query: 347 LSCRKGYPVRVVRSHKEKRSSYAPESGVRYDGVYRIEKCWRKNGIQ----VCRYLFVRCD 402
LSC+ GYPVRVVRSHKEKRS+YAPE GVRYDGVYRIEKCWRK G+Q VCRYLFVRCD
Sbjct: 365 LSCKLGYPVRVVRSHKEKRSAYAPEEGVRYDGVYRIEKCWRKVGVQGSFKVCRYLFVRCD 424
Query: 403 NEPAPWTTDDVGDRPRPLPKIDELKGAVDITERKGDPSWDFDEEKGSWLWKKPPPASKKS 462
NEPAPWT+D+ GDRPRP+P I EL A D+ ERK PSWDFDE +G W W KPPPASKKS
Sbjct: 425 NEPAPWTSDENGDRPRPIPNIPELNMATDLFERKETPSWDFDEGEGCWKWMKPPPASKKS 484
Query: 463 IDVINPKDGTRVR--VKKTKNVSLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEG 520
++V+ P++ +R +K + +++ +LLKEF C IC +VL+ P+TTPCAHNFCK CLE
Sbjct: 485 VNVLAPEERKNLRKAIKAAHSNTMRARLLKEFKCQICQQVLTLPVTTPCAHNFCKACLEA 544
Query: 521 AFSGKSFIRKRACEGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIA 580
F+GK+ +R+R+ GGR+LR++KNV+ CP C DI+DFLQNPQVNRE+ V+E L++
Sbjct: 545 KFAGKTLVRERS-TGGRTLRSRKNVLNCPCCPTDISDFLQNPQVNREVAEVIEKLKT--- 600
Query: 581 QDENSEQVENSEELSDKSDEIVKNVPDEVEVSKPCDDSTESDENVKNVTD 630
Q+E++ ++E+ +E E + P +DS + + +K TD
Sbjct: 601 QEEDTAELEDEDE-------------GECSGTTPEEDSEQPKKRIKLDTD 637
>At1g66040 hypothetical protein
Length = 622
Score = 758 bits (1958), Expect = 0.0
Identities = 371/612 (60%), Positives = 441/612 (71%), Gaps = 21/612 (3%)
Query: 5 LPCDADGTCMLCKLKPPPSETLQCHTCITPWHLPCL-PTPPASFV-NWDCPDCSSSISDF 62
LPCD DG CM C++ PP ETL C TC+TPWH+ CL P AS +W+CPDCS +
Sbjct: 7 LPCDGDGVCMRCQVTPPSEETLTCGTCVTPWHVSCLLPESLASSTGDWECPDCSGVVVPS 66
Query: 63 HAPAPA----PSAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGPSSGEKPPAAGGILD 118
AP S+G+ LV+AIRAI +D +LT+ +KAKK+Q L+ G L+
Sbjct: 67 AAPGTGISGPESSGSVLVAAIRAIQADVTLTEAEKAKKRQRLMSGGGDDGVDDEEKKKLE 126
Query: 119 IFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKW-IGQGKRTCANCRRDIPAKIASQPR 177
IF CS C+QLP+RPVTTPCGHNFCLKCFEKW +GQGK TC CR IP +A PR
Sbjct: 127 IF-----CSICIQLPERPVTTPCGHNFCLKCFEKWAVGQGKLTCMICRSKIPRHVAKNPR 181
Query: 178 INSQLAIAIRLAKLAKSEGKVSGEPKIYHFMRNQDRPDKAFTTDRAKKTGKANACSGKIF 237
IN L AIRLA + K G+ + K++H +RNQDRPDKAFTT+RA KTGKANA SGK F
Sbjct: 182 INLALVSAIRLANVTKCSGEATAA-KVHHIIRNQDRPDKAFTTERAVKTGKANAASGKFF 240
Query: 238 VTVPPDHFGPIPAENDPTRNQGVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYGAQSVA 297
VT+P DHFGPIPA ND TRNQGVLVGE+WEDR ECRQWG H PHVAGIAGQ+ GAQSVA
Sbjct: 241 VTIPRDHFGPIPAANDVTRNQGVLVGESWEDRQECRQWGVHFPHVAGIAGQAAVGAQSVA 300
Query: 298 LSGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRV 357
LSGGY DDEDHGEWFLYTGSGGRDLSGNKR NK+QS DQ F+NMNEALRLSC+ GYPVRV
Sbjct: 301 LSGGYDDDEDHGEWFLYTGSGGRDLSGNKRVNKIQSSDQAFKNMNEALRLSCKMGYPVRV 360
Query: 358 VRSHKEKRSSYAPESGVRYDGVYRIEKCWRKNGIQ----VCRYLFVRCDNEPAPWTTDDV 413
VRS KEKRS+YAP GVRYDGVYRIEKCW G+Q +CRYLFVRCDNEPAPWT+D+
Sbjct: 361 VRSWKEKRSAYAPAEGVRYDGVYRIEKCWSNVGVQGLHKMCRYLFVRCDNEPAPWTSDEH 420
Query: 414 GDRPRPLPKIDELKGAVDITERKGDPSWDFDEEKGSWLWKKPPPASKKSIDVINPKDGTR 473
GDRPRPLP + EL+ A D+ RK PSW FDE +G W W K PP S+ ++D K R
Sbjct: 421 GDRPRPLPDVPELENATDLFVRKESPSWGFDEAEGRWKWMKSPPVSRMALDTEERKKNKR 480
Query: 474 VRVKKTKNVSLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRAC 533
K N ++K +LLKEF C IC KVLS P+TTPCAHNFCK CLE F+G + +R R+
Sbjct: 481 A---KKGNNAMKARLLKEFSCQICRKVLSLPVTTPCAHNFCKACLEAKFAGITQLRDRS- 536
Query: 534 EGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIAQDENSEQVENSEE 593
G R LRA+KN+M CP C+ D+++FLQNPQVNREMM ++E+ + + E +E SEE
Sbjct: 537 NGVRKLRAKKNIMTCPCCTTDLSEFLQNPQVNREMMEIIENFKKSEEEAEVAESSNISEE 596
Query: 594 LSDKSDEIVKNV 605
++S+ K +
Sbjct: 597 EEEESEPPTKKI 608
>At5g39550 zinc finger -like protein
Length = 617
Score = 756 bits (1953), Expect = 0.0
Identities = 371/608 (61%), Positives = 446/608 (73%), Gaps = 24/608 (3%)
Query: 5 LPCDADGTCMLCKLKPPPSETLQCHTCITPWHLPCL-PTPPASFVN-WDCPDCSSSISDF 62
LPCD DG CM C++ PP ETL C TC+TPWH+PCL P AS W+CPDCS +
Sbjct: 7 LPCDGDGVCMRCQVNPPSEETLTCGTCVTPWHVPCLLPESLASSTGEWECPDCSGVVVPS 66
Query: 63 HAP----APAPSAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGPSSGEKPPAAGGILD 118
AP A S+G+ LV+AIRAI +D +LT+ +KAKK+Q+L+ G L+
Sbjct: 67 AAPGTGNARPESSGSVLVAAIRAIQADETLTEAEKAKKRQKLMSGGGDDGVDEEEKKKLE 126
Query: 119 IFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKW-IGQGKRTCANCRRDIPAKIASQPR 177
IF CS C+QLP+RP+TTPCGHNFCLKCFEKW +GQGK TC CR IP +A PR
Sbjct: 127 IF-----CSICIQLPERPITTPCGHNFCLKCFEKWAVGQGKLTCMICRSKIPRHVAKNPR 181
Query: 178 INSQLAIAIRLAKLAKSEGKVSGEPKIYHFMRNQDRPDKAFTTDRAKKTGKANACSGKIF 237
IN L AIRLA + K + + K++H +RNQDRP+KAFTT+RA KTGKANA SGK F
Sbjct: 182 INLALVSAIRLANVTKCSVEATAA-KVHHIIRNQDRPEKAFTTERAVKTGKANAASGKFF 240
Query: 238 VTVPPDHFGPIPAENDPTRNQGVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYGAQSVA 297
VT+P DHFGPIPAEND TR QGVLVGE+WEDR ECRQWGAH PH+AGIAGQS GAQSVA
Sbjct: 241 VTIPRDHFGPIPAENDVTRKQGVLVGESWEDRQECRQWGAHFPHIAGIAGQSAVGAQSVA 300
Query: 298 LSGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRV 357
LSGGY DDEDHGEWFLYTGSGGRDLSGNKR NK QS DQ F+NMNE+LRLSC+ GYPVRV
Sbjct: 301 LSGGYDDDEDHGEWFLYTGSGGRDLSGNKRINKKQSSDQAFKNMNESLRLSCKMGYPVRV 360
Query: 358 VRSHKEKRSSYAPESGVRYDGVYRIEKCWRKNGIQ----VCRYLFVRCDNEPAPWTTDDV 413
VRS KEKRS+YAP GVRYDGVYRIEKCW G+Q VCRYLFVRCDNEPAPWT+D+
Sbjct: 361 VRSWKEKRSAYAPAEGVRYDGVYRIEKCWSNVGVQGSFKVCRYLFVRCDNEPAPWTSDEH 420
Query: 414 GDRPRPLPKIDELKGAVDITERKGDPSWDFDEEKGSWLWKKPPPASKKSIDVINPKDGTR 473
GDRPRPLP + EL+ A D+ RK PSWDFDE +G W W K PP S+ ++D P++ R
Sbjct: 421 GDRPRPLPNVPELETAADLFVRKESPSWDFDEAEGRWKWMKSPPVSRMALD---PEE--R 475
Query: 474 VRVKKTKNVSLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRAC 533
+ K+ KN ++K +LLKEF C IC +VLS P+TTPCAHNFCK CLE F+G + +R+R+
Sbjct: 476 KKNKRAKN-TMKARLLKEFSCQICREVLSLPVTTPCAHNFCKACLEAKFAGITQLRERS- 533
Query: 534 EGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIAQDENSEQVENSEE 593
GGR LRA+KN+M CP C+ D+++FLQNPQVNREMM ++E+ + + + S E EE
Sbjct: 534 NGGRKLRAKKNIMTCPCCTTDLSEFLQNPQVNREMMEIIENFKKSEEEADASISEEEEEE 593
Query: 594 LSDKSDEI 601
+ +I
Sbjct: 594 SEPPTKKI 601
>At1g57800 hypothetical protein
Length = 650
Score = 704 bits (1818), Expect = 0.0
Identities = 346/633 (54%), Positives = 443/633 (69%), Gaps = 64/633 (10%)
Query: 2 AHHLPCDADGTCMLCKLKPPPSETLQCHTCITPWHLPCLPTPPASF---VNWDCPDCSSS 58
A PCD +G CM CK PPP E+L C TC+TPWH+ CL +PP + + W CPDCS
Sbjct: 4 ATQYPCDPEGVCMRCKSMPPPEESLTCGTCVTPWHVSCLLSPPETLSATLQWLCPDCSGE 63
Query: 59 ISDFHAPAPAP---SAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGP----------- 104
+ A S G+DLV+AI +I++D +L+ ++KAKKKQ+L+ G
Sbjct: 64 TNPLPVSGVAAGYGSVGSDLVAAIHSIEADETLSAEEKAKKKQQLLSGKGVVDEDDEEEK 123
Query: 105 ---SSGEKPPAAGGILDIFDGSINCSFCMQLPDRPVT-----------------TPCGHN 144
S G+KP +D+ CSFCMQ +PV+ TPCGHN
Sbjct: 124 KKTSKGKKP------IDVLS-HFECSFCMQSLQKPVSVRVLFALALMLVWFLESTPCGHN 176
Query: 145 FCLKCFEKWIGQGKRTCANCRRDIPAKIASQPRINSQLAIAIRLAKLA-KSEGKVSGEPK 203
CLKCF KW+GQG R+C CR IP + + PRIN + AIRLA+++ K++ + S K
Sbjct: 177 ACLKCFLKWMGQGHRSCGTCRSVIPESMVTNPRINLSIVSAIRLARVSEKADARTS---K 233
Query: 204 IYHFMRNQDRPDKAFTTDRAKKTGKANACSGKIFVTVPPDHFGPIPAENDPTRNQGVLVG 263
+ H++ N+DRPDKAFTT+RAKKTG ANA SGKIFVT+P DHFGPIPAENDP RNQG+LVG
Sbjct: 234 VVHYVDNEDRPDKAFTTERAKKTGNANASSGKIFVTIPRDHFGPIPAENDPVRNQGLLVG 293
Query: 264 ETWEDRMECRQWGAHLPHVAGIAGQSTYGAQSVALSGGYIDDEDHGEWFLYTGSGGRDLS 323
E+W+ R+ CRQWGAH PHV+GIAGQ++YGAQSV L+GGY DDEDHGEWFLYTG
Sbjct: 294 ESWKGRLACRQWGAHFPHVSGIAGQASYGAQSVVLAGGYDDDEDHGEWFLYTG------- 346
Query: 324 GNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPESG-VRYDGVYRI 382
RTN +Q+FDQ F N NEALRLSC+ GYPVRVVRS K+KRS YAP+ G +RYDGVYRI
Sbjct: 347 ---RTNTVQAFDQVFLNFNEALRLSCKLGYPVRVVRSTKDKRSPYAPQGGLLRYDGVYRI 403
Query: 383 EKCWRKNGIQVCRYLFVRCDNEPAPWTTDDVGDRPRPLPKIDELKGAVDITERKGDPSWD 442
EKCWR GIQ+CR+LFVRCDNEPAPWT+D+ GDRPRPLP + EL A D+ ERK PSWD
Sbjct: 404 EKCWRIVGIQMCRFLFVRCDNEPAPWTSDEHGDRPRPLPNVPELNMATDLFERKESPSWD 463
Query: 443 FDEEKGSWLWKKPPPASKKSI-DVINPKDGTRVR--VKKTKNVSLKEKLLKEFGCNICHK 499
FDE + W W KPPPASKK++ +V++P++ +R +K +++ +LLKEF C IC K
Sbjct: 464 FDEGEDRWRWMKPPPASKKAVKNVLDPEERKLLREAIKSANPNTMRARLLKEFKCQICQK 523
Query: 500 VLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNVMKCPSCSNDIADFL 559
V+++P+TTPCAHNFCK CLE F+G + +R+R GGR LR+QK+VMKCP C DIA+F+
Sbjct: 524 VMTNPVTTPCAHNFCKACLESKFAGTALVRERG-SGGRKLRSQKSVMKCPCCPTDIAEFV 582
Query: 560 QNPQVNREMMGVVESLQSQIAQDENSEQVENSE 592
QNPQVNRE+ V+E L+ Q ++EN++ ++ +
Sbjct: 583 QNPQVNREVAEVIEKLKKQ-EEEENAKSLDEGQ 614
>At1g66050 hypothetical protein
Length = 598
Score = 700 bits (1806), Expect = 0.0
Identities = 350/607 (57%), Positives = 417/607 (68%), Gaps = 46/607 (7%)
Query: 5 LPCDADGTCMLCKLKPPPSETLQCHTCITPWHLPCL-PTPPASFV-NWDCPDCSSSISDF 62
LPCD DG CM C++ PP ETL C TC+TPWH+ CL P AS +W+CPDCS +
Sbjct: 7 LPCDGDGVCMRCQVNPPSEETLTCGTCVTPWHVSCLLPESLASSTGDWECPDCSGVVVPS 66
Query: 63 HAPAPA----PSAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGPSSGEKPPAAGGILD 118
AP S+G+ LV+AIRAI +D +LT+ +KAKK+Q L+ G L+
Sbjct: 67 AAPGTGISGPESSGSVLVTAIRAIQADVTLTEAEKAKKRQRLMSGGGDDGVDDEEKKKLE 126
Query: 119 IFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKW-IGQGKRTCANCRRDIPAKIASQPR 177
IF CS C+QLP+RPVTTPCGHNFCLKCFEKW +GQGK TC CR IP +A PR
Sbjct: 127 IF-----CSICIQLPERPVTTPCGHNFCLKCFEKWAVGQGKLTCMICRSKIPRHVAKNPR 181
Query: 178 INSQLAIAIRLAKLAKSEGKVSGEPKIYHFMRNQDRPDKAFTTDRAKKTGKANACSGKIF 237
IN L AIRLA + K G+ + K++H +RNQDRPDKAFTT+RA KTGKANA SG
Sbjct: 182 INLALVSAIRLANVTKCSGEATAA-KVHHIIRNQDRPDKAFTTERAVKTGKANAASG--- 237
Query: 238 VTVPPDHFGPIPAENDPTRNQGVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYGAQSVA 297
VLVGE+WEDR ECRQWG H PHVAGIAGQ+ GAQSVA
Sbjct: 238 ----------------------VLVGESWEDRQECRQWGVHFPHVAGIAGQAAVGAQSVA 275
Query: 298 LSGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRV 357
LSGGY DDEDHGEWFLYTGSGGRDLSGNKR NK+QS DQ F+NMNEALRLSC+ GYPVRV
Sbjct: 276 LSGGYDDDEDHGEWFLYTGSGGRDLSGNKRVNKIQSSDQAFKNMNEALRLSCKMGYPVRV 335
Query: 358 VRSHKEKRSSYAPESGVRYDGVYRIEKCWRKNGIQ----VCRYLFVRCDNEPAPWTTDDV 413
VRS KEKRS+YAP GVRYDGVYRIEKCW G+Q +CRYLFVRCDNEPAPWT+D+
Sbjct: 336 VRSWKEKRSAYAPAEGVRYDGVYRIEKCWSNVGVQGLHKMCRYLFVRCDNEPAPWTSDEH 395
Query: 414 GDRPRPLPKIDELKGAVDITERKGDPSWDFDEEKGSWLWKKPPPASKKSIDVINPKDGTR 473
GDRPRPLP + EL+ A D+ RK PSW FDE +G W W K PP S+ ++D K R
Sbjct: 396 GDRPRPLPDVPELENATDLFVRKESPSWGFDEAEGRWKWMKSPPVSRMALDTEERKKNKR 455
Query: 474 VRVKKTKNVSLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRAC 533
K N ++K +LLKEF C IC KVLS P+TTPCAHNFCK CLE F+G + +R R+
Sbjct: 456 A---KKGNNAMKARLLKEFSCQICRKVLSLPVTTPCAHNFCKACLEAKFAGITQLRDRS- 511
Query: 534 EGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIAQDENSEQVENSEE 593
G R LRA+KN+M CP C+ D+++FLQNPQVNREMM ++E+ + + E +E SEE
Sbjct: 512 NGVRKLRAKKNIMTCPCCTTDLSEFLQNPQVNREMMEIIENFKKSEEEAEVAESSNISEE 571
Query: 594 LSDKSDE 600
++ E
Sbjct: 572 EGEEESE 578
>At4g08590 unknown protein with zinc finger motive
Length = 465
Score = 479 bits (1232), Expect = e-135
Identities = 251/472 (53%), Positives = 304/472 (64%), Gaps = 58/472 (12%)
Query: 3 HHLPCDADGTCMLCKLKPPPSETLQCHTCITPWHLPCLPTPPASFVNWDCPDCSSSISDF 62
+ LPCD T E+L TCITP H+ L +P D S +
Sbjct: 5 NQLPCDCVSTA---------EESLTSGTCITPTHVTSLSSPL---------DRSGDVDPL 46
Query: 63 HAPAPAPSAGADLVSAIRAIDSDASLTDQQKAKKKQELVGGP-SSGEKPPAAGGILDIFD 121
P S G+ +D S+TD + KK++ ++ G + E + G I + D
Sbjct: 47 --PVSDESGGSK---------ADESMTDADETKKRKRILSGDCEADENNKSDGEIASLND 95
Query: 122 G---------SINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRTCANCRRDIPAKI 172
G +NCS C QLPDRPVT CGHNFCLKCF+KWI QG + CA CR IP K+
Sbjct: 96 GVDAFTAICEDLNCSLCNQLPDRPVTILCGHNFCLKCFDKWIDQGNQICATCRSTIPDKM 155
Query: 173 ASQPRINSQLAIAIRLAKLAKSEGKVSGEPKIYHFMRNQDRPDKAFTTDRAKKTGKANAC 232
A+ PR+NS L IR K+AK+ G G + F NQD P+ AF T RAK G+ NA
Sbjct: 156 AANPRVNSSLVSVIRYVKVAKTAGV--GTANFFPFTSNQDGPENAFRTKRAK-IGEENAA 212
Query: 233 SGKIFVTVPPDHFGPIPAENDPTRNQGVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYG 292
+I+VTVP DHFGPIPAE+DP RNQGVLVGE+WE+R+ECRQWG HLPHV+ IAGQ YG
Sbjct: 213 --RIYVTVPFDHFGPIPAEHDPVRNQGVLVGESWENRVECRQWGVHLPHVSCIAGQEDYG 270
Query: 293 AQSVALSGGYIDDEDHGEWFLYTG-SGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRK 351
AQSV +SGGY DDEDHGEWFLYTG S GR + DQ+FE++NEALR+SC
Sbjct: 271 AQSVVISGGYKDDEDHGEWFLYTGRSRGRHFANE---------DQEFEDLNEALRVSCEM 321
Query: 352 GYPVRVVRSHKEKRSSYAPESGVRYDGVYRIEKCWRK----NGIQVCRYLFVRCDNEPAP 407
GYPVRVVRS+K++ S+YAP+ GVRYDGVYRIEKCWRK + +VCRYLFVRCDNEPAP
Sbjct: 322 GYPVRVVRSYKDRYSAYAPKEGVRYDGVYRIEKCWRKARFPDSFKVCRYLFVRCDNEPAP 381
Query: 408 WTTDDVGDRPRPLPKIDELKGAVDITERKGDPSWDFDEEKGSWLWKKPPPAS 459
W +D+ GDRPRPLP I EL+ A D+ ERK PSWDFDE +G W W KPPPA+
Sbjct: 382 WNSDESGDRPRPLPNIPELETASDLFERKESPSWDFDEAEGRWRWMKPPPAN 433
Score = 41.2 bits (95), Expect = 0.002
Identities = 39/156 (25%), Positives = 62/156 (39%), Gaps = 33/156 (21%)
Query: 419 PLPKIDELKGAVDITERKGDPSW-DFDEEKGSWLWKKPPPASKKSIDVINPKDGTRVRVK 477
PLP DE G+ K D S D DE K +K + D N DG +
Sbjct: 45 PLPVSDESGGS------KADESMTDADETKK----RKRILSGDCEADENNKSDGEIASLN 94
Query: 478 KTKNVSLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGR 537
V + ++ C++C+++ P+T C HNFC C + + + G
Sbjct: 95 D--GVDAFTAICEDLNCSLCNQLPDRPVTILCGHNFCLKCFD-----------KWIDQGN 141
Query: 538 SLRAQKNVMKCPSCSNDIAD-FLQNPQVNREMMGVV 572
+ C +C + I D NP+VN ++ V+
Sbjct: 142 QI--------CATCRSTIPDKMAANPRVNSSLVSVI 169
>At5g13960 unknown protein (At5g13960)
Length = 624
Score = 83.2 bits (204), Expect = 4e-16
Identities = 62/157 (39%), Positives = 83/157 (52%), Gaps = 16/157 (10%)
Query: 293 AQSVALSGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKG 352
A S+ +SG Y DD D+ + YTG GG +L+GNKR K DQ E N AL+ C
Sbjct: 198 AVSIVMSGQYEDDLDNADTVTYTGQGGHNLTGNKRQIK----DQLLERGNLALKHCCEYN 253
Query: 353 YPVRVVRSHKEKRSSYAPESGVRYDGVYRIEKCWRKNGIQ---VCRYLFVRCDNEPAPWT 409
PVRV R H K SSY + YDG+Y++EK W + G+ V +Y R + +P T
Sbjct: 254 VPVRVTRGHNCK-SSYT-KRVYTYDGLYKVEKFWAQKGVSGFTVYKYRLKRLEGQP-ELT 310
Query: 410 TDDVGDRPRPLP-KIDELKGAV--DIT---ERKGDPS 440
TD V +P E++G V DI+ E KG P+
Sbjct: 311 TDQVNFVAGRIPTSTSEIEGLVCEDISGGLEFKGIPA 347
>At2g22740 similar to mammalian MHC III region protein G9a
Length = 790
Score = 79.0 bits (193), Expect = 8e-15
Identities = 54/155 (34%), Positives = 76/155 (48%), Gaps = 9/155 (5%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYG----AQSVALSGGYIDDEDHGEWFLY 314
GV VG+ ++ RME G H P AGI YG A S+ SGGY D D+ + Y
Sbjct: 334 GVEVGDEFQYRMELNILGIHKPSQAGI-DYMKYGKAKVATSIVASGGYDDHLDNSDVLTY 392
Query: 315 TGSGGRDLSGNKRTNKLQS-FDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPESG 373
TG GG + K+ +L+ DQK N AL S K PVRV+R + +
Sbjct: 393 TGQGGNVMQVKKKGEELKEPEDQKLITGNLALATSIEKQTPVRVIRGKHKSTHDKSKGGN 452
Query: 374 VRYDGVYRIEKCWRK---NGIQVCRYLFVRCDNEP 405
YDG+Y +EK W++ +G+ V ++ R +P
Sbjct: 453 YVYDGLYLVEKYWQQVGSHGMNVFKFQLRRIPGQP 487
>At2g35160 similar to mammalian MHC III region protein G9a
Length = 794
Score = 73.6 bits (179), Expect = 4e-13
Identities = 57/160 (35%), Positives = 80/160 (49%), Gaps = 14/160 (8%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYG----AQSVALSGGYIDDEDHGEWFLY 314
GV VG+ ++ RME G H P +GI G A S+ SGGY D D+ + +Y
Sbjct: 369 GVEVGDEFQYRMELNLLGIHRPSQSGIDYMKDDGGELVATSIVSSGGYNDVLDNSDVLIY 428
Query: 315 TGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEK--RSSYAPES 372
TG GG G K+ N+ DQ+ N AL+ S K PVRV+R K +SS ++
Sbjct: 429 TGQGGN--VGKKKNNEPPK-DQQLVTGNLALKNSINKKNPVRVIRGIKNTTLQSSVVAKN 485
Query: 373 GVRYDGVYRIEKCWRKNGIQ---VCRYLFVRCDNEP-APW 408
V YDG+Y +E+ W + G V ++ R +P PW
Sbjct: 486 YV-YDGLYLVEEYWEETGSHGKLVFKFKLRRIPGQPELPW 524
>At5g04940 SET-domain protein-like
Length = 670
Score = 70.9 bits (172), Expect = 2e-12
Identities = 59/164 (35%), Positives = 81/164 (48%), Gaps = 22/164 (13%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYG-------AQSVALSGGYIDDEDHGEW 311
GV +G+ + R E G H P +AGI G A S+ SG Y +DE + +
Sbjct: 215 GVEIGDVFFFRFEMCLVGLHSPSMAGIDYLVVKGETEEEPIATSIVSSGYYDNDEGNPDV 274
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPE 371
+YTG GG N +K QS DQK E N AL S R+ VRV+R KE +S+ +
Sbjct: 275 LIYTGQGG-----NADKDK-QSSDQKLERGNLALEKSLRRDSAVRVIRGLKE--ASHNAK 326
Query: 372 SGVRYDGVYRIEKCW---RKNGIQVCRYLFVRCDNEP---APWT 409
+ YDG+Y I++ W K+G +Y VR +P A WT
Sbjct: 327 IYI-YDGLYEIKESWVEKGKSGHNTFKYKLVRAPGQPPAFASWT 369
>At2g33290 similar to mammalian MHC III region protein G9a
Length = 651
Score = 68.6 bits (166), Expect = 1e-11
Identities = 55/157 (35%), Positives = 75/157 (47%), Gaps = 18/157 (11%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI----AGQSTYG---AQSVALSGGYIDDEDHGEW 311
GV VG+ + RME G H AGI A +S G A S+ +SGGY DDED G+
Sbjct: 211 GVEVGDIFFYRMELCVLGLHGQTQAGIDCLTAERSATGEPIATSIVVSGGYEDDEDTGDV 270
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPE 371
+YTG GG+D + N Q+ N + S G VRV+R K + S
Sbjct: 271 LVYTGHGGQDHQHKQCDN------QRLVGGNLGMERSMHYGIEVRVIRGIKYENS--ISS 322
Query: 372 SGVRYDGVYRIEKCW---RKNGIQVCRYLFVRCDNEP 405
YDG+Y+I W K+G V ++ VR + +P
Sbjct: 323 KVYVYDGLYKIVDWWFAVGKSGFGVFKFRLVRIEGQP 359
>At4g13460 putative protein
Length = 650
Score = 66.6 bits (161), Expect = 4e-11
Identities = 56/159 (35%), Positives = 75/159 (46%), Gaps = 24/159 (15%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI----AGQSTYG---AQSVALSGGYIDDEDHGEW 311
GV VG+ + R E G H +GI S+ G A SV +SGGY DD+D G+
Sbjct: 209 GVQVGDIFFFRFELCVMGLHGHPQSGIDFLTGSLSSNGEPIATSVIVSGGYEDDDDQGDV 268
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPE 371
+YTG GG+D G Q+ Q+ E N A+ S G VRV+R K Y E
Sbjct: 269 IMYTGQGGQDRLGR------QAEHQRLEGGNLAMERSMYYGIEVRVIRGLK-----YENE 317
Query: 372 SGVR---YDGVYRIEKCW---RKNGIQVCRYLFVRCDNE 404
R YDG++RI W K+G V +Y R + +
Sbjct: 318 VSSRVYVYDGLFRIVDSWFDVGKSGFGVFKYRLERIEGQ 356
>At1g73100 putative protein
Length = 669
Score = 64.3 bits (155), Expect = 2e-10
Identities = 54/157 (34%), Positives = 73/157 (46%), Gaps = 18/157 (11%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI------AGQSTYG-AQSVALSGGYIDDEDHGEW 311
G+ VG+ + R+E G H+ +AGI AG A S+ SG Y + E
Sbjct: 212 GIEVGDIFFSRIEMCLVGLHMQTMAGIDYIISKAGSDEESLATSIVSSGRYEGEAQDPES 271
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPE 371
+Y+G GG N N+ Q+ DQK E N AL S RKG VRVVR ++ S
Sbjct: 272 LIYSGQGG-----NADKNR-QASDQKLERGNLALENSLRKGNGVRVVRGEEDAASKTG-- 323
Query: 372 SGVRYDGVYRIEKCW---RKNGIQVCRYLFVRCDNEP 405
YDG+Y I + W K+G +Y VR +P
Sbjct: 324 KIYIYDGLYSISESWVEKGKSGCNTFKYKLVRQPGQP 360
>At1g17770 putative protein
Length = 693
Score = 60.8 bits (146), Expect = 2e-09
Identities = 54/186 (29%), Positives = 81/186 (43%), Gaps = 27/186 (14%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI----AGQST---YGAQSVALSGGYIDDEDHGEW 311
G+ VG+ + E G H + GI A +S + A V +G Y + + +
Sbjct: 231 GIHVGDIFYYWGEMCLVGLHKSNYGGIDFFTAAESAVEGHAAMCVVTAGQYDGETEGLDT 290
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRS----HKEKRSS 367
+Y+G GG D+ GN R DQ+ + N AL S KG VRVVR H+ +
Sbjct: 291 LIYSGQGGTDVYGNAR-------DQEMKGGNLALEASVSKGNDVRVVRGVIHPHENNQKI 343
Query: 368 YAPESGVRYDGVYRIEKCWR---KNGIQVCRYLFVRCDNEPAPWTTDDVGDRPRPLPKID 424
Y YDG+Y + K W K+G + R+ VR N+P + + R ID
Sbjct: 344 YI------YDGMYLVSKFWTVTGKSGFKEFRFKLVRKPNQPPAYAIWKTVENLRNHDLID 397
Query: 425 ELKGAV 430
+G +
Sbjct: 398 SRQGFI 403
>At5g47150 putative protein
Length = 328
Score = 52.8 bits (125), Expect = 6e-07
Identities = 44/139 (31%), Positives = 66/139 (46%), Gaps = 14/139 (10%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI----AGQSTYGAQSVALSG-GYIDDEDHGEWFL 313
G+ +G+ ++ + E R G H + GI G VA G GY D + G +
Sbjct: 180 GINIGDVFQYKTELRVVGLHSKPMCGIDYIKLGDDRITTSIVASEGYGYNDTYNSGV-MV 238
Query: 314 YTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPESG 373
YTG GG ++ K+T DQK N AL S R+ VRV+R E+R +
Sbjct: 239 YTGEGGNVINKQKKTE-----DQKLVKGNLALATSMRQKSQVRVIRG--EERLDRKGKRY 291
Query: 374 VRYDGVYRIEKCWRKNGIQ 392
V YDG+Y +E+ W + ++
Sbjct: 292 V-YDGLYMVEEYWVERDVR 309
>At5g47160 putative protein
Length = 415
Score = 51.6 bits (122), Expect = 1e-06
Identities = 45/139 (32%), Positives = 66/139 (47%), Gaps = 13/139 (9%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYGAQSVALSGGYIDDEDHGEWFL----- 313
G+ VG+ + + G H ++GI G + VA S + D+G+ F+
Sbjct: 266 GIKVGDKIQFKAALSVIGLHFGIMSGI-DYMYKGNKEVATSIVSSEGNDYGDRFINDVMI 324
Query: 314 YTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPESG 373
Y G GG S + + K DQK N AL S ++ PVRV+R E+R +
Sbjct: 325 YCGQGGNMRSKDHKAIK----DQKLVGGNLALANSIKEKTPVRVIRG--ERRLDNRGKDY 378
Query: 374 VRYDGVYRIEKCWRKNGIQ 392
V YDG+YR+EK W + G Q
Sbjct: 379 V-YDGLYRVEKYWEERGPQ 396
>At2g24740 pseudogene
Length = 429
Score = 50.8 bits (120), Expect = 2e-06
Identities = 39/141 (27%), Positives = 61/141 (42%), Gaps = 20/141 (14%)
Query: 293 AQSVALSGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKG 352
A SV SG Y ++ + E +Y+G GG+ DQ + N AL S R+
Sbjct: 29 ATSVVTSGKYDNETEDLETLIYSGHGGKPC------------DQVLQRGNRALEASVRRR 76
Query: 353 YPVRVVRSHKEKRSSYAPESGVRYDGVYRIEKCWR---KNGIQVCRYLFVRCDNEPAPWT 409
VRV+R Y E YDG+Y + CW+ K+G + R+ +R +P +
Sbjct: 77 NEVRVIRG-----ELYNNEKVYIYDGLYLVSDCWQVTGKSGFKEYRFKLLRKPGQPPGYA 131
Query: 410 TDDVGDRPRPLPKIDELKGAV 430
+ + R ID +G +
Sbjct: 132 IWKLVENLRNHELIDPRQGFI 152
>At2g05900 hypothetical protein
Length = 312
Score = 47.0 bits (110), Expect = 4e-05
Identities = 39/118 (33%), Positives = 56/118 (47%), Gaps = 12/118 (10%)
Query: 293 AQSVALSGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKG 352
A SV SG D + + ++TG GG D+ + N QK E +N L + RK
Sbjct: 30 AVSVISSGKNADKTEDPDSLIFTGFGGTDMYHGQPCN------QKLERLNIPLEAAFRKK 83
Query: 353 YPVRVVRSHKEKRSSYAPESGVRYDGVYRIEKCWR---KNGIQVCRYLFVR-CDNEPA 406
VRVVR K+++ + + YDG Y I W +NG V ++ VR D +PA
Sbjct: 84 SIVRVVRCMKDEKRTNG--NIYIYDGTYMITNRWEEEGQNGFIVFKFKLVREPDQKPA 139
>At3g27330 hypothetical protein
Length = 913
Score = 45.8 bits (107), Expect = 8e-05
Identities = 17/50 (34%), Positives = 27/50 (54%)
Query: 117 LDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRTCANCRR 166
+D ++C+ C+++ P TT CGH+FC KC + R C CR+
Sbjct: 715 IDKLRDELSCAICLEICFEPSTTTCGHSFCKKCLRSAADKCGRKCPKCRQ 764
Score = 43.1 bits (100), Expect = 5e-04
Identities = 23/66 (34%), Positives = 27/66 (40%), Gaps = 19/66 (28%)
Query: 486 EKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNV 545
+KL E C IC ++ P TT C H+FCK CL A A K
Sbjct: 716 DKLRDELSCAICLEICFEPSTTTCGHSFCKKCLRSA-------------------ADKCG 756
Query: 546 MKCPSC 551
KCP C
Sbjct: 757 RKCPKC 762
>At4g27470 putative RING zinc finger protein
Length = 243
Score = 45.4 bits (106), Expect = 1e-04
Identities = 24/84 (28%), Positives = 36/84 (42%), Gaps = 18/84 (21%)
Query: 94 AKKKQELVGGPSSGEKPPAAGGILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKW 153
AK+K L P++G+ + G +C+ C+ PV T CGH FC C KW
Sbjct: 21 AKQKPNLTTAPTAGQANES---------GCFDCNICLDTAHDPVVTLCGHLFCWPCIYKW 71
Query: 154 ---------IGQGKRTCANCRRDI 168
+ Q + C C+ +I
Sbjct: 72 LHVQLSSVSVDQHQNNCPVCKSNI 95
Score = 32.3 bits (72), Expect = 0.89
Identities = 12/27 (44%), Positives = 14/27 (51%)
Query: 492 FGCNICHKVLSSPLTTPCAHNFCKVCL 518
F CNIC P+ T C H FC C+
Sbjct: 42 FDCNICLDTAHDPVVTLCGHLFCWPCI 68
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,716,165
Number of Sequences: 26719
Number of extensions: 871338
Number of successful extensions: 3783
Number of sequences better than 10.0: 325
Number of HSP's better than 10.0 without gapping: 120
Number of HSP's successfully gapped in prelim test: 206
Number of HSP's that attempted gapping in prelim test: 3351
Number of HSP's gapped (non-prelim): 552
length of query: 680
length of database: 11,318,596
effective HSP length: 106
effective length of query: 574
effective length of database: 8,486,382
effective search space: 4871183268
effective search space used: 4871183268
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0191.7


