

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0182.2
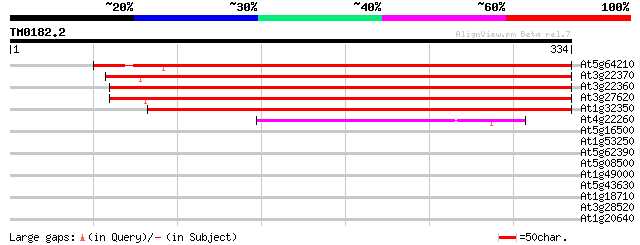
(334 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g64210 alternative oxidase 2 (sp|O22049) 456 e-128
At3g22370 alternative oxidase 1a precursor 442 e-125
At3g22360 alternative oxidase 1b precursor 427 e-120
At3g27620 alternative oxidase 1c precursor 424 e-119
At1g32350 oxidase like protein 412 e-115
At4g22260 IMMUTANS (IM) 76 3e-14
At5g16500 protein kinase-like protein 31 1.1
At1g53250 hypothetical protein; similar to ESTs gb|T22311.1, gb|... 30 1.4
At5g62390 KED - like protein 30 2.4
At5g08500 cleft lip and palate associated transmembrane protein-... 30 2.4
At1g49000 hypothetical protein; similar to ESTs gb|R64892.1, gb|... 29 3.1
At5g43630 unknown protein 29 4.1
At1g18710 Putative MYB47 transcription factor 28 5.4
At3g28520 hypothetical protein 28 9.2
At1g20640 unknown protein 28 9.2
>At5g64210 alternative oxidase 2 (sp|O22049)
Length = 282
Score = 456 bits (1172), Expect = e-128
Identities = 218/286 (76%), Positives = 248/286 (86%), Gaps = 6/286 (2%)
Query: 51 MSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGI--SRPKITREDGTEWPWNCF 108
M +A EKK+E + + + GSV V SYWGI ++ KITR+DG++WPWNCF
Sbjct: 1 MGMSSASAMEKKDENLTVK----KGQNGGGSVAVPSYWGIETAKMKITRKDGSDWPWNCF 56
Query: 109 MPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAV 168
MPWETY+ +LSIDL KHHVPKN DKVAYR VKLLRIPTD+FFQRRYGCRAMMLETVAAV
Sbjct: 57 MPWETYQANLSIDLKKHHVPKNIADKVAYRIVKLLRIPTDIFFQRRYGCRAMMLETVAAV 116
Query: 169 PGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQG 228
PGMVGGMLLHL+S+RKF+ SGGWIKALLEEAENERMHLMTM+ELV+PKWYER LV+ VQG
Sbjct: 117 PGMVGGMLLHLKSIRKFEHSGGWIKALLEEAENERMHLMTMMELVKPKWYERLLVMLVQG 176
Query: 229 VFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLP 288
+FFN+FFV Y++SP++AHRVVGYLEEEAIHSYTE+LKDI++G IENV APAIAIDYWRLP
Sbjct: 177 IFFNSFFVCYVISPRLAHRVVGYLEEEAIHSYTEFLKDIDNGKIENVAAPAIAIDYWRLP 236
Query: 289 KDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
KDATLKDV+TVIRADEAHHRDVNHFASDI GKELREA AP+GYH
Sbjct: 237 KDATLKDVVTVIRADEAHHRDVNHFASDIRNQGKELREAAAPIGYH 282
>At3g22370 alternative oxidase 1a precursor
Length = 354
Score = 442 bits (1138), Expect = e-125
Identities = 204/279 (73%), Positives = 241/279 (86%), Gaps = 2/279 (0%)
Query: 58 EEEKKEEKAEKESLRTEAK--KNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
EE+ ++K E ES +A N G ++SYWG+ KIT+EDG+EW WNCF PWETY+
Sbjct: 76 EEDANQKKTENESTGGDAAGGNNKGDKGIASYWGVEPNKITKEDGSEWKWNCFRPWETYK 135
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
D++IDL KHHVP FLD++AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGM
Sbjct: 136 ADITIDLKKHHVPTTFLDRIAYWTVKSLRWPTDLFFQRRYGCRAMMLETVAAVPGMVGGM 195
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
LLH +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +PKWYER LV+TVQGVFFNA+F
Sbjct: 196 LLHCKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPKWYERALVITVQGVFFNAYF 255
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
+ YL+SPK AHR+VGYLEEEAIHSYTE+LK+++ G IENVPAPAIAIDYWRLP DATL+D
Sbjct: 256 LGYLISPKFAHRMVGYLEEEAIHSYTEFLKELDKGNIENVPAPAIAIDYWRLPADATLRD 315
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+ V+RADEAHHRDVNHFASDIH+ G+EL+EAPAP+GYH
Sbjct: 316 VVMVVRADEAHHRDVNHFASDIHYQGRELKEAPAPIGYH 354
>At3g22360 alternative oxidase 1b precursor
Length = 325
Score = 427 bits (1097), Expect = e-120
Identities = 198/276 (71%), Positives = 236/276 (84%), Gaps = 1/276 (0%)
Query: 60 EKKEEKAEKESLRTEAKK-NDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDL 118
EKK+ EK S +A + N G ++ SYWG+ KIT+EDGTEW W+CF PWETY+ DL
Sbjct: 50 EKKKTTEEKGSSGGKADQGNKGEQLIVSYWGVKPMKITKEDGTEWKWSCFRPWETYKSDL 109
Query: 119 SIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLH 178
+IDL KHHVP DK+AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGML+H
Sbjct: 110 TIDLKKHHVPSTLPDKLAYWTVKSLRWPTDLFFQRRYGCRAMMLETVAAVPGMVGGMLVH 169
Query: 179 LRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLY 238
+SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +P WYER LV+ VQG+FFNA+F+ Y
Sbjct: 170 CKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPNWYERALVIAVQGIFFNAYFLGY 229
Query: 239 LLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVIT 298
L+SPK AHR+VGYLEEEAIHSYTE+LK++++G IENVPAPAIAIDYWRL DATL+DV+
Sbjct: 230 LISPKFAHRMVGYLEEEAIHSYTEFLKELDNGNIENVPAPAIAIDYWRLEADATLRDVVM 289
Query: 299 VIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+RADEAHHRDVNH+ASDIH+ G+EL+EAPAP+GYH
Sbjct: 290 VVRADEAHHRDVNHYASDIHYQGRELKEAPAPIGYH 325
>At3g27620 alternative oxidase 1c precursor
Length = 329
Score = 424 bits (1089), Expect = e-119
Identities = 195/279 (69%), Positives = 234/279 (82%), Gaps = 4/279 (1%)
Query: 60 EKKEEKAEKESLRTEAKKND----GSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
EKK+ E+E K ND G ++ SYWG+ KIT+EDGTEW W+CF PWETY+
Sbjct: 51 EKKKTSEEEEGSGDGVKVNDQGNKGEQLIVSYWGVKPMKITKEDGTEWKWSCFRPWETYK 110
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
DL+IDL KHHVP DK+AY VK LR PTD+FFQRRYGCRA+MLETVAAVPGMVGGM
Sbjct: 111 ADLTIDLKKHHVPSTLPDKIAYWMVKSLRWPTDLFFQRRYGCRAIMLETVAAVPGMVGGM 170
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
L+H +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +PKWYER LV++VQGVFFNA+
Sbjct: 171 LMHFKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPKWYERALVISVQGVFFNAYL 230
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
+ Y++SPK AHR+VGYLEEEAIHSYTE+LK++++G IENVPAPAIA+DYWRL DATL+D
Sbjct: 231 IGYIISPKFAHRMVGYLEEEAIHSYTEFLKELDNGNIENVPAPAIAVDYWRLEADATLRD 290
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+ V+RADEAHHRDVNH+ASDIH+ G EL+EAPAP+GYH
Sbjct: 291 VVMVVRADEAHHRDVNHYASDIHYQGHELKEAPAPIGYH 329
>At1g32350 oxidase like protein
Length = 318
Score = 412 bits (1058), Expect = e-115
Identities = 184/252 (73%), Positives = 221/252 (87%)
Query: 83 VVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKL 142
V+S+YWGI KIT+ DG+ W WNCF PW++Y+PD+SID+TKHH P NF DK AY TV+
Sbjct: 67 VISTYWGIPPTKITKPDGSAWKWNCFQPWDSYKPDVSIDVTKHHKPSNFTDKFAYWTVQT 126
Query: 143 LRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENE 202
L+IP +FFQR++ C AM+LETVAAVPGMVGGMLLHL+SLR+F+ SGGWIKALLEEAENE
Sbjct: 127 LKIPVQLFFQRKHMCHAMLLETVAAVPGMVGGMLLHLKSLRRFEHSGGWIKALLEEAENE 186
Query: 203 RMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTE 262
RMHLMT +EL QPKWYER +V TVQGVFFNA+F+ Y++SPK+AHR+ GYLEEEA++SYTE
Sbjct: 187 RMHLMTFIELSQPKWYERAIVFTVQGVFFNAYFLAYVISPKLAHRITGYLEEEAVNSYTE 246
Query: 263 YLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGK 322
+LKDI++G EN PAPAIAIDYWRLPKDATL+DV+ VIRADEAHHRD+NH+ASDI F G
Sbjct: 247 FLKDIDAGKFENSPAPAIAIDYWRLPKDATLRDVVYVIRADEAHHRDINHYASDIQFKGH 306
Query: 323 ELREAPAPLGYH 334
EL+EAPAP+GYH
Sbjct: 307 ELKEAPAPIGYH 318
>At4g22260 IMMUTANS (IM)
Length = 351
Score = 75.9 bits (185), Expect = 3e-14
Identities = 53/180 (29%), Positives = 83/180 (45%), Gaps = 21/180 (11%)
Query: 148 DVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLM 207
D ++ R R +LET+A VP +LH+ + + ++K E+ NE HL+
Sbjct: 121 DTLYRDRTYARFFVLETIARVPYFAFMSVLHMYETFGWWRRADYLKVHFAESWNEMHHLL 180
Query: 208 TMVEL-VQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKD 266
M EL W++RFL + ++ LY+LSP++A+ +E A +Y ++LK
Sbjct: 181 IMEELGGNSWWFDRFLAQHIATFYYFMTVFLYILSPRMAYHFSECVESHAYETYDKFLK- 239
Query: 267 IESGAIENVPAPAIAIDYW-------------------RLPKDATLKDVITVIRADEAHH 307
++N+PAP IA+ Y+ R P L DV IR DEA H
Sbjct: 240 ASGEELKNMPAPDIAVKYYTGGDLYLFDEFQTSRTPNTRRPVIENLYDVFVNIRDDEAEH 299
>At5g16500 protein kinase-like protein
Length = 636
Score = 30.8 bits (68), Expect = 1.1
Identities = 14/28 (50%), Positives = 19/28 (67%)
Query: 52 SSQAAPEEEKKEEKAEKESLRTEAKKND 79
SS + EEE+KE+KAEKE T K+ +
Sbjct: 403 SSDSEDEEEEKEQKAEKEEESTSKKRQE 430
>At1g53250 hypothetical protein; similar to ESTs gb|T22311.1,
gb|AA585734.1, emb|F19896.1
Length = 363
Score = 30.4 bits (67), Expect = 1.4
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 19/98 (19%)
Query: 49 RMMSSQAAPEE-EKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNC 107
+M++ +AA EK AEK+ R E + +G I +PK RE+
Sbjct: 231 KMIAKEAAKARTEKMRRAAEKKKEREEKDRREGK--------IRKPKQEREN-------- 274
Query: 108 FMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRI 145
P R L LTK H K L K+A T +++ +
Sbjct: 275 --PTIASRSKLKKRLTKIHKKKTSLGKIAIGTDRVVSV 310
>At5g62390 KED - like protein
Length = 446
Score = 29.6 bits (65), Expect = 2.4
Identities = 15/45 (33%), Positives = 27/45 (59%)
Query: 40 ENGVFSCWNRMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVV 84
ENG S + ++ +++K EEK +KE + T++KK + + VV
Sbjct: 216 ENGEVSHTYIIKATTGGEKKKKHEEKEKKEKIETKSKKKEKTRVV 260
>At5g08500 cleft lip and palate associated transmembrane
protein-like
Length = 590
Score = 29.6 bits (65), Expect = 2.4
Identities = 19/55 (34%), Positives = 25/55 (44%), Gaps = 3/55 (5%)
Query: 73 TEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCF---MPWETYRPDLSIDLTK 124
+EA KN+GS+ ++ +S I D P NCF P TY P D K
Sbjct: 121 SEALKNNGSLYAHVFFALSGYPIDLSDPEYQPLNCFGRTHPVATYLPKRKADKKK 175
>At1g49000 hypothetical protein; similar to ESTs gb|R64892.1,
gb|T04316.1
Length = 156
Score = 29.3 bits (64), Expect = 3.1
Identities = 18/78 (23%), Positives = 35/78 (44%), Gaps = 1/78 (1%)
Query: 2 KHLALSYALRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSC-WNRMMSSQAAPEEE 60
K L++S+ + + C RH ++L +R GE G WN + SS ++P +
Sbjct: 15 KRLSVSFLVSMMVLCARHANRLSKKLKLKSKKRTCSGGEGGGERFRWNMISSSMSSPRPK 74
Query: 61 KKEEKAEKESLRTEAKKN 78
+ +++ +KN
Sbjct: 75 ELFTTLSNKAMTMVRRKN 92
>At5g43630 unknown protein
Length = 831
Score = 28.9 bits (63), Expect = 4.1
Identities = 18/70 (25%), Positives = 34/70 (47%), Gaps = 1/70 (1%)
Query: 36 LVSGENGVFSCW-NRMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPK 94
L + +G+ S W +R S + P+++ E E + ++A K S + + + RP
Sbjct: 484 LTNRNSGLKSLWISRFSSKGSFPQKKASETAKEANASASDAAKTRDSRKMLADKNVIRPS 543
Query: 95 ITREDGTEWP 104
I+ DG + P
Sbjct: 544 ISSVDGPDKP 553
>At1g18710 Putative MYB47 transcription factor
Length = 267
Score = 28.5 bits (62), Expect = 5.4
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 2/30 (6%)
Query: 52 SSQAAPEEEKKEEKAEKESLRTEAKKNDGS 81
S QA EEEK+EE+ E++S+ +K DGS
Sbjct: 195 SDQAKEEEEKEEEEEERDSMM--GQKIDGS 222
>At3g28520 hypothetical protein
Length = 478
Score = 27.7 bits (60), Expect = 9.2
Identities = 12/28 (42%), Positives = 19/28 (67%)
Query: 58 EEEKKEEKAEKESLRTEAKKNDGSVVVS 85
E+E KEEK E E+L+ + N+ +V +S
Sbjct: 304 EDEDKEEKKEAENLKRVSGNNESNVTLS 331
>At1g20640 unknown protein
Length = 844
Score = 27.7 bits (60), Expect = 9.2
Identities = 18/76 (23%), Positives = 36/76 (46%)
Query: 31 EVRRFLVSGENGVFSCWNRMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGI 90
E +R +++ + + + R + + E E++ E E+E + T +N + +S W
Sbjct: 429 EEQRKMLNALSTIMAHVPRSLRTVTDKELEEESEVIEREEIVTPKIENASELHGNSPWNA 488
Query: 91 SRPKITREDGTEWPWN 106
S +I R + T P N
Sbjct: 489 SLEEIQRSNNTSNPQN 504
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.136 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,583,701
Number of Sequences: 26719
Number of extensions: 326569
Number of successful extensions: 1628
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1607
Number of HSP's gapped (non-prelim): 25
length of query: 334
length of database: 11,318,596
effective HSP length: 100
effective length of query: 234
effective length of database: 8,646,696
effective search space: 2023326864
effective search space used: 2023326864
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0182.2


