

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0189.12
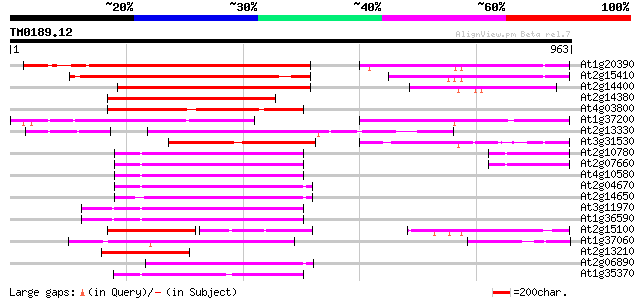
(963 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 423 e-118
At2g15410 putative retroelement pol polyprotein 379 e-105
At2g14400 putative retroelement pol polyprotein 372 e-103
At2g14380 putative retroelement pol polyprotein 337 3e-92
At4g03800 312 5e-85
At1g37200 hypothetical protein 312 5e-85
At2g13330 F14O4.9 288 1e-77
At3g31530 hypothetical protein 221 2e-57
At2g10780 pseudogene 204 1e-52
At2g07660 putative retroelement pol polyprotein 204 3e-52
At4g10580 putative reverse-transcriptase -like protein 200 4e-51
At2g04670 putative retroelement pol polyprotein 195 9e-50
At2g14650 putative retroelement pol polyprotein 188 1e-47
At3g11970 hypothetical protein 181 2e-45
At1g36590 hypothetical protein 181 2e-45
At2g15100 putative retroelement pol polyprotein 174 2e-43
At1g37060 Athila retroelment ORF 1, putative 171 1e-42
At2g13210 pseudogene 162 6e-40
At2g06890 putative retroelement integrase 162 8e-40
At1g35370 hypothetical protein 162 1e-39
>At1g20390 hypothetical protein
Length = 1791
Score = 423 bits (1088), Expect = e-118
Identities = 217/492 (44%), Positives = 308/492 (62%), Gaps = 32/492 (6%)
Query: 25 VKYLILETEGSYNMIIGRITLNRLCAVISTAQLAVKYPLSNGHIGGIVVDQRIARECYCN 84
VK++++ YN+I+G ++++ A+ ST VK+P NG + R +E
Sbjct: 645 VKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFPTHNG-----IFTLRAPKEA--- 696
Query: 85 AVDRYGKKNASVGHRCSEVETPEESLDPRGEGRVNRPTPIEETKDLKFGEKILKIGTKLS 144
+TP S + R I+E+ + + +G ++S
Sbjct: 697 -------------------KTPSRSYEESELCRTEMVN-IDESDPTR----CVGVGAEIS 732
Query: 145 EEQEQRISKLLGDNLDLFAWSHKYMPGIDPNFICHRLALNPGVEPITQTRRRMGNEKEKA 204
+ LL N FAWS + M GIDP H L ++P +P+ Q RR++G E+ +A
Sbjct: 733 PSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKPVKQKRRKLGPERARA 792
Query: 205 IQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSID 264
+ +EV KLL A I E+KYP WL N V+VKK NGKWR+CVD TDLNK CPKDSYPLP ID
Sbjct: 793 VNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKDSYPLPHID 852
Query: 265 KLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQ 324
+LV+ GN LLS MDA+SGY+QI MH+ D++KT+F+T R YCY+ M FGLKNAGATYQ
Sbjct: 853 RLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQ 912
Query: 325 RLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIR 384
R ++++ Q+GR +EVY+DDM+VKS+ +H E L + F + + M+LNP KC+FG+
Sbjct: 913 RFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCTFGVT 972
Query: 385 GGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPF 444
G+FLG+++T R IE NP + +AI E+ SP N +EVQRL GRI AL+RF+ S DK PF
Sbjct: 973 SGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTDKCLPF 1032
Query: 445 IKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQGLPLHLYFSVGDHAISSVILQE 504
LK+ F+W+ + EEAF +LK+ LS PP+L KP G L+LY +V DHA+SSV+++E
Sbjct: 1033 YNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHAVSSVLVRE 1092
Query: 505 VDGEQKIVYFVS 516
GEQ+ +++ S
Sbjct: 1093 DRGEQRPIFYTS 1104
Score = 247 bits (631), Expect = 2e-65
Identities = 144/379 (37%), Positives = 207/379 (53%), Gaps = 19/379 (5%)
Query: 601 FLVELTPEEGEKVST-----QWILFVDGSSNDNGSEAGVTLQGPGELVLEQSLRFQFKTS 655
FL+EL + E+ + +W L+VDGSS+ GS G+ L P VLEQS R +F +
Sbjct: 1189 FLIELPLQSAERAVSGNRGEEWSLYVDGSSSARGSGIGIRLVSPTAEVLEQSFRLRFVAT 1248
Query: 656 NNQAEYEALIAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMG 715
NN AEYE LIAGL+LA +QI ++ TDS L+A Q++GE++ K + Y++ V+L+
Sbjct: 1249 NNVAEYEVLIAGLRLAAGMQITTIHAFTDSQLIAGQLSGEYEAKNEKMDAYLKIVQLMTK 1308
Query: 716 RLQQVVVEFVPRAQNQRADALAKLASTRKPGNNRSVIQETLANPNIEG----EFVASVDR 771
+ + +PR N ADALA LA T R + E++ P+I+ E V ++
Sbjct: 1309 DFENFKLSKIPRGDNAPADALAALALTSDSDLRRIIPVESIDKPSIDSTDAVEIVNTIRS 1368
Query: 772 QET--------WMNPIIDILAGEPSDVVKYSKAQRR-EAGHYTLLDGILFRRGFSSPLLR 822
W I D L+ K++ + R +A YTL+ L + +L
Sbjct: 1369 SNAPDPADPTDWRVEIRDYLSDGTLPSDKWTARRLRIKAAKYTLMKEHLLKVSAFGAMLN 1428
Query: 823 CLPPEKYEAVMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFT 882
CL + +M E HEG +H GGR+LA K+ + GFYWPT+ +C F KC++CQ
Sbjct: 1429 CLHGTEINEIMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHA 1488
Query: 883 DLPRAPPGQLVTISSPWPFAMWGVDLVGPFPTARSQMKFILVAVDYFTKWVEAEPLASIT 942
P L +P+PF W +D+VGP P +R Q +FILV DYFTKWVEAE A+I
Sbjct: 1489 PTIHQPTELLRAGVAPYPFMRWAMDIVGPMPASR-QKRFILVMTDYFTKWVEAESYATIR 1547
Query: 943 AAKIISFYWKRIVCRFGVP 961
A + +F WK I+CR G+P
Sbjct: 1548 ANDVQNFVWKFIICRHGLP 1566
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 379 bits (972), Expect = e-105
Identities = 190/414 (45%), Positives = 264/414 (62%), Gaps = 26/414 (6%)
Query: 103 VETPEESLDPRGEGRVNRPTPIEETKDLKFGEKILKIGTKLSEEQEQRISKLLGDNLDLF 162
V T +E +P+ + T ++ D + + I L E + + L N F
Sbjct: 702 VPTQDERPNPQ------KGTVVQVNIDESDPSRCVGIRIDLPSELQNELVNFLRQNAATF 755
Query: 163 AWSHKYMPGIDPNFICHRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIK 222
AWS + MPGID CH L ++P +P+ Q RR++G ++ K + +EV KLL A I E++
Sbjct: 756 AWSVEDMPGIDSAVTCHELNVDPTYKPLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVR 815
Query: 223 YPTWLVNVVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAY 282
YP WL N V+VKK NGKWR+C+D TDLNK CPKDS+PLP ID+LV+ GNELLS MDA+
Sbjct: 816 YPDWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAF 875
Query: 283 SGYHQIKMHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVY 342
SGY+QI MHQ+D +KT F+T + YCY+ MPFGLKNAGATY RL++++F Q+ +MEVY
Sbjct: 876 SGYNQILMHQNDREKTVFITDQGTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVY 935
Query: 343 VDDMIVKSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINP 402
+DDM+VKS+ H L + F + ++NM+LNP KC+FG+ G+FLG+L+T R IE NP
Sbjct: 936 IDDMLVKSLRAEEHITHLRQCFQVLNRYNMKLNPSKCTFGVTSGEFLGYLVTRRGIEANP 995
Query: 403 DKCKAIQEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEE 462
+ AI ++ SP N +EVQRLIGRI AL+RF+ S DK PF + L+ N FEW+ +CEE
Sbjct: 996 KQISAIIDLPSPRNTREVQRLIGRIAALNRFISRSTDKCLPFYQLLRANKRFEWDEKCEE 1055
Query: 463 AFTRLKEMLSAPPVLSKPVQGLPLHLYFSVGDHAISSVILQEVDGEQKIVYFVS 516
G L+LY +V A+S V+++E GEQ +++VS
Sbjct: 1056 --------------------GETLYLYIAVSTSAVSGVLVREDRGEQHPIFYVS 1089
Score = 197 bits (502), Expect = 2e-50
Identities = 125/375 (33%), Positives = 179/375 (47%), Gaps = 65/375 (17%)
Query: 651 QFKTSNNQAEYEALIAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERV 710
+F SNN+AEYEALIAGL+LA +++ + DS LVA+Q +G ++ K + Y++ V
Sbjct: 1189 RFPASNNEAEYEALIAGLRLAHGIEVKKIQAYCDSQLVASQFSGNYEAKNERMDAYLKVV 1248
Query: 711 RLLMGRLQQVVVEFVPRAQNQRADALAKLASTRKPGNNRSV-----------IQETLANP 759
R L + + +PR+ N ADALA LAST P R + IQ T+ P
Sbjct: 1249 RELSYNFEVFELTKIPRSDNAPADALAVLASTSDPDLRRVIPVECIDVPSIKIQGTITKP 1308
Query: 760 NI------------------------------EGEFVASVDRQET--------------- 774
+ G V S D ++
Sbjct: 1309 SEYTVIDLTAEQLSIVMVILPPATDRSEDAGPSGNLVCSSDPGQSSGCDRSDDSNRSADS 1368
Query: 775 -------WMNPIIDILAGEPSDVVKYSKAQ-RREAGHYTLLDGILFRRGFSSPLLRCLPP 826
W+ I +A K++ + R + YTL L R + LL CL
Sbjct: 1369 DSPASTNWIEEIRSYIADRIVPKDKWAARRLRARSAQYTLPHEHLLRWSATGVLLSCLDD 1428
Query: 827 EKYEAVMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPR 886
E+ + VM E+HEG +H GGR+LA K+ + G YWPT+ +C +FV +C+KCQ
Sbjct: 1429 EEAQQVMREIHEGAGGNHSGGRALALKIRKHGQYWPTMNADCEKFVARCEKCQGHAPFIH 1488
Query: 887 APPGQLVTISSPWPFAMWGVDLVGPFPTARSQMKFILVAVDYFTKWVEAEPLASITAAKI 946
P L T P+PF W +D++ PFP +R Q K+ILV DYFTKW EAE A I + ++
Sbjct: 1489 IPSEVLQTAMPPYPFMRWAMDIIRPFPASR-QKKYILVMTDYFTKWFEAESYARIQSKEV 1547
Query: 947 ISFYWKRIVCRFGVP 961
+F WK I+C G+P
Sbjct: 1548 QNFVWKNIICHHGLP 1562
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 372 bits (956), Expect = e-103
Identities = 176/330 (53%), Positives = 238/330 (71%)
Query: 186 GVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKWRMCVD 245
G+ RR++G E+ KA+ EV+KLL IRE++YP WL N V+VKK NGK R+C+D
Sbjct: 461 GISRADVNRRKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCID 520
Query: 246 NTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTFMTARV 305
TDLNK CPKDS+PLP ID+LV+ GNELLS MDA+SGY+QI M+ D++KT F+T R
Sbjct: 521 FTDLNKACPKDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRG 580
Query: 306 NYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDLMEAFG 365
YCY+ MPFGL+NAGATY RL++++F VG+ MEVY+DDM++KS+ +H + L E F
Sbjct: 581 IYCYKVMPFGLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFA 640
Query: 366 RIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEVQRLIG 425
+ ++ M+LNP KC+FG+ G+FLG+++T R IE NP++ A M SP N KEVQRL G
Sbjct: 641 ILNQYQMKLNPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTG 700
Query: 426 RITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQGLP 485
RI AL+RF+ S DKS PF + LK N F W+ +CEEAF +LK L++PPVL KP
Sbjct: 701 RIAALNRFISRSTDKSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEK 760
Query: 486 LHLYFSVGDHAISSVILQEVDGEQKIVYFV 515
L+LY SV +HA+S V+++E GEQK +Y++
Sbjct: 761 LYLYVSVSNHAVSGVLIREDRGEQKPIYYI 790
Score = 145 bits (367), Expect = 8e-35
Identities = 89/291 (30%), Positives = 147/291 (49%), Gaps = 39/291 (13%)
Query: 687 LVANQVNGEFQVKELALIKYVERVRLLMGRLQQVVVEFVPRAQNQRADALAKLASTRKPG 746
++ NQ NG+++ K+ + Y+ V+ L ++ + +PR QN ADALA LAST P
Sbjct: 790 IIVNQFNGDYEAKDSRMEAYLHVVKDLAKNFRKFELIRIPRGQNTTADALAALASTSDPE 849
Query: 747 NNRSVIQETLANPNIEGEFVASV---------DRQETWMN-PIIDILAGEPSDVVKYSKA 796
N + E ++ +I+ E A V DR +T + P + + + V K +
Sbjct: 850 VNSIIPVECISERSIKEEKEAFVVTRSRAAARDRGDTAVELPPVKRRKSKEATVPKPAVQ 909
Query: 797 Q--------------RREAGHYTLLD---------------GILFRRGFSSPLLRCLPPE 827
+ + EA +D L+RRG S P L +
Sbjct: 910 ENIGIELIEEVISDAKIEAETEPWVDEDILAPNEHPEPEALNSLYRRGVSDPYLLSIFGP 969
Query: 828 KYEAVMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRA 887
E VM+EVHEG+C SH GR++A K+ + +YWPT+ +C ++ ++CK+CQ+ L
Sbjct: 970 VVEIVMSEVHEGLCGSHSSGRAMAFKIKKMCYYWPTMITDCVKYAQRCKRCQLHAPLIHQ 1029
Query: 888 PPGQLVTISSPWPFAMWGVDLVGPFPTARSQMKFILVAVDYFTKWVEAEPL 938
P +IS+P+PF W +D++GP + ++++LV DYF+KW+EAE +
Sbjct: 1030 PSELFSSISAPYPFMRWSMDIIGPLHRSTRGVQYLLVLTDYFSKWIEAEAI 1080
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 337 bits (863), Expect = 3e-92
Identities = 157/288 (54%), Positives = 210/288 (72%), Gaps = 1/288 (0%)
Query: 169 MPGIDPNFICHRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLV 228
M GIDP CH L ++P + + Q RR++G E+ KA+ EV+KLL A I E+KYP WL
Sbjct: 477 MVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWLA 536
Query: 229 NVVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQI 288
N V+VKK N KWR+C+D TDLNK CPKDS+PLP ID++V+ GNELLS MDA+SGY+QI
Sbjct: 537 NPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQI 596
Query: 289 KMHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIV 348
MH+ D++KT+F+ R YCY+ MPFGLKN GA YQRL++++F Q+G+ MEVY+DDM+V
Sbjct: 597 PMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDMLV 656
Query: 349 KSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAI 408
KS ++H + L F + K+NM+LNP KC FG+ G+FLG+++T R IE NP + +AI
Sbjct: 657 KSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQIRAI 716
Query: 409 QEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKK-NTTFE 455
+++SP N KEVQRL GRI L+RF+ S DKS PF + L+ N TFE
Sbjct: 717 LDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLRSANKTFE 764
>At4g03800
Length = 637
Score = 312 bits (800), Expect = 5e-85
Identities = 157/337 (46%), Positives = 222/337 (65%), Gaps = 21/337 (6%)
Query: 168 YMPGIDPNFICHRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWL 227
YMP IDP+ ICH L ++P +P+ Q RR++G E+ KA+ +++KLL IRE++YP W+
Sbjct: 320 YMPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWV 379
Query: 228 VNVVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQ 287
V+VKK NGK R+C+D TDLNK CPKDS+PLP ID+LV+ GNELL+ MDA+ GY+Q
Sbjct: 380 AITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQ 439
Query: 288 IKMHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMI 347
I M+ D++KT+F+T R GATYQ L++++F + + MEV +DD +
Sbjct: 440 IMMNPEDQEKTSFITDR---------------GATYQWLVNKMFNEHLRKTMEVSIDDTL 484
Query: 348 VKSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKA 407
VKS+ +H + L E F + ++ M+LN KC+FG+ G+FLG+++T R IE NP++ A
Sbjct: 485 VKSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQINA 544
Query: 408 IQEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRL 467
+ S N KEVQRL GRI + S DKS PF + LK N F W+ +CEEAF +L
Sbjct: 545 FLKTPSLRNFKEVQRLTGRIAS------RSTDKSLPFYQILKGNNGFLWDEKCEEAFRQL 598
Query: 468 KEMLSAPPVLSKPVQGLPLHLYFSVGDHAISSVILQE 504
K L+ PPVLSKP L+LY V +HA+S V+++E
Sbjct: 599 KAYLTTPPVLSKPEADEKLYLYVFVSNHAVSGVLVRE 635
Score = 30.8 bits (68), Expect = 3.8
Identities = 14/42 (33%), Positives = 26/42 (61%)
Query: 25 VKYLILETEGSYNMIIGRITLNRLCAVISTAQLAVKYPLSNG 66
V +++ + +YN+I+G + ++ AV ST +K+P SNG
Sbjct: 263 VDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQWLKFPTSNG 304
>At1g37200 hypothetical protein
Length = 1564
Score = 312 bits (800), Expect = 5e-85
Identities = 176/430 (40%), Positives = 247/430 (56%), Gaps = 19/430 (4%)
Query: 2 VWVKDALDLDTTFGEEENAR----TLSVKYLILETEGS-----YNMIIGRITLNRLCAVI 52
+W + DLD + R + ++++T S +N I+GR L+ + AV
Sbjct: 489 LWESETTDLDKPHDDAIVIRIDVGNYKLSRIMIDTGSSVDKAPFNAILGRPWLHAMKAVP 548
Query: 53 STAQLAVKYPLSNGHIGGIVVDQRIARECYCNAVDRYGKKNASV---GHRCSEVETPEES 109
ST +K+P G I I QR +R CY + + K V + +E++T S
Sbjct: 549 STYHQCIKFPSEKG-IAVIYGSQRSSRRCYMGSYELIKKAELVVLMIEDKLAEMKTVRSS 607
Query: 110 LDPRGEGRVNRPTPIEETKDLKFGEKILKIGTKLSEEQEQRISKLLGDNLDLFAWSHKYM 169
DP G + + + D ++ + +G L + L +N D FAWS +
Sbjct: 608 -DPSQCGP-QKSSITQVYIDESDPKQCVGVGQDLDPAIREDFITFLKENKDSFAWSSANL 665
Query: 170 PGIDPNFICHRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVN 229
GI H L ++P PI Q RR++G E+ KA+Q EV++LL IRE+KYP WL N
Sbjct: 666 QGISLEVTSHELNVDPTYRPIKQKRRKLGPERAKAVQDEVDRLLKIGSIREVKYPDWLAN 725
Query: 230 VVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIK 289
V+VK NGKWR+C+D DLNK CPKDS+PLP ID+LV G+ELLS MDA+SGY+QI
Sbjct: 726 PVVVKNKNGKWRVCIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGYNQIL 785
Query: 290 MHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVK 349
M D++KT F+T CY+ MPFGLKN GATYQRL++R+F Q+G+ ME+Y+DDM+VK
Sbjct: 786 MRPDDQEKTAFITD----CYKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDDMLVK 841
Query: 350 SVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQ 409
S +H L E F + K M+LNPEKCSFG+ G+FLG+L+T R IE NP + A
Sbjct: 842 SAHEKDHLPQLRECFKILNKFEMKLNPEKCSFGVPSGEFLGYLVTQRGIEANPKQIAAFI 901
Query: 410 EMKSPSNVKE 419
+M SP ++
Sbjct: 902 DMPSPKMARD 911
Score = 194 bits (492), Expect = 3e-49
Identities = 124/415 (29%), Positives = 201/415 (47%), Gaps = 64/415 (15%)
Query: 601 FLVEL--TPEEGEKVSTQWILFVDGSSNDNGSEAGVTLQGPGELVLEQSLRFQFKTSNNQ 658
F++EL +G + +W+L VDGSSN GS G+ L P V+EQS R F SNN+
Sbjct: 1023 FVIELPLADLDGTNSNKKWLLHVDGSSNRQGSGEGIQLTSPTGEVIEQSFRLGFNASNNE 1082
Query: 659 AEYEALIAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQ 718
+EYEALI G+KLA ++I + +DS LV +Q +GE++ K+ + Y+E V+ L + +
Sbjct: 1083 SEYEALIDGIKLAQGMRIRDIHAHSDSQLVTSQFHGEYEAKDERMEAYLELVKTLTQQFE 1142
Query: 719 QVVVEFVPRAQNQRADALAKLASTRKPGNNRSVIQETLANPNIE---------------- 762
+ +PR +N DALA LAST P R + E + +P+I+
Sbjct: 1143 SFELTRIPRGENTSTDALAALASTLDPFVKRIIPVEGIEHPSIDLTVKHAGMETSATCNF 1202
Query: 763 -------------------------------GEFVASVDRQ--ETWMNPIIDILAGEPSD 789
E +AS + + W PIID + +
Sbjct: 1203 TRVTRSTTAAARALAAAAEAGAAEEEAKLRTPEQLASYEPEPYNDWRIPIIDYIERGITP 1262
Query: 790 VVKY-SKAQRREAGHYTLLDGILFRRGFSSPLLRCLPPEKYEAVMTEVHEGVCASHIGGR 848
K+ ++ + ++ Y +++G L +R + P + C ++ + +M +HEG C SH GR
Sbjct: 1263 PDKWETRKLKAQSARYCIMEGRLMKRSVAGPYMVCTYGQQTKDLMKSMHEGQCGSHYSGR 1322
Query: 849 SLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPFAMWGVDL 908
+L +L +C +C KCQ PP ++ +ISSP+PF W +D+
Sbjct: 1323 TLRCIML----------ADCIAHSLRCDKCQRHAPTLHQPPEEMSSISSPYPFMKWSMDV 1372
Query: 909 VGPFPTA--RSQMKFILVAVDYFTKWVEAEPLASITAAKIISFYWKRIVCRFGVP 961
V P + + ++K +LV DY TKW+E + +T ++ F + IV R G+P
Sbjct: 1373 VRPMEASGGKKRLKNLLVLTDYSTKWIEGKAFQQVTEKQVEVFLLENIVYRHGIP 1427
>At2g13330 F14O4.9
Length = 889
Score = 288 bits (737), Expect = 1e-77
Identities = 193/541 (35%), Positives = 282/541 (51%), Gaps = 58/541 (10%)
Query: 237 NGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDED 296
NGKWR+C+D DLNK CPKDS+PLP ID+LV+ + +E LS MDA+ GY+QI M + ++
Sbjct: 291 NGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQE 350
Query: 297 KTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNH 356
KT F+T R Y Y+ MPFGLKNAG TYQRL++R+F Q+G+ +EVY+DDM+VKS +H
Sbjct: 351 KTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKDH 410
Query: 357 HEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSN 416
L E F + K M+LNPEKCSF ++ +FL +L+T R IE NP + A EM SP
Sbjct: 411 VPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSPKM 470
Query: 417 VKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPV 476
+EVQRL GRI AL+ F+ S +K PF + L+K F+WN +CE+AF +LK L+ PP+
Sbjct: 471 AREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFKQLKAYLTEPPI 530
Query: 477 LSKPVQGLPLHLYFSVGDH--AISSVILQEVDGEQKI-VYFVSYHFRGRRSGIRR----- 528
L+KP +G PL+LY + A+ + L V +K+ +YF S+ S R
Sbjct: 531 LAKPEKGEPLYLYTNRDKRCPAMEKLALTVVMSVRKLRLYFQSHPIVVMTSQPIRTILHS 590
Query: 529 --*KKRCSPSSSPPDAYDRIFKAFR*K*ELTFL*DRCCRSQTFQ-EGSSRDQWSCRNMDC 585
R + + YD ++ R + L D + +G++ ++ ++D
Sbjct: 591 LTQSGRLAKWAIELREYDIEYRT-RTSLKAQVLADFVIKLPLADLDGTNSNKKWLLHVDG 649
Query: 586 SMTKGGRSTHIP*QIFLVELTPEEGEKVSTQWILFVDGSSNDNGSE---AGVTL-QGPGE 641
S + G I +LT GE + L + S+N++ E AG+ L Q G
Sbjct: 650 SSNRQGSGVGI-------QLTSPTGEVIEQSLQLGFNASNNESEYEALIAGIKLAQEKGI 702
Query: 642 LVLEQSLRFQFKTSNNQAEYEALIAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKEL 701
+ Q TS EYEA K+
Sbjct: 703 REIHAYSDSQLVTSQFHGEYEA-----------------------------------KDE 727
Query: 702 ALIKYVERVRLLMGRLQQVVVEFVPRAQNQRADALAKLASTRKPGNNRSVIQETLANPNI 761
+ Y+E V+ L + + + +PR +N AD LA LAST P R + E + + +I
Sbjct: 728 RMEAYLELVKTLAQQFESFKLTRIPRGENTSADTLAALASTSDPFVKRIIPVEGIEHTSI 787
Query: 762 E 762
+
Sbjct: 788 D 788
Score = 49.3 bits (116), Expect = 1e-05
Identities = 40/149 (26%), Positives = 70/149 (46%), Gaps = 6/149 (4%)
Query: 27 YLILETEGSYNMIIGRITLNRLCAVISTAQLAVKYPLSNGHIGGIVVDQRIARECYCNAV 86
+L+++ + +N I+GR L+ + AV ST +K+P G I + QR +R+CY +
Sbjct: 141 FLVVDKKAPFNAILGRPWLHVMKAVPSTYHQCIKFPSYKG-IAVVYGSQRSSRKCYMGSY 199
Query: 87 DRYGKKN---ASVGHRCSEVETPEESLDPRGEGRVNRPTPIEETKDLKFGEKILKIGTKL 143
+ K + + +E++T SLDP G + + D ++ + IG L
Sbjct: 200 EDIKKADPVVLMIEDELAEMKT-VRSLDPSQRG-TRKSLITQVCIDESDPKRCVGIGHDL 257
Query: 144 SEEQEQRISKLLGDNLDLFAWSHKYMPGI 172
+ + L +N D FAWS + GI
Sbjct: 258 DLTVREDLITFLKENKDSFAWSSANLQGI 286
>At3g31530 hypothetical protein
Length = 831
Score = 221 bits (563), Expect = 2e-57
Identities = 110/254 (43%), Positives = 164/254 (64%), Gaps = 13/254 (5%)
Query: 273 NELL--SLMDAYSGYHQIKMHQSDEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRV 330
N+LL S +++ + + M+ D++KT F T + +CY+ MPFGLKNAGATY+R ++++
Sbjct: 77 NQLLETSYCHSWTLFVVMMMNPEDQEKTAFYTEQGIFCYRVMPFGLKNAGATYERFVNKI 136
Query: 331 FEGQVGRNMEVYVDDMIVKSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLG 390
F Q+G+ MEVY+DDM+VKS+ +H L E F ++ +N++LNP KC FG+R G
Sbjct: 137 FALQIGKTMEVYIDDMLVKSMTEKDHISHLRECFKQLNLYNVKLNPAKCRFGVRSG---- 192
Query: 391 FLITSRRIEINPDKCKAIQEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKK 450
IE NP + +A+ M SP N +EVQ L GR+ AL+RF+ S +K F L++
Sbjct: 193 -------IEANPKQIEALLGMASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRR 245
Query: 451 NTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQGLPLHLYFSVGDHAISSVILQEVDGEQK 510
N FEW + CEEAF LK+ L+ PP+L+KPV G P +LY +V D A+S +++E GEQK
Sbjct: 246 NKKFEWTTRCEEAFQELKKYLATPPILAKPVIGEPQYLYVAVSDTAVSGELVREDRGEQK 305
Query: 511 IVYFVSYHFRGRRS 524
++++VS F S
Sbjct: 306 LIFYVSQTFTSAES 319
Score = 178 bits (451), Expect = 1e-44
Identities = 128/379 (33%), Positives = 183/379 (47%), Gaps = 72/379 (18%)
Query: 601 FLVELTPEEGEK--VSTQWILFVDGSSNDNGSEAGVTLQGP-GELVLEQSLRFQFKTSNN 657
F+VEL +E + + T W+L VDGSS+ GS G+ L P GE +NN
Sbjct: 396 FIVELPTKEARENPLDTTWLLHVDGSSSKQGSGVGIRLTSPTGE------------ATNN 443
Query: 658 QAEYEALIAGLKLAIEVQIDSLPVRTDSPLVANQVNGEFQVKELALIKYVERVRLLMGRL 717
AEYEAL+AGL LA ++I DS L+ANQ NGE+ ++ + Y+ V+ L
Sbjct: 444 VAEYEALVAGLNLAWGLKIGKTRAFCDSQLIANQFNGEYTTQDKKMEAYLIHVQNLAKNF 503
Query: 718 QQVVVEFVPRAQNQRADALAKLASTRKPGNNRSVIQETLANPNIE---GEFVASV----- 769
+ + +PR +N ADALA LAST R + E + P+IE E V +
Sbjct: 504 DEFELTRIPRGENTSADALAALASTSDTSLKRVIPVEFIEKPSIELGKEEHVLPIQISAD 563
Query: 770 -----DRQETWMNPIIDILAGE--PSDVVKYSKAQRREAGHYTLLDGILFRRGFSSPLLR 822
D WM PII ++ PSD K K + +A + L+D L++ S PL+
Sbjct: 564 QDDPDDCNSEWMEPIISYISEGKLPSDKWKARKL-KAQAARFVLVDTKLYKWRLSGPLMT 622
Query: 823 CLPPEKYEAVMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFT 882
C+ E +M E+H G SH PTI +
Sbjct: 623 CVEAEAICKIMKEIHSG---SHA----------------PTIHQ---------------- 647
Query: 883 DLPRAPPGQLVTISSPWPFAMWGVDLVGPFPTARSQMKFILVAVDYFTKWVEAEPLASIT 942
P L +I+SP+PF W +D++GP ++ Q K +LV DYF+KW+EAE ASI
Sbjct: 648 -----PAELLSSIASPYPFMRWSMDIIGPMHPSK-QKKLVLVLTDYFSKWIEAESYASIK 701
Query: 943 AAKIISFYWKRIVCRFGVP 961
A++ +F K I+CR G+P
Sbjct: 702 DAQVENFVLKHILCRHGIP 720
Score = 32.3 bits (72), Expect = 1.3
Identities = 22/72 (30%), Positives = 34/72 (46%), Gaps = 8/72 (11%)
Query: 22 TLSVKYLILETEGSYNMIIGRITLNRLCAVISTAQLAVKYPLSN------GH--IGGIVV 73
T V + I + YN IIG LN+ AV ST L +K+P S+ GH +
Sbjct: 16 TKIVDFSITDQPTVYNAIIGTPWLNQFRAVASTYHLCLKFPTSDSVWEKLGHERAEAVKA 75
Query: 74 DQRIARECYCNA 85
+ ++ YC++
Sbjct: 76 ENQLLETSYCHS 87
>At2g10780 pseudogene
Length = 1611
Score = 204 bits (520), Expect = 1e-52
Identities = 108/324 (33%), Positives = 177/324 (54%), Gaps = 1/324 (0%)
Query: 181 LALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKW 240
+ L PG PI++ RM + +++++ +LL FIR P W V+ VKK +G +
Sbjct: 651 IELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSF 709
Query: 241 RMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTF 300
R+C+D LNK K+ YPLP ID+L+D G + S +D SGYHQI + +D KT F
Sbjct: 710 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 769
Query: 301 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDL 360
T ++ + MPFGL NA A + ++M+ VF + + ++++D++V S H E L
Sbjct: 770 RTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHL 829
Query: 361 MEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEV 420
R+++H + KCSF R FLG +I+ + + ++P+K ++I+E P N E+
Sbjct: 830 RAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEI 889
Query: 421 QRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKP 480
+ +G RF++ + P + K+T F W+ ECE++F LK ML+ PVL P
Sbjct: 890 RSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLP 949
Query: 481 VQGLPLHLYFSVGDHAISSVILQE 504
+G P +Y + V++Q+
Sbjct: 950 EEGEPYTVYTDASIVGLGCVLMQK 973
Score = 51.6 bits (122), Expect = 2e-06
Identities = 39/144 (27%), Positives = 68/144 (47%), Gaps = 6/144 (4%)
Query: 823 CLPPEKY--EAVMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQV 880
C+P ++ E ++ E H+ + H G + + L+ ++W ++K+ A +V KC CQ+
Sbjct: 1152 CVPNDRALKEEILREAHQSKFSIHPGSNKMY-RDLKRYYHWVGMKKDVARWVAKCPTCQL 1210
Query: 881 FTDLPRAPPGQLVTISSP-WPFAMWGVDLVGPFPTA-RSQMKFILVAVDYFTKWVEAEPL 938
+ P G L + P W + +D V PT +S+ + V VD TK +
Sbjct: 1211 VKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAI 1270
Query: 939 ASITAAKIIS-FYWKRIVCRFGVP 961
+ A+II+ Y IV G+P
Sbjct: 1271 SDKDGAEIIAEKYIDEIVRLHGIP 1294
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 204 bits (518), Expect = 3e-52
Identities = 109/324 (33%), Positives = 176/324 (53%), Gaps = 1/324 (0%)
Query: 181 LALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKW 240
+ L PG PI++ RM + +++++ +LLA FIR P W V+ VKK +G +
Sbjct: 146 IELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSP-WGAPVLFVKKKDGSF 204
Query: 241 RMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTF 300
R+C+D LNK K+ YPLP ID+L+D G + S +D SGYHQI + +D KT F
Sbjct: 205 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 264
Query: 301 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDL 360
T ++ + MPFGL NA A + ++M+ VF + + +++DD++V S H E L
Sbjct: 265 RTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHL 324
Query: 361 MEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEV 420
R+++H + K SF R FLG +I+ + + ++P+K ++I+E P N E+
Sbjct: 325 RAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEI 384
Query: 421 QRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKP 480
+ +G RF++ + P + K+T F W+ ECE++F LK ML PVL P
Sbjct: 385 RSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLP 444
Query: 481 VQGLPLHLYFSVGDHAISSVILQE 504
+G P +Y + V++Q+
Sbjct: 445 EEGEPYTVYTDASIVGLGCVLMQK 468
Score = 50.8 bits (120), Expect = 4e-06
Identities = 39/144 (27%), Positives = 68/144 (47%), Gaps = 6/144 (4%)
Query: 823 CLPPEKY--EAVMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQV 880
C+P ++ E ++ E H+ + H G + + L+ ++W +RK+ A +V KC CQ+
Sbjct: 544 CVPNDRALKEEILREAHQSKFSIHPGSNKMY-RDLKRYYHWVGMRKDVARWVAKCPTCQL 602
Query: 881 FTDLPRAPPGQLVTIS-SPWPFAMWGVDLVGPFPTA-RSQMKFILVAVDYFTKWVEAEPL 938
+ P G L + S W + +D V PT +S+ + V VD TK +
Sbjct: 603 VKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSAHFMAI 662
Query: 939 ASITAAKIIS-FYWKRIVCRFGVP 961
+ A+II+ Y I+ G+P
Sbjct: 663 SDKDGAEIIAEKYIDEIMRLHGIP 686
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 200 bits (508), Expect = 4e-51
Identities = 106/323 (32%), Positives = 174/323 (53%), Gaps = 1/323 (0%)
Query: 181 LALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKW 240
+ L PG P+++ RM + +++++ LL FIR P W V+ VKK +G +
Sbjct: 483 IELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 541
Query: 241 RMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTF 300
R+C+D +LN+ K+ YPLP ID+L+D G S +D SGYHQI + ++D KT F
Sbjct: 542 RLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 601
Query: 301 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDL 360
T ++ + MPFGL NA A + RLM+ VF+ + + +++DD++V S L
Sbjct: 602 RTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHL 661
Query: 361 MEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEV 420
++++ + KCSF R FLG ++++ + ++P+K +AI++ P+N E+
Sbjct: 662 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 721
Query: 421 QRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKP 480
+ +G RF+ + P K K+ F W+ ECEE F LKEML++ PVL+ P
Sbjct: 722 RSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALP 781
Query: 481 VQGLPLHLYFSVGDHAISSVILQ 503
G P +Y + V++Q
Sbjct: 782 EHGQPYMVYTDASRVGLGCVLMQ 804
Score = 29.6 bits (65), Expect = 8.5
Identities = 27/109 (24%), Positives = 48/109 (43%), Gaps = 3/109 (2%)
Query: 855 LRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSP-WPFAMWGVDLVGPFP 913
L+ + W ++ + A +V +C CQ+ + G L ++ P W + +DLV
Sbjct: 965 LKRYYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGLR 1024
Query: 914 TARSQMKFILVAVDYFTKWVEAEPLASITAAKIIS-FYWKRIVCRFGVP 961
+R++ I V VD TK + A +++ + IV GVP
Sbjct: 1025 VSRTK-DAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVP 1072
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 195 bits (496), Expect = 9e-50
Identities = 108/340 (31%), Positives = 177/340 (51%), Gaps = 5/340 (1%)
Query: 181 LALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKW 240
+ L PG P+++ RM + +++++ LL FIR P W V+ VKK +G +
Sbjct: 509 IELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 567
Query: 241 RMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTF 300
R+C+D LN K+ YPLP ID+L+D G S +D SGYHQI + ++D KT F
Sbjct: 568 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 627
Query: 301 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDL 360
T ++ + MPF L NA A + RLM+ VF+ + + +++DD++V S H L
Sbjct: 628 RTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHL 687
Query: 361 MEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEV 420
++++ + KCSF R FLG ++++ + ++P+K +AI++ P+N E+
Sbjct: 688 RRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 747
Query: 421 QRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKP 480
+ + RF+ + P K K+ F W+ ECEE F LKEML++ PVL+ P
Sbjct: 748 RSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 807
Query: 481 VQGLPLHLYFSVGDHAISSVILQEVDGEQKIVYFVSYHFR 520
G P +Y + V++Q K++ + S R
Sbjct: 808 EHGQPYMVYTDASRVGLGCVLMQ----RGKVIAYASRQLR 843
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 188 bits (477), Expect = 1e-47
Identities = 105/340 (30%), Positives = 175/340 (50%), Gaps = 17/340 (5%)
Query: 181 LALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKW 240
+ L PG P+++ RM + +++++ LL A V+ VKK +G +
Sbjct: 470 IELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAP-------------VLFVKKKDGSF 516
Query: 241 RMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTF 300
R+C+D LN K+ YPLP ID+L+D G S +D SGYH I + ++D KT F
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAF 576
Query: 301 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDL 360
T ++ + MPFGL NA A + RLM+ VF+ + + +++DD++V S H L
Sbjct: 577 RTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHL 636
Query: 361 MEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEV 420
++++ + KCSF R FLG ++++ + ++P+K +AI++ +P+N E+
Sbjct: 637 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEI 696
Query: 421 QRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKP 480
+ +G RF+ + P K K+ F W+ ECEE F LKEML++ PVL+ P
Sbjct: 697 RSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALP 756
Query: 481 VQGLPLHLYFSVGDHAISSVILQEVDGEQKIVYFVSYHFR 520
G P +Y + V++Q K++ + S R
Sbjct: 757 EHGEPYMVYTDASGVGLGCVLMQ----RGKVIAYASRQLR 792
Score = 33.1 bits (74), Expect = 0.77
Identities = 27/98 (27%), Positives = 44/98 (44%), Gaps = 3/98 (3%)
Query: 866 KECAEFVRKCKKCQVFTDLPRAPPGQLVTISSP-WPFAMWGVDLVGPFPTARSQMKFILV 924
++ A +V +C CQ+ + P G L ++ P W + +D V P +R++ I V
Sbjct: 941 EDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTK-DAIWV 999
Query: 925 AVDYFTKWVEAEPLASITAAKIIS-FYWKRIVCRFGVP 961
VD TK + A +++ Y IV GVP
Sbjct: 1000 IVDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVP 1037
>At3g11970 hypothetical protein
Length = 1499
Score = 181 bits (459), Expect = 2e-45
Identities = 117/382 (30%), Positives = 196/382 (50%), Gaps = 6/382 (1%)
Query: 124 IEETKDLKFGEKILKIGTKLSE-EQEQRISKLLGDNLDLFAWSHKYMPGIDPNFICHRLA 182
++E + GE + I SE +E + ++L + D+F P + + H++
Sbjct: 538 VQEVSESTEGE-LCTINALTSELGEESVVEEVLNEYPDIFIEPTALPPFREKHN--HKIK 594
Query: 183 LNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKWRM 242
L G P+ Q R ++ I + V LL ++ P + VV+VKK +G WR+
Sbjct: 595 LLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDGTWRL 653
Query: 243 CVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTFMT 302
CVD +LN KDS+P+P I+ L+D G + S +D +GYHQ++M D KT F T
Sbjct: 654 CVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKT 713
Query: 303 ARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDLME 362
++ Y MPFGL NA AT+Q LM+ +F+ + + + V+ DD++V S H + L +
Sbjct: 714 HSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQ 773
Query: 363 AFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEVQR 422
F ++ + + KC+F + ++LG I+++ IE +P K KA++E P+ +K+++
Sbjct: 774 VFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRG 833
Query: 423 LIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQ 482
+G RF+ G + P + L K FEW + ++AF LK L PVLS P+
Sbjct: 834 FLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLF 892
Query: 483 GLPLHLYFSVGDHAISSVILQE 504
+ I +V++QE
Sbjct: 893 DKQFVVETDACGQGIGAVLMQE 914
Score = 40.8 bits (94), Expect = 0.004
Identities = 28/105 (26%), Positives = 48/105 (45%), Gaps = 10/105 (9%)
Query: 847 GRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPFAMW-- 904
GR + + ++ FYW + K+ ++R C CQ P A PG L + P P +W
Sbjct: 1098 GRDVTHQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPL--PIPDTIWSE 1155
Query: 905 -GVDLVGPFPTARSQMKFILVAVDYFTKWVE----AEPLASITAA 944
+D + P + + I+V VD +K + P +++T A
Sbjct: 1156 VSMDFIEGLPVSGGK-TVIMVVVDRLSKAAHFIALSHPYSALTVA 1199
>At1g36590 hypothetical protein
Length = 1499
Score = 181 bits (459), Expect = 2e-45
Identities = 117/382 (30%), Positives = 196/382 (50%), Gaps = 6/382 (1%)
Query: 124 IEETKDLKFGEKILKIGTKLSE-EQEQRISKLLGDNLDLFAWSHKYMPGIDPNFICHRLA 182
++E + GE + I SE +E + ++L + D+F P + + H++
Sbjct: 538 VQEVSESTEGE-LCTINALTSELGEESVVEEVLNEYPDIFIEPTALPPFREKHN--HKIK 594
Query: 183 LNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANGKWRM 242
L G P+ Q R ++ I + V LL ++ P + VV+VKK +G WR+
Sbjct: 595 LLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDGTWRL 653
Query: 243 CVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTFMT 302
CVD +LN KDS+P+P I+ L+D G + S +D +GYHQ++M D KT F T
Sbjct: 654 CVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKT 713
Query: 303 ARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDLME 362
++ Y MPFGL NA AT+Q LM+ +F+ + + + V+ DD++V S H + L +
Sbjct: 714 HSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHLKQ 773
Query: 363 AFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEVQR 422
F ++ + + KC+F + ++LG I+++ IE +P K KA++E P+ +K+++
Sbjct: 774 VFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQLRG 833
Query: 423 LIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQ 482
+G RF+ G + P + L K FEW + ++AF LK L PVLS P+
Sbjct: 834 FLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFEDLKAALCQAPVLSLPLF 892
Query: 483 GLPLHLYFSVGDHAISSVILQE 504
+ I +V++QE
Sbjct: 893 DKQFVVETDACGQGIGAVLMQE 914
Score = 35.8 bits (81), Expect = 0.12
Identities = 27/106 (25%), Positives = 48/106 (44%), Gaps = 10/106 (9%)
Query: 847 GRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPFAMW-- 904
GR + + ++ FY + K+ ++R C CQ P A PG L + P P +W
Sbjct: 1098 GRDVTHQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPL--PIPDTIWSE 1155
Query: 905 -GVDLVGPFPTARSQMKFILVAVDYFTKWVE----AEPLASITAAK 945
+D + P + + I+V VD +K + P +++T A+
Sbjct: 1156 VSMDFIEGLPVSGGK-TVIMVVVDRLSKAAHFIALSHPYSALTVAQ 1200
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 174 bits (442), Expect = 2e-43
Identities = 81/151 (53%), Positives = 108/151 (70%)
Query: 169 MPGIDPNFICHRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLV 228
M GI I H L ++P +P+ Q RR++G ++ +A+ EV +LL IRE+KYP WL
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 229 NVVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQI 288
N V+VKK NGKWR+CVD TDLNK C KD +PLP ID+LV+ G+E+LS MDA+SGY+QI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 289 KMHQSDEDKTTFMTARVNYCYQTMPFGLKNA 319
M+ D++KT+F+T Y Y+ MPFGLKNA
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKNA 562
Score = 139 bits (349), Expect = 1e-32
Identities = 92/336 (27%), Positives = 150/336 (44%), Gaps = 72/336 (21%)
Query: 683 TDSPLVANQVNGEFQVKELALIKYVERVRLLMGRLQQVVVEFVPR--------------- 727
+D+P AN+ +GE++ K+ + Y+ VR + GR +Q + +PR
Sbjct: 784 SDAP-TANRYSGEYEAKDACMEAYLNLVREVSGRFEQFELTRIPRAENSAANALAALAST 842
Query: 728 ------------------------------AQNQRADALAKLASTRKPGNNRSVIQ---- 753
A +R DA + + G++ +
Sbjct: 843 FEVTLPRVIPVETISQPSIRLDEISFVTTRAMRRRLDAQSAENGLHQLGDDEEISDAVHP 902
Query: 754 -ETLANPNIEGEFVASVDRQET------WMNPIIDILAGEPSDVVKYSKAQ-RREAGHYT 805
E + N ++ A + Q W PI D + K++ + + +
Sbjct: 903 TEIVENQSLPDNHNAPLPDQPPHDWGADWREPIRDYILNGTLPAEKWAARKLKATCARFC 962
Query: 806 LLDGILFRRGFSSPLLRCLPPEKYEAVMTEVHEGVCASHIGGRSLASKVLRAGFYWPTIR 865
+ + IL+RR FS+P C+ E+ VM EVH+G C +H GGRSLA KV + G+YWPT+
Sbjct: 963 IANDILYRRIFSAPDAVCIFGEQTRTVMKEVHDGTCGNHTGGRSLAFKVRKYGYYWPTLV 1022
Query: 866 KECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPFAMWGVDLVGPFPTARSQMKFILVA 925
+C + RKC++CQ L P L T+S+P+PF W +D+VGP +
Sbjct: 1023 ADCEAYARKCEQCQKHAPLILQPAELLTTVSAPYPFMKWLMDIVGPLHVS---------- 1072
Query: 926 VDYFTKWVEAEPLASITAAKIISFYWKRIVCRFGVP 961
T+ VEA ++IT ++ +F WK I+CR G+P
Sbjct: 1073 ----TRGVEAAAYSNITHVQVWNFIWKDIICRHGLP 1104
Score = 52.0 bits (123), Expect = 2e-06
Identities = 44/195 (22%), Positives = 82/195 (41%), Gaps = 8/195 (4%)
Query: 327 MDRVFEGQVGRNMEVYVDDMI-VKSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRG 385
+DR+ E G M ++D +L + ++ + ++ P FG++
Sbjct: 506 IDRLVESTTGHEMLSFMDAFSGYNQILMNPEDQEKTSFITECGTYYYKVMP----FGLKN 561
Query: 386 GKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSAPFI 445
+ T R E DK + ++ S + T ++ + PF+
Sbjct: 562 AVYFRCHRTRNRSEFEADKRFPVNDLTSQYKGSPTIDMSSGSTEQIHIPIYR--QVFPFL 619
Query: 446 KCLKK-NTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQGLPLHLYFSVGDHAISSVILQE 504
K + F WN CEEAF +LK LS P VL+K G L LY +V + A++ V ++
Sbjct: 620 HFTAKISKGFIWNESCEEAFKQLKRYLSEPSVLAKREFGEQLFLYIAVSESAVTGVQVRV 679
Query: 505 VDGEQKIVYFVSYHF 519
+++ +++V +
Sbjct: 680 ERSDKRPIFYVKTRY 694
Score = 30.0 bits (66), Expect = 6.5
Identities = 15/47 (31%), Positives = 25/47 (52%)
Query: 25 VKYLILETEGSYNMIIGRITLNRLCAVISTAQLAVKYPLSNGHIGGI 71
V++ + + +YN+I+G L + V ST VK+P G + GI
Sbjct: 369 VEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGKMTGI 415
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 171 bits (434), Expect = 1e-42
Identities = 113/406 (27%), Positives = 186/406 (44%), Gaps = 25/406 (6%)
Query: 101 SEVETPEESLDPRGEGRVNRPTPIEETKDLKFGEKILKIGTKLSEEQEQRISKLLGDNLD 160
SE++ P+ L P +G + T + +++ L + + K +G +LD
Sbjct: 818 SEIKAPKVDLKPLPKGLRYVFLGLNSTYPVIVNDELTADQVNLLITELMKYRKAIGYSLD 877
Query: 161 LFAWSHKYMPGIDPNFICHRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIRE 220
+ GI P HR+ L + +RR+ ++ +++E+ KLL A I
Sbjct: 878 -------DIKGISPTLCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYP 930
Query: 221 IKYPTWLVNVVIVKKANGKW------------------RMCVDNTDLNKTCPKDSYPLPS 262
I TW+ V V K G RMC++ LN K+ +PLP
Sbjct: 931 ISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLPF 990
Query: 263 IDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKTTFMTARVNYCYQTMPFGLKNAGAT 322
ID +++ + +D+YSG+ QI +H +D+ KTTF + Y+ MPFGL NA AT
Sbjct: 991 IDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPAT 1050
Query: 323 YQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHEDLMEAFGRIQKHNMRLNPEKCSFG 382
+QR M +F + +EV++DD V S+ +L R ++ N+ LN EKC F
Sbjct: 1051 FQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFM 1110
Query: 383 IRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVKEVQRLIGRITALSRFLLHSGDKSA 442
+R G LG I+ IE++ K + +++ P VK+++ +G F+ +
Sbjct: 1111 VREGIVLGRKISEEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLAR 1170
Query: 443 PFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLSKPVQGLPLHL 488
P + L K T F ++ EC AF +KE L P++ P P +
Sbjct: 1171 PLTRLLCKETEFAFDDECLTAFKLIKEALITAPIVQAPNWDFPFEI 1216
Score = 87.0 bits (214), Expect = 5e-17
Identities = 51/178 (28%), Positives = 84/178 (46%), Gaps = 22/178 (12%)
Query: 786 EPSDVVKYSKAQR-REAGHYTLLDGILFRRGFSSPLLRCLPPEKYEAVMTEVHEGVCASH 844
EP ++ Y K + ++ H+ + L+ C+ ++ E ++ H H
Sbjct: 1348 EPPNLSSYEKKKFFKDINHFYWDEPYLYTLCKDKIYRTCVSEDEIEGILLHCHGFAYGGH 1407
Query: 845 IGGRSLASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPFAMW 904
SK+L+AGF+WP++ K+ EF+ KC + F +W
Sbjct: 1408 FATFKTMSKILQAGFWWPSMFKDAQEFISKCDSFE--------------------NFDVW 1447
Query: 905 GVDLVGPFPTARSQMKFILVAVDYFTKWVEAEPLASITAAKIISFYWKRIVCRFGVPR 962
G+D +GPFP++ K+ILVA+DY +KWVEA + A ++ + I RFGVPR
Sbjct: 1448 GIDFMGPFPSSYGN-KYILVAIDYVSKWVEAIASHTNDARVVLKLFKTIIFPRFGVPR 1504
>At2g13210 pseudogene
Length = 152
Score = 162 bits (411), Expect = 6e-40
Identities = 71/151 (47%), Positives = 104/151 (68%)
Query: 158 NLDLFAWSHKYMPGIDPNFICHRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADF 217
++ F WS + +PG+ + H L ++P +P+ Q RR++G E+ +A++ EV KLL
Sbjct: 2 DVSTFVWSPEDLPGVSVEIVAHELNVDPTFKPVKQKRRKLGRERAEAVKAEVEKLLRIGS 61
Query: 218 IREIKYPTWLVNVVIVKKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLS 277
I + KYP WL N V+VKK NGKWR+C+D TDLNK CPKDS+P+ I +LV+ G++LLS
Sbjct: 62 ITKAKYPEWLANPVVVKKKNGKWRVCIDFTDLNKACPKDSFPMSHIGRLVESTSGDKLLS 121
Query: 278 LMDAYSGYHQIKMHQSDEDKTTFMTARVNYC 308
MDA++GY+QI M+ D++KTTF T + C
Sbjct: 122 FMDAFAGYNQIMMNSEDQEKTTFYTEQGIVC 152
>At2g06890 putative retroelement integrase
Length = 1215
Score = 162 bits (410), Expect = 8e-40
Identities = 95/288 (32%), Positives = 149/288 (50%), Gaps = 4/288 (1%)
Query: 234 KKANGKWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQS 293
++ +G WRMC D +N K +P+P +D ++D G+ + S +D SGYHQI+M++
Sbjct: 463 RQKDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKIDLKSGYHQIRMNEG 522
Query: 294 DEDKTTFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLG 353
DE KT F T Y + MPFGL +A +T+ RLM+ V +G + VY DD++V S
Sbjct: 523 DEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFVIVYFDDILVYSESL 582
Query: 354 SNHHEDLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKS 413
H E L ++K + N +KC+F FLGF++++ ++++ +K KAI++ S
Sbjct: 583 REHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVKVDEEKVKAIRDWPS 642
Query: 414 PSNVKEVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSA 473
P V EV+ G RF AP + +KK+ F+W EEAF LK+ L+
Sbjct: 643 PKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKAQEEAFQSLKDKLTN 702
Query: 474 PPVLSKPVQGLPLHLYFSVGDHAISSVILQEVDGEQKIVYFVSYHFRG 521
PVL + I +V++Q +QK++ F S G
Sbjct: 703 APVLILSEFLKTFEIECDASGIGIGAVLMQ----DQKLIAFFSEKLGG 746
>At1g35370 hypothetical protein
Length = 1447
Score = 162 bits (409), Expect = 1e-39
Identities = 104/326 (31%), Positives = 168/326 (50%), Gaps = 10/326 (3%)
Query: 179 HRLALNPGVEPITQTRRRMGNEKEKAIQQEVNKLLAADFIREIKYPTWLVNVVIVKKANG 238
H++ L G P+ Q R ++ I + V ++ + I+ P + VV+VKK +G
Sbjct: 570 HKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSP-FASPVVLVKKKDG 628
Query: 239 KWRMCVDNTDLNKTCPKDSYPLPSIDKLVDGALGNELLSLMDAYSGYHQIKMHQSDEDKT 298
WR+CVD T+LN KD + +P I+ L+D G+ + S +D +GYHQ++M D KT
Sbjct: 629 TWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKT 688
Query: 299 TFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSNHHE 358
F T ++ Y M FGL NA AT+Q LM+ VF + + + V+ DD+++ S H E
Sbjct: 689 AFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKE 748
Query: 359 DLMEAFGRIQKHNMRLNPEKCSFGIRGGKFLGFLITSRRIEINPDKCKAIQEMKSPSNVK 418
L F ++ H + F + LG I++R IE +P K +A++E +P+ VK
Sbjct: 749 HLRLVFEVMRLHKL--------FAKGSKEHLGHFISAREIETDPAKIQAVKEWPTPTTVK 800
Query: 419 EVQRLIGRITALSRFLLHSGDKSAPFIKCLKKNTTFEWNSECEEAFTRLKEMLSAPPVLS 478
+V+ +G RF+ + G + P + L K F W+ E + AF LK +L PVL+
Sbjct: 801 QVRGFLGFAGYYRRFVRNFGVIAGP-LHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLA 859
Query: 479 KPVQGLPLHLYFSVGDHAISSVILQE 504
PV + I +V++Q+
Sbjct: 860 LPVFDKQFMVETDACGQGIRAVLMQK 885
Score = 40.4 bits (93), Expect = 0.005
Identities = 41/171 (23%), Positives = 74/171 (42%), Gaps = 20/171 (11%)
Query: 785 GEPSDVVKYSKAQRREAGHYTLLDGILFRRGFSSPLLRCLPPEKYEAVMTEVHEGVCASH 844
G D++ + HY+ IL R+ + + P E + ++ + + S
Sbjct: 1011 GVLKDIISALQQHPDAKKHYSWSQDILRRKS------KIVVPNDVE-ITNKLLQWLHCSG 1063
Query: 845 IGGRS---LASKVLRAGFYWPTIRKECAEFVRKCKKCQVFTDLPRAPPGQLVTISSPWPF 901
+GGRS + + +++ FYW + K+ F+R C CQ A PG L + P P
Sbjct: 1064 MGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPL--PIPD 1121
Query: 902 AMW---GVDLVGPFPTARSQMKFILVAVDYFTKWVE----AEPLASITAAK 945
+W +D + P + + I+V VD +K A P +++T A+
Sbjct: 1122 KIWCDVSMDFIEGLPNSGGK-SVIMVVVDRLSKAAHFVALAHPYSALTVAQ 1171
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,594,853
Number of Sequences: 26719
Number of extensions: 927363
Number of successful extensions: 2274
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 2089
Number of HSP's gapped (non-prelim): 149
length of query: 963
length of database: 11,318,596
effective HSP length: 109
effective length of query: 854
effective length of database: 8,406,225
effective search space: 7178916150
effective search space used: 7178916150
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0189.12


