

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0181.18
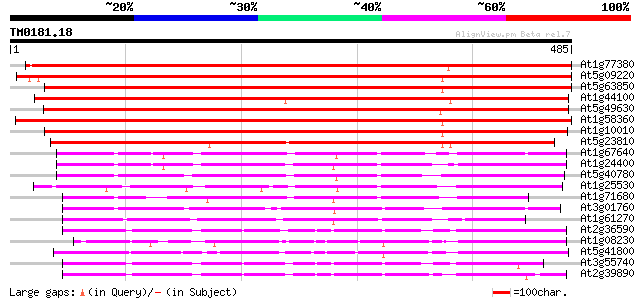
(485 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g77380 amino acid permease AAP3 763 0.0
At5g09220 amino acid transport protein AAP2 735 0.0
At5g63850 amino acid transporter AAP4 (pir||S51169) 716 0.0
At1g44100 amino acid permease AAP5 709 0.0
At5g49630 amino acid permease 6 (emb|CAA65051.1) 575 e-164
At1g58360 unknown protein 563 e-161
At1g10010 putative amino acid permease 553 e-158
At5g23810 amino acid transporter 467 e-132
At1g67640 putative amino acid permease 193 2e-49
At1g24400 amino acid transporter-like protein 2 (AATL2) 190 1e-48
At5g40780 amino acid permease 176 3e-44
At1g25530 hypothetical protein 166 3e-41
At1g71680 amino acid permease, putative 162 3e-40
At3g01760 putative amino acid permease 156 3e-38
At1g61270 amino acid permease 147 2e-35
At2g36590 putative proline transporter 114 2e-25
At1g08230 unknown protein 108 9e-24
At5g41800 amino acid permease-like protein; proline transporter-... 106 3e-23
At3g55740 proline transporter 2 105 4e-23
At2g39890 proline transporter 1 102 5e-22
>At1g77380 amino acid permease AAP3
Length = 476
Score = 763 bits (1971), Expect = 0.0
Identities = 358/474 (75%), Positives = 422/474 (88%), Gaps = 3/474 (0%)
Query: 14 HHQAFDVSLDMQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGW 73
+HQ +++DM Q GGSK DDDG+ KRTG+VWTASAHIITAVIGSGVLSLAWA AQLGW
Sbjct: 4 NHQTV-LAVDMPQTGGSKYLDDDGKNKRTGSVWTASAHIITAVIGSGVLSLAWATAQLGW 62
Query: 74 VAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQY 133
+AGP VM+LFS VTY+TS LL ACYR+GDP++GKRNYTYMD V SN+GG++V LCGIVQY
Sbjct: 63 LAGPVVMLLFSAVTYFTSSLLAACYRSGDPISGKRNYTYMDAVRSNLGGVKVTLCGIVQY 122
Query: 134 LNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDF 193
LN+FGVAIGYTIAS+ISM+AI+RSNCFHKS GKDPCHMN N YMI+FGLV+I+ SQIPDF
Sbjct: 123 LNIFGVAIGYTIASAISMMAIKRSNCFHKSGGKDPCHMNSNPYMIAFGLVQILFSQIPDF 182
Query: 194 DQLWWLSILAAVMSFTYSTIGLGLGFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQAL 253
DQLWWLSILAAVMSFTYS+ GL LG +V+ NG V GSLTGI++G VT+TQK+WRT QAL
Sbjct: 183 DQLWWLSILAAVMSFTYSSAGLALGIAQVVVNGKVKGSLTGISIGAVTETQKIWRTFQAL 242
Query: 254 GDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSS 313
GDIAFAYSYS+ILIEIQDTVKSPPSE KTMKKA+ +SV VTT+FYMLCGC GYAAFGD S
Sbjct: 243 GDIAFAYSYSIILIEIQDTVKSPPSEEKTMKKATLVSVSVTTMFYMLCGCMGYAAFGDLS 302
Query: 314 PGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKT 373
PGNLLTGFGFYNP+WL+DIANAAIVIHL+GAYQVY QPLFAF+E A+ +FPDS+F+ K
Sbjct: 303 PGNLLTGFGFYNPYWLLDIANAAIVIHLIGAYQVYCQPLFAFIEKQASIQFPDSEFIAKD 362
Query: 374 VQIPL--FNAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVE 431
++IP+ F ++N+FRL+WRT+FVIITT+ISMLLPFFND+VGLLGALGFWPLTVYFPVE
Sbjct: 363 IKIPIPGFKPLRLNVFRLIWRTVFVIITTVISMLLPFFNDVVGLLGALGFWPLTVYFPVE 422
Query: 432 MYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFKAIY 485
MYI QK+IP+WS +W+C Q+FSL CL+V++AAAAGS+AG+ LDL+ YKPF++ Y
Sbjct: 423 MYIAQKKIPRWSTRWVCLQVFSLGCLVVSIAAAAGSIAGVLLDLKSYKPFRSEY 476
>At5g09220 amino acid transport protein AAP2
Length = 493
Score = 735 bits (1897), Expect = 0.0
Identities = 345/486 (70%), Positives = 421/486 (85%), Gaps = 7/486 (1%)
Query: 7 NGSKNNHHH--QAFDVSLD--MQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVL 62
N +++HHH Q FDV+ + Q KCFDDDGRLKRTGTVWTASAHIITAVIGSGVL
Sbjct: 8 NNHRHHHHHGHQVFDVASHDFVPPQPAFKCFDDDGRLKRTGTVWTASAHIITAVIGSGVL 67
Query: 63 SLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGG 122
SLAWAIAQLGW+AGPAVM+LFSLVT Y+S LL CYR GD V+GKRNYTYMD V S +GG
Sbjct: 68 SLAWAIAQLGWIAGPAVMLLFSLVTLYSSTLLSDCYRTGDAVSGKRNYTYMDAVRSILGG 127
Query: 123 IQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGL 182
+ K+CG++QYLNLFG+AIGYTIA+SISM+AI+RSNCFHKS GKDPCHM+ N YMI FG+
Sbjct: 128 FKFKICGLIQYLNLFGIAIGYTIAASISMMAIKRSNCFHKSGGKDPCHMSSNPYMIVFGV 187
Query: 183 VEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENGAVGGSLTGITVGTVTQ 242
EI+LSQ+PDFDQ+WW+SI+AAVMSFTYS IGL LG +V NG GSLTGI++GTVTQ
Sbjct: 188 AEILLSQVPDFDQIWWISIVAAVMSFTYSAIGLALGIVQVAANGVFKGSLTGISIGTVTQ 247
Query: 243 TQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCG 302
TQK+WRT QALGDIAFAYSYS++LIEIQDTV+SPP+ESKTMKKA+ IS+ VTT+FYMLCG
Sbjct: 248 TQKIWRTFQALGDIAFAYSYSVVLIEIQDTVRSPPAESKTMKKATKISIAVTTIFYMLCG 307
Query: 303 CFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAE 362
GYAAFGD++PGNLLTGFGFYNPFWL+DIANAAIV+HLVGAYQV++QP+FAF+E AE
Sbjct: 308 SMGYAAFGDAAPGNLLTGFGFYNPFWLLDIANAAIVVHLVGAYQVFAQPIFAFIEKSVAE 367
Query: 363 KFPDSDFVTK--TVQIPLFNA-YKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGAL 419
++PD+DF++K ++IP F + YK+N+FR+V+R+ FV+ TT+ISML+PFFND+VG+LGAL
Sbjct: 368 RYPDNDFLSKEFEIRIPGFKSPYKVNVFRMVYRSGFVVTTTVISMLMPFFNDVVGILGAL 427
Query: 420 GFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYK 479
GFWPLTVYFPVEMYI Q+++ KWS +W+C Q+ S+ACL+++V A GS+AG+ LDL+VYK
Sbjct: 428 GFWPLTVYFPVEMYIKQRKVEKWSTRWVCLQMLSVACLVISVVAGVGSIAGVMLDLKVYK 487
Query: 480 PFKAIY 485
PFK+ Y
Sbjct: 488 PFKSTY 493
>At5g63850 amino acid transporter AAP4 (pir||S51169)
Length = 466
Score = 716 bits (1849), Expect = 0.0
Identities = 332/458 (72%), Positives = 399/458 (86%), Gaps = 3/458 (0%)
Query: 31 KCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYT 90
KCFDDDGRLKR+GTVWTASAHIITAVIGSGVLSLAWAI QLGW+AGP VM+LFS VTYY+
Sbjct: 9 KCFDDDGRLKRSGTVWTASAHIITAVIGSGVLSLAWAIGQLGWIAGPTVMLLFSFVTYYS 68
Query: 91 SILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSIS 150
S LL CYR GDPV+GKRNYTYMD V S +GG + K+CG++QYLNLFG+ +GYTIA+SIS
Sbjct: 69 STLLSDCYRTGDPVSGKRNYTYMDAVRSILGGFRFKICGLIQYLNLFGITVGYTIAASIS 128
Query: 151 MIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTY 210
M+AI+RSNCFH+S GK+PCHM+ N YMI FG+ EI+LSQI DFDQ+WWLSI+AA+MSFTY
Sbjct: 129 MMAIKRSNCFHESGGKNPCHMSSNPYMIMFGVTEILLSQIKDFDQIWWLSIVAAIMSFTY 188
Query: 211 STIGLGLGFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQ 270
S IGL LG +V NG V GSLTGI++G VTQTQK+WRT QALGDIAFAYSYS++LIEIQ
Sbjct: 189 SAIGLALGIIQVAANGVVKGSLTGISIGAVTQTQKIWRTFQALGDIAFAYSYSVVLIEIQ 248
Query: 271 DTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLI 330
DTV+SPP+ESKTMK A+ IS+ VTT FYMLCGC GYAAFGD +PGNLLTGFGFYNPFWL+
Sbjct: 249 DTVRSPPAESKTMKIATRISIAVTTTFYMLCGCMGYAAFGDKAPGNLLTGFGFYNPFWLL 308
Query: 331 DIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTK--TVQIPLFNA-YKINLF 387
D+ANAAIVIHLVGAYQV++QP+FAF+E AA +FPDSD VTK ++IP F + YK+N+F
Sbjct: 309 DVANAAIVIHLVGAYQVFAQPIFAFIEKQAAARFPDSDLVTKEYEIRIPGFRSPYKVNVF 368
Query: 388 RLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWI 447
R V+R+ FV++TT+ISML+PFFND+VG+LGALGFWPLTVYFPVEMYI Q+++ +WS+KW+
Sbjct: 369 RAVYRSGFVVLTTVISMLMPFFNDVVGILGALGFWPLTVYFPVEMYIRQRKVERWSMKWV 428
Query: 448 CFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFKAIY 485
C Q+ S CL++T+ A GS+AG+ LDL+VYKPFK Y
Sbjct: 429 CLQMLSCGCLMITLVAGVGSIAGVMLDLKVYKPFKTTY 466
>At1g44100 amino acid permease AAP5
Length = 480
Score = 709 bits (1831), Expect = 0.0
Identities = 333/470 (70%), Positives = 399/470 (84%), Gaps = 8/470 (1%)
Query: 22 LDMQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMI 81
LD+ + S FDDDGR KRTGTVWTASAHIITAVIGSGVLSLAWA+AQ+GW+ GP M+
Sbjct: 9 LDVLPKHSSDSFDDDGRPKRTGTVWTASAHIITAVIGSGVLSLAWAVAQIGWIGGPVAML 68
Query: 82 LFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAI 141
LFS VT+YTS LLC+CYR+GD V GKRNYTYMD +HSN+GGI+VK+CG+VQY+NLFG AI
Sbjct: 69 LFSFVTFYTSTLLCSCYRSGDSVTGKRNYTYMDAIHSNLGGIKVKVCGVVQYVNLFGTAI 128
Query: 142 GYTIASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSI 201
GYTIAS+IS++AI+R++C + DPCH+NGN+YMI+FG+V+I+ SQIPDFDQLWWLSI
Sbjct: 129 GYTIASAISLVAIQRTSCQQMNGPNDPCHVNGNVYMIAFGIVQIIFSQIPDFDQLWWLSI 188
Query: 202 LAAVMSFTYSTIGLGLGFGKVIENGAVGGSLTGITV------GTVTQTQKVWRTLQALGD 255
+AAVMSF YS IGLGLG KV+EN + GSLTG+TV GTVT +QK+WRT Q+LG+
Sbjct: 189 VAAVMSFAYSAIGLGLGVSKVVENKEIKGSLTGVTVGTVTLSGTVTSSQKIWRTFQSLGN 248
Query: 256 IAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPG 315
IAFAYSYSMILIEIQDTVKSPP+E TM+KA+F+SV VTT+FYMLCGC GYAAFGD++PG
Sbjct: 249 IAFAYSYSMILIEIQDTVKSPPAEVNTMRKATFVSVAVTTVFYMLCGCVGYAAFGDNAPG 308
Query: 316 NLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQ 375
NLL GF NP+WL+DIAN AIVIHLVGAYQVY QPLFAFVE A+ +FP+S+FVTK ++
Sbjct: 309 NLLAHGGFRNPYWLLDIANLAIVIHLVGAYQVYCQPLFAFVEKEASRRFPESEFVTKEIK 368
Query: 376 IPLF--NAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMY 433
I LF + +NLFRLVWRT FV+ TTLISML+PFFND+VGLLGA+GFWPLTVYFPVEMY
Sbjct: 369 IQLFPGKPFNLNLFRLVWRTFFVMTTTLISMLMPFFNDVVGLLGAIGFWPLTVYFPVEMY 428
Query: 434 IIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFKA 483
I QK +P+W KW+C Q+ S+ CL V+VAAAAGSV GI DL+VYKPF++
Sbjct: 429 IAQKNVPRWGTKWVCLQVLSVTCLFVSVAAAAGSVIGIVSDLKVYKPFQS 478
>At5g49630 amino acid permease 6 (emb|CAA65051.1)
Length = 481
Score = 575 bits (1482), Expect = e-164
Identities = 274/459 (59%), Positives = 353/459 (76%), Gaps = 5/459 (1%)
Query: 30 SKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYY 89
+K FD+DGR KRTGT T SAHIITAVIGSGVLSLAWAIAQLGWVAGPAV++ FS +TY+
Sbjct: 22 NKNFDEDGRDKRTGTWMTGSAHIITAVIGSGVLSLAWAIAQLGWVAGPAVLMAFSFITYF 81
Query: 90 TSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSI 149
TS +L CYR+ DPV GKRNYTYM+VV S +GG +V+LCG+ QY NL G+ IGYTI +SI
Sbjct: 82 TSTMLADCYRSPDPVTGKRNYTYMEVVRSYLGGRKVQLCGLAQYGNLIGITIGYTITASI 141
Query: 150 SMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFT 209
SM+A++RSNCFHK+ C + +MI F +++I+LSQIP+F L WLSILAAVMSF
Sbjct: 142 SMVAVKRSNCFHKNGHNVKCATSNTPFMIIFAIIQIILSQIPNFHNLSWLSILAAVMSFC 201
Query: 210 YSTIGLGLGFGKVIENGA-VGGSLTGITVG-TVTQTQKVWRTLQALGDIAFAYSYSMILI 267
Y++IG+GL K G V +LTG+TVG V+ +K+WRT QA+GDIAFAY+YS +LI
Sbjct: 202 YASIGVGLSIAKAAGGGEHVRTTLTGVTVGIDVSGAEKIWRTFQAIGDIAFAYAYSTVLI 261
Query: 268 EIQDTVKS-PPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNP 326
EIQDT+K+ PPSE+K MK+AS + V TT FYMLCGC GYAAFG+ +PGN LTGFGFY P
Sbjct: 262 EIQDTLKAGPPSENKAMKRASLVGVSTTTFFYMLCGCVGYAAFGNDAPGNFLTGFGFYEP 321
Query: 327 FWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVT--KTVQIPLFNAYKI 384
FWLID AN I +HL+GAYQV+ QP+F FVE+ +A+++PD+ F+T + +P + I
Sbjct: 322 FWLIDFANVCIAVHLIGAYQVFCQPIFQFVESQSAKRWPDNKFITGEYKIHVPCCGDFSI 381
Query: 385 NLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSI 444
N RLVWRT +V++T +++M+ PFFND +GL+GA FWPLTVYFP+EM+I QK+IPK+S
Sbjct: 382 NFLRLVWRTSYVVVTAVVAMIFPFFNDFLGLIGAASFWPLTVYFPIEMHIAQKKIPKFSF 441
Query: 445 KWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFKA 483
W ++ S C IV++ AAAGSV G+ L+ +KPF+A
Sbjct: 442 TWTWLKILSWTCFIVSLVAAAGSVQGLIQSLKDFKPFQA 480
>At1g58360 unknown protein
Length = 485
Score = 563 bits (1451), Expect = e-161
Identities = 265/483 (54%), Positives = 356/483 (72%), Gaps = 3/483 (0%)
Query: 6 KNGSKNNHHHQAFDVSLDMQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLA 65
K+ + H+H + +K D+DGR KRTGT TASAHIITAVIGSGVLSLA
Sbjct: 2 KSFNTEGHNHSTAESGDAYTVSDPTKNVDEDGREKRTGTWLTASAHIITAVIGSGVLSLA 61
Query: 66 WAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQV 125
WAIAQLGW+AG +++++FS +TY+TS +L CYR DPV GKRNYTYMDVV S +GG +V
Sbjct: 62 WAIAQLGWIAGTSILLIFSFITYFTSTMLADCYRAPDPVTGKRNYTYMDVVRSYLGGRKV 121
Query: 126 KLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEI 185
+LCG+ QY NL GV +GYTI +SIS++A+ +SNCFH C ++ YM FG++++
Sbjct: 122 QLCGVAQYGNLIGVTVGYTITASISLVAVGKSNCFHDKGHTADCTISNYPYMAVFGIIQV 181
Query: 186 VLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENGAVGGSLTGITVGT-VTQTQ 244
+LSQIP+F +L +LSI+AAVMSFTY+TIG+GL V S+TG VG VT Q
Sbjct: 182 ILSQIPNFHKLSFLSIMAAVMSFTYATIGIGLAIATVAGGKVGKTSMTGTAVGVDVTAAQ 241
Query: 245 KVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCF 304
K+WR+ QA+GDIAFAY+Y+ +LIEIQDT++S P+E+K MK+AS + V TT FY+LCGC
Sbjct: 242 KIWRSFQAVGDIAFAYAYATVLIEIQDTLRSSPAENKAMKRASLVGVSTTTFFYILCGCI 301
Query: 305 GYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKF 364
GYAAFG+++PG+ LT FGF+ PFWLID ANA I +HL+GAYQV++QP+F FVE +
Sbjct: 302 GYAAFGNNAPGDFLTDFGFFEPFWLIDFANACIAVHLIGAYQVFAQPIFQFVEKKCNRNY 361
Query: 365 PDSDFVTK--TVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFW 422
PD+ F+T +V +P + I+LFRLVWRT +V+ITT+++M+ PFFN I+GL+GA FW
Sbjct: 362 PDNKFITSEYSVNVPFLGKFNISLFRLVWRTAYVVITTVVAMIFPFFNAILGLIGAASFW 421
Query: 423 PLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFK 482
PLTVYFPVEM+I Q +I K+S +WI + CLIV++ AAAGS+AG+ ++ YKPF+
Sbjct: 422 PLTVYFPVEMHIAQTKIKKYSARWIALKTMCYVCLIVSLLAAAGSIAGLISSVKTYKPFR 481
Query: 483 AIY 485
++
Sbjct: 482 TMH 484
>At1g10010 putative amino acid permease
Length = 475
Score = 553 bits (1425), Expect = e-158
Identities = 260/455 (57%), Positives = 342/455 (75%), Gaps = 3/455 (0%)
Query: 31 KCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYT 90
K DDDGR KRTGT WTASAHIITAVIGSGVLSLAWAIAQLGWVAG V++ F+++TYYT
Sbjct: 18 KSVDDDGREKRTGTFWTASAHIITAVIGSGVLSLAWAIAQLGWVAGTTVLVAFAIITYYT 77
Query: 91 SILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSIS 150
S LL CYR+ D + G RNY YM VV S +GG +V+LCG+ QY+NL GV IGYTI +SIS
Sbjct: 78 STLLADCYRSPDSITGTRNYNYMGVVRSYLGGKKVQLCGVAQYVNLVGVTIGYTITASIS 137
Query: 151 MIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTY 210
++AI +SNC+H K C ++ YM +FG+V+I+LSQ+P+F +L +LSI+AAVMSF+Y
Sbjct: 138 LVAIGKSNCYHDKGHKAKCSVSNYPYMAAFGIVQIILSQLPNFHKLSFLSIIAAVMSFSY 197
Query: 211 STIGLGLGFGKVIENGAVGGSLTGITVGT-VTQTQKVWRTLQALGDIAFAYSYSMILIEI 269
++IG+GL V LTG +G VT ++KVW+ QA+GDIAF+Y+++ ILIEI
Sbjct: 198 ASIGIGLAIATVASGKIGKTELTGTVIGVDVTASEKVWKLFQAIGDIAFSYAFTTILIEI 257
Query: 270 QDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWL 329
QDT++S P E+K MK+AS + V TT+FY+LCGC GYAAFG+ +PG+ LT FGFY P+WL
Sbjct: 258 QDTLRSSPPENKVMKRASLVGVSTTTVFYILCGCIGYAAFGNQAPGDFLTDFGFYEPYWL 317
Query: 330 IDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTK--TVQIPLFNAYKINLF 387
ID ANA I +HL+GAYQVY+QP F FVE +K+P S+F+ K + ++PL ++NLF
Sbjct: 318 IDFANACIALHLIGAYQVYAQPFFQFVEENCNKKWPQSNFINKEYSSKVPLLGKCRVNLF 377
Query: 388 RLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWI 447
RLVWRT +V++TT ++M+ PFFN I+GLLGA FWPLTVYFPV M+I Q ++ K+S +W+
Sbjct: 378 RLVWRTCYVVLTTFVAMIFPFFNAILGLLGAFAFWPLTVYFPVAMHIAQAKVKKYSRRWL 437
Query: 448 CFQLFSLACLIVTVAAAAGSVAGIFLDLQVYKPFK 482
L L CLIV+ AA GS+ G+ ++ YKPFK
Sbjct: 438 ALNLLVLVCLIVSALAAVGSIIGLINSVKSYKPFK 472
>At5g23810 amino acid transporter
Length = 467
Score = 467 bits (1202), Expect = e-132
Identities = 227/443 (51%), Positives = 312/443 (70%), Gaps = 9/443 (2%)
Query: 36 DGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLC 95
D RTGT+WTA AHIIT VIG+GVLSLAWA A+LGW+AGPA +I F+ VT ++ LL
Sbjct: 21 DSVTARTGTLWTAVAHIITGVIGAGVLSLAWATAELGWIAGPAALIAFAGVTLLSAFLLS 80
Query: 96 ACYRNGDPVNGK-RNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAI 154
CYR DP NG R +Y V +G +CG+V Y++LFG I YTI + AI
Sbjct: 81 DCYRFPDPNNGPLRLNSYSQAVKLYLGKKNEIVCGVVVYISLFGCGIAYTIVIATCSRAI 140
Query: 155 ERSNCFHKSEGKDPCHM--NGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYST 212
+SNC+H++ C N N +M+ FGL +I +SQIP+F + WLS++AA+MSFTYS
Sbjct: 141 MKSNCYHRNGHNATCSYGDNNNYFMVLFGLTQIFMSQIPNFHNMVWLSLVAAIMSFTYSF 200
Query: 213 IGLGLGFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDT 272
IG+GL GK+IEN + GS+ GI + +KVW QALG+IAF+Y +S+IL+EIQDT
Sbjct: 201 IGIGLALGKIIENRKIEGSIRGIPAEN--RGEKVWIVFQALGNIAFSYPFSIILLEIQDT 258
Query: 273 VKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDI 332
++SPP+E +TMKKAS ++V + T F+ CGCFGYAAFGDS+PGNLLTGFGFY PFWL+D
Sbjct: 259 LRSPPAEKQTMKKASTVAVFIQTFFFFCCGCFGYAAFGDSTPGNLLTGFGFYEPFWLVDF 318
Query: 333 ANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTK--TVQIPLF--NAYKINLFR 388
ANA IV+HLVG YQVYSQP+FA E +K+P++ F+ + ++PL ++N R
Sbjct: 319 ANACIVLHLVGGYQVYSQPIFAAAERSLTKKYPENKFIARFYGFKLPLLRGETVRLNPMR 378
Query: 389 LVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWIC 448
+ RT++V+ITT ++++ P+FN+++G++GAL FWPL VYFPVEM I+QK+I W+ W+
Sbjct: 379 MCLRTMYVLITTGVAVMFPYFNEVLGVVGALAFWPLAVYFPVEMCILQKKIRSWTRPWLL 438
Query: 449 FQLFSLACLIVTVAAAAGSVAGI 471
+ FS CL+V + + GS+ G+
Sbjct: 439 LRGFSFVCLLVCLLSLVGSIYGL 461
>At1g67640 putative amino acid permease
Length = 441
Score = 193 bits (490), Expect = 2e-49
Identities = 134/446 (30%), Positives = 219/446 (49%), Gaps = 39/446 (8%)
Query: 41 RTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRN 100
R W ++ H +TA++G+GVLSL +A++ LGW G +MI+ L+T+YT L +
Sbjct: 29 RNAKWWYSAFHNVTAMVGAGVLSLPYAMSNLGWGPGVTIMIMSWLITFYT---LWQMVQM 85
Query: 101 GDPVNGKRNYTYMDVVHSNMGGIQVKLCGIV--QYLNLFGVAIGYTIASSISMIAIERSN 158
+ V GKR Y ++ G ++ L +V Q + GV I Y + S+ I
Sbjct: 86 HEMVPGKRFDRYHELGQHAFGE-KLGLWIVVPQQLIVEVGVDIVYMVTGGKSLKKIHDLL 144
Query: 159 CFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLG 218
C D ++ +++ F + VL+ +P+F+ + +S+ AAVMS +YSTI
Sbjct: 145 C------TDCKNIRTTYWIMIFASIHFVLAHLPNFNSISIVSLAAAVMSLSYSTIAWATS 198
Query: 219 FGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPS 278
K + S T G V + L ALGD+AFAY+ +++EIQ T+ S P
Sbjct: 199 VKKGVHPNVDYSSRASTTSGNV------FNFLNALGDVAFAYAGHNVVLEIQATIPSTPE 252
Query: 279 ESK--TMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAA 336
+ M K ++ +V + Y Y FG+S N+L P WLI IANA
Sbjct: 253 KPSKIAMWKGVVVAYIVVAICYFPVAFVCYYIFGNSVDDNIL--MTLEKPIWLIAIANAF 310
Query: 337 IVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFV 396
+V+H++G+YQ+Y+ P+F +E + V K + P F R + RT++V
Sbjct: 311 VVVHVIGSYQIYAMPVFDMLETF---------LVKKMMFAPSFK------LRFITRTLYV 355
Query: 397 IITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLAC 456
T +++ +PFF ++G G F P T Y P M++ K+ K+ + W C F +
Sbjct: 356 AFTMFVAICIPFFGGLLGFFGGFAFAPTTYYLPCIMWLCIKKPKKYGLSW-CINWFCIVV 414
Query: 457 -LIVTVAAAAGSVAGIFLDLQVYKPF 481
+I+T+ A G + I + + Y+ F
Sbjct: 415 GVILTILAPIGGLRTIIISAKNYEFF 440
>At1g24400 amino acid transporter-like protein 2 (AATL2)
Length = 441
Score = 190 bits (483), Expect = 1e-48
Identities = 127/446 (28%), Positives = 219/446 (48%), Gaps = 39/446 (8%)
Query: 41 RTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRN 100
R W ++ H +TA++G+GVLSL +A++ LGW G +M++ ++T YT L
Sbjct: 29 RNAKWWYSAFHNVTAMVGAGVLSLPYAMSNLGWGPGVTIMVMSWIITLYT---LWQMVEM 85
Query: 101 GDPVNGKRNYTYMDVVHSNMGGIQVKLCGIV--QYLNLFGVAIGYTIASSISMIAIERSN 158
+ V GKR Y ++ G ++ L +V Q + GV I Y + S+ + +
Sbjct: 86 HEIVPGKRLDRYHELGQHAFGE-KLGLWIVVPQQLIVEVGVDIVYMVTGGASLKKVHQLV 144
Query: 159 CFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLG 218
C D + +++ F V V+S +P+F+ + +S+ AAVMS TYSTI
Sbjct: 145 C------PDCKEIRTTFWIMIFASVHFVISHLPNFNSISIISLAAAVMSLTYSTIAWAAS 198
Query: 219 FGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPS 278
K + VG KV+ L ALGD+AFAY+ +++EIQ T+ S P
Sbjct: 199 VHKGVHPDVDYSPRASTDVG------KVFNFLNALGDVAFAYAGHNVVLEIQATIPSTPE 252
Query: 279 --ESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAA 336
M + ++ +V + Y GY FG+S N+L P WLI +AN
Sbjct: 253 MPSKVPMWRGVIVAYIVVAICYFPVAFLGYYIFGNSVDDNIL--ITLEKPIWLIAMANMF 310
Query: 337 IVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFN-AYKINLFRLVWRTIF 395
+VIH++G+YQ+++ P+F +E +K FN ++K+ R + R+++
Sbjct: 311 VVIHVIGSYQIFAMPVFDMLETVLVKKMN-------------FNPSFKL---RFITRSLY 354
Query: 396 VIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLA 455
V T ++++ +PFF ++G G F P T Y P M+++ K+ ++ + W +
Sbjct: 355 VAFTMIVAICVPFFGGLLGFFGGFAFAPTTYYLPCIMWLVLKKPKRFGLSWTANWFCIIV 414
Query: 456 CLIVTVAAAAGSVAGIFLDLQVYKPF 481
+++T+ A G + I ++ + YK F
Sbjct: 415 GVLLTILAPIGGLRTIIINAKTYKFF 440
>At5g40780 amino acid permease
Length = 446
Score = 176 bits (445), Expect = 3e-44
Identities = 124/442 (28%), Positives = 202/442 (45%), Gaps = 35/442 (7%)
Query: 41 RTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRN 100
R W ++ H +TA++G+GVL L +A++QLGW G AV++L ++T YT L
Sbjct: 34 RNAKWWYSAFHNVTAMVGAGVLGLPYAMSQLGWGPGIAVLVLSWVITLYT---LWQMVEM 90
Query: 101 GDPVNGKRNYTYMDVVHSNMG-GIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNC 159
+ V GKR Y ++ G + + + Q + GV I Y + S+
Sbjct: 91 HEMVPGKRFDRYHELGQHAFGEKLGLYIVVPQQLIVEIGVCIVYMVTGGKSL------KK 144
Query: 160 FHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGF 219
FH+ D + +++ F V VLS +P+F+ + +S+ AAVMS +YSTI
Sbjct: 145 FHELVCDDCKPIKLTYFIMIFASVHFVLSHLPNFNSISGVSLAAAVMSLSYSTIAWASSA 204
Query: 220 GKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSE 279
K ++ G T GT V+ LGD+AFAY+ +++EIQ T+ S P +
Sbjct: 205 SKGVQEDVQYGYKAKTTAGT------VFNFFSGLGDVAFAYAGHNVVLEIQATIPSTPEK 258
Query: 280 SK--TMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAI 337
M + ++ +V L Y GY FG+ N+L P WLI AN +
Sbjct: 259 PSKGPMWRGVIVAYIVVALCYFPVALVGYYIFGNGVEDNIL--MSLKKPAWLIATANIFV 316
Query: 338 VIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVI 397
VIH++G+YQ+Y+ P+F +E T+ + N R R +V
Sbjct: 317 VIHVIGSYQIYAMPVFDMME---------------TLLVKKLNFRPTTTLRFFVRNFYVA 361
Query: 398 ITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACL 457
T + M PFF ++ G F P T + P +++ + K+S+ W + + L
Sbjct: 362 ATMFVGMTFPFFGGLLAFFGGFAFAPTTYFLPCVIWLAIYKPKKYSLSWWANWVCIVFGL 421
Query: 458 IVTVAAAAGSVAGIFLDLQVYK 479
+ V + G + I + + YK
Sbjct: 422 FLMVLSPIGGLRTIVIQAKGYK 443
>At1g25530 hypothetical protein
Length = 440
Score = 166 bits (420), Expect = 3e-41
Identities = 133/469 (28%), Positives = 211/469 (44%), Gaps = 53/469 (11%)
Query: 21 SLDMQQQGGSKCFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVM 80
S + ++ G K +D R W ++ H +TA+IG+GVLSL +A+A LGW G V+
Sbjct: 10 SKETDRKSGEKWTAEDP--SRPAKWWYSTFHTVTAMIGAGVLSLPYAMAYLGWGPGTFVL 67
Query: 81 IL---FSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNL- 136
+ +L T + + L C V G R Y+D+ G + Q L +
Sbjct: 68 AMTWGLTLNTMWQMVQLHEC------VPGTRFDRYIDLGRYAFGPKLGPWIVLPQQLIVQ 121
Query: 137 FGVAIGYTIASSISM---IAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDF 193
G I Y + + + I S C + + +++ FG V +LSQ+P+F
Sbjct: 122 VGCNIVYMVTGGKCLKQFVEITCSTC---------TPVRQSYWILGFGGVHFILSQLPNF 172
Query: 194 DQLWWLSILAAVMSFTYSTIGLG--LGFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQ 251
+ + +S+ AAVMS YSTI G + G+V + V G T +R
Sbjct: 173 NSVAGVSLAAAVMSLCYSTIAWGGSIAHGRVPD---VSYDYKATNPGDFT-----FRVFN 224
Query: 252 ALGDIAFAYSYSMILIEIQDTVKSPPSESKT--MKKASFISVVVTTLFYMLCGCFGYAAF 309
ALG I+FA++ + +EIQ T+ S P M + + VV + Y Y AF
Sbjct: 225 ALGQISFAFAGHAVALEIQATMPSTPERPSKVPMWQGVIGAYVVNAVCYFPVALICYWAF 284
Query: 310 GDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDF 369
G N+L P WLI AN +V+H++G+YQV++ P+F +E KF
Sbjct: 285 GQDVDDNVL--MNLQRPAWLIAAANLMVVVHVIGSYQVFAMPVFDLLERMMVNKFGFKHG 342
Query: 370 VTKTVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFP 429
V + R RTI+V T I + PFF D++G G GF P + + P
Sbjct: 343 V---------------VLRFFTRTIYVAFTLFIGVSFPFFGDLLGFFGGFGFAPTSFFLP 387
Query: 430 VEMYIIQKRIPKWSIKWICFQLFSLACLIVTVAAAAGSVAGIFLDLQVY 478
M++I K+ ++S+ W + + + + +A+ G + I D Y
Sbjct: 388 SIMWLIIKKPRRFSVTWFVNWISIIVGVFIMLASTIGGLRNIIADSSTY 436
>At1g71680 amino acid permease, putative
Length = 434
Score = 162 bits (411), Expect = 3e-40
Identities = 116/418 (27%), Positives = 195/418 (45%), Gaps = 57/418 (13%)
Query: 46 WTASA-HIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPV 104
W SA H +TA++G+GVL L +A++QLGW G +I+ +T+Y+ L + + V
Sbjct: 41 WYYSAFHNVTAMVGAGVLGLPFAMSQLGWGPGLVAIIMSWAITFYS---LWQMVQLHEAV 97
Query: 105 NGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSE 164
GKR Y ++ FG +GY I ++ S+ +
Sbjct: 98 PGKRLDRYPELGQE-----------------AFGPKLGYWIVMPQQLLVQIASDIVYNVT 140
Query: 165 GKDPC------------HMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYST 212
G H+ Y++ F +++VLSQ PDF+ + +S+LAA+MSF YS
Sbjct: 141 GGKSLKKFVELLFPNLEHIRQTYYILGFAALQLVLSQSPDFNSIKIVSLLAALMSFLYSM 200
Query: 213 IGLGLGFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDT 272
I K E+ + G TV ++ V+ +G IAFA++ +++EIQ T
Sbjct: 201 IASVASIAKGTEHRPSTYGVRGDTVASM-----VFDAFNGIGTIAFAFAGHSVVLEIQAT 255
Query: 273 VKSPPS--ESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLI 330
+ S P K M K ++ ++ + Y+ GY AFG ++L P WLI
Sbjct: 256 IPSTPEVPSKKPMWKGVVVAYIIVIICYLFVAISGYWAFGAHVEDDVL--ISLERPAWLI 313
Query: 331 DIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLV 390
AN + IH++G+YQV++ +F +E+Y + + T RLV
Sbjct: 314 AAANFMVFIHVIGSYQVFAMIVFDTIESYLVKTLKFTPSTT---------------LRLV 358
Query: 391 WRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWIC 448
R+ +V + L+++ +PFF ++G G L F + + P +++I KR ++S W C
Sbjct: 359 ARSTYVALICLVAVCIPFFGGLLGFFGGLVFSSTSYFLPCIIWLIMKRPKRFSAHWWC 416
>At3g01760 putative amino acid permease
Length = 455
Score = 156 bits (394), Expect = 3e-38
Identities = 122/437 (27%), Positives = 214/437 (48%), Gaps = 37/437 (8%)
Query: 46 WTASA-HIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPV 104
W SA H +TA++G+GVL L +A+++LGW G V+IL ++T YT L +
Sbjct: 36 WYYSAFHNVTAIVGAGVLGLPYAMSELGWGPGVVVLILSWVITLYT---LWQMIEMHEMF 92
Query: 105 NGKRNYTYMDVVHSNMGG-IQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKS 163
G+R Y ++ + G + + + +Q L V I Y + S+ + +
Sbjct: 93 EGQRFDRYHELGQAAFGKKLGLYIIVPLQLLVEISVCIVYMVTGGKSL---KNVHDLALG 149
Query: 164 EGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVI 223
+G + +++ F + VLS + +F+ + +S++AAVMS +YSTI K
Sbjct: 150 DGDKCTKLRIQHFILIFASSQFVLSLLKNFNSISGVSLVAAVMSVSYSTIAWVASLRK-- 207
Query: 224 ENGAVGGSLTGITVGTVTQTQKV-WRTLQALGDIAFAYSYSMILIEIQDTVKSPPSE--S 280
GA GS + G +T V L ALG++AFAY+ +++EIQ T+ S P
Sbjct: 208 --GATTGS---VEYGYRKRTTSVPLAFLSALGEMAFAYAGHNVVLEIQATIPSTPENPSK 262
Query: 281 KTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIH 340
+ M K + ++ ++ Y G+ FG+S ++L P L+ +AN +VIH
Sbjct: 263 RPMWKGAVVAYIIVAFCYFPVALVGFKTFGNSVEESILESLT--KPTALVIVANMFVVIH 320
Query: 341 LVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITT 400
L+G+YQVY+ P+F +E+ V I +++ + R R FV T
Sbjct: 321 LLGSYQVYAMPVFDMIES---------------VMIRIWHFSPTRVLRFTIRWTFVAATM 365
Query: 401 LISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLAC-LIV 459
I++ LP+++ ++ G F P T + P M++I K+ ++S+ W C F + L++
Sbjct: 366 GIAVGLPYYSALLSFFGGFVFAPTTYFIPCIMWLILKKPKRFSLSW-CMNWFCIIFGLVL 424
Query: 460 TVAAAAGSVAGIFLDLQ 476
+ A G +A + ++Q
Sbjct: 425 MIIAPIGGLAKLIYNIQ 441
>At1g61270 amino acid permease
Length = 418
Score = 147 bits (370), Expect = 2e-35
Identities = 113/406 (27%), Positives = 196/406 (47%), Gaps = 36/406 (8%)
Query: 46 WTASA-HIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPV 104
W SA H +TA++G+GVL L +A+++LGW G V+IL ++T YT + + +
Sbjct: 28 WYYSAFHNVTAIVGAGVLGLPYAMSELGWGPGVVVLILSWVITLYTFWQMIEMH---EMF 84
Query: 105 NGKRNYTYMDVVHSNMG-GIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKS 163
GKR Y ++ + G + + + +Q L I Y + S+ I + S
Sbjct: 85 EGKRFDRYHELGQAAFGKKLGLYIVVPLQLLVETSACIVYMVTGGESLKKIHQ-----LS 139
Query: 164 EGKDPCH-MNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKV 222
G C + +++ F + VLS + +F+ + +S++AAVMS +YSTI K
Sbjct: 140 VGDYECRKLKVRHFILIFASSQFVLSLLKNFNSISGVSLVAAVMSMSYSTIAWVASLTKG 199
Query: 223 IENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPS--ES 280
+ N G T L ALG++AFAY+ +++EIQ T+ S P
Sbjct: 200 VANNVEYG------YKRRNNTSVPLAFLGALGEMAFAYAGHNVVLEIQATIPSTPENPSK 253
Query: 281 KTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIH 340
+ M K + ++ ++ Y G+ FG++ N+L P LI +AN ++IH
Sbjct: 254 RPMWKGAIVAYIIVAFCYFPVALVGFWTFGNNVEENILK--TLRGPKGLIIVANIFVIIH 311
Query: 341 LVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITT 400
L+G+YQVY+ P+F +E+ +K+ F+ ++ F + W FV T
Sbjct: 312 LMGSYQVYAMPVFDMIESVMIKKWH-------------FSPTRVLRFTIRW--TFVAATM 356
Query: 401 LISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKW 446
I++ LP F+ ++ G F P T + P +++I K+ ++S+ W
Sbjct: 357 GIAVALPHFSALLSFFGGFIFAPTTYFIPCIIWLILKKPKRFSLSW 402
>At2g36590 putative proline transporter
Length = 436
Score = 114 bits (284), Expect = 2e-25
Identities = 112/440 (25%), Positives = 197/440 (44%), Gaps = 40/440 (9%)
Query: 46 WTASAHIITAVIGSG-VLSLAWAI-AQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDP 103
W +A ++T I S VL + + LGW+ G +IL + ++ Y + L+ + G
Sbjct: 30 WFQAAFVLTTSINSAYVLGYSGTVMVPLGWIGGVVGLILATAISLYANTLVAKLHEFG-- 87
Query: 104 VNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKS 163
GKR+ Y D+ G L ++QY+NLF + G+ I + ++ A+
Sbjct: 88 --GKRHIRYRDLAGFIYGRKAYCLTWVLQYVNLFMINCGFIILAGSALKAVYVLF----- 140
Query: 164 EGKDPCHMNGNIYMISFGLVEIVLS-QIPDFDQLW-WLSILAAVMSFTYSTIGLGLGFGK 221
+D M ++ GL+ V + IP L WL++ + ++S Y + + L
Sbjct: 141 --RDDHAMKLPHFIAIAGLICAVFAIGIPHLSALGIWLAV-STILSLIYIVVAIVLSVKD 197
Query: 222 VIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESK 281
++ + + G + K++ A + F ++ M L EIQ TVK P K
Sbjct: 198 GVKAPSRDYEIQG------SPLSKLFTITGAAATLVFVFNTGM-LPEIQATVKQPVV--K 248
Query: 282 TMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHL 341
M KA + V L GY A+G S+ LL P W+ +AN + ++
Sbjct: 249 NMMKALYFQFTVGVLPMFAVVFIGYWAYGSSTSPYLLNNVN--GPLWVKALANISAILQS 306
Query: 342 VGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTL 401
V + +++ P + ++ D+ F K + L N LFR++ R ++ ++TL
Sbjct: 307 VISLHIFASPTYEYM---------DTKFGIKGNPLALKNL----LFRIMARGGYIAVSTL 353
Query: 402 ISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLACLIVTV 461
+S LLPF D + L GA+ +PLT MY K +++ +C L + +++V
Sbjct: 354 LSALLPFLGDFMSLTGAVSTFPLTFILANHMYYKAKNNKLNTLQKLCHWLNVVFFSLMSV 413
Query: 462 AAAAGSVAGIFLDLQVYKPF 481
AAA ++ I LD + + F
Sbjct: 414 AAAIAALRLIALDSKNFHVF 433
>At1g08230 unknown protein
Length = 422
Score = 108 bits (269), Expect = 9e-24
Identities = 116/445 (26%), Positives = 189/445 (42%), Gaps = 61/445 (13%)
Query: 56 VIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDV 115
V+ +G L + + GW AG + ++ + VT+Y+ LL + + G R + D+
Sbjct: 17 VVDAGSL---FVLKSKGWAAGISCLVGGAAVTFYSYTLLSLTLEHHASL-GNRYLRFRDM 72
Query: 116 VHSNM---------GGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGK 166
H + G IQ+ +C +GV I + + A+
Sbjct: 73 AHHILSPKWGRYYVGPIQMAVC--------YGVVIANALLGGQCLKAMYLV--------- 115
Query: 167 DPCHMNGNI----YMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKV 222
NG + ++I FG + +VL+Q P F L +++ L+ ++ YS
Sbjct: 116 --VQPNGEMKLFEFVIIFGCLLLVLAQFPSFHSLRYINSLSLLLCLLYSASAAAASIYIG 173
Query: 223 IENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKT 282
E A T VG +V+ A+ IA Y I+ EIQ T+ S P + K
Sbjct: 174 KEPNAPEKDYT--IVGD--PETRVFGIFNAMAIIATTYGNG-IIPEIQATI-SAPVKGKM 227
Query: 283 MKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGF------GFYNPFWLIDIANAA 336
MK +VV F+ + GY AFG + G + T F ++ P W I + N
Sbjct: 228 MKGLCMCYLVVIMTFFTVA-ITGYWAFGKKANGLIFTNFLNAETNHYFVPTWFIFLVNLF 286
Query: 337 IVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFV 396
V+ L VY QP+ +E+ ++ +F + V IP RLV R++FV
Sbjct: 287 TVLQLSAVAVVYLQPINDILESVISDP-TKKEFSIRNV-IP----------RLVVRSLFV 334
Query: 397 IITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLFSLAC 456
++ T+++ +LPFF D+ LLGA GF PL PV + + K S + + ++
Sbjct: 335 VMATIVAAMLPFFGDVNSLLGAFGFIPLDFVLPVVFFNFTFKPSKKSFIFWINTVIAVVF 394
Query: 457 LIVTVAAAAGSVAGIFLDLQVYKPF 481
+ V A +V I +D YK F
Sbjct: 395 SCLGVIAMVAAVRQIIIDANTYKLF 419
>At5g41800 amino acid permease-like protein; proline
transporter-like protein
Length = 452
Score = 106 bits (264), Expect = 3e-23
Identities = 107/451 (23%), Positives = 198/451 (43%), Gaps = 33/451 (7%)
Query: 39 LKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACY 98
L+ G W A H+ TA++G +L+L +A LGW G + LVT+Y L+
Sbjct: 25 LQSKGEWWHAGFHLTTAIVGPTILTLPYAFRGLGWWLGFVCLTTMGLVTFYAYYLMSKVL 84
Query: 99 RNGDPVNGKRNYTYMDVVHSNMG-GIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERS 157
+ + +G+R+ + ++ +G G+ + +Q G+ IG + + + I S
Sbjct: 85 DHCEK-SGRRHIRFRELAADVLGSGLMFYVVIFIQTAINTGIGIGAILLAG-QCLDIMYS 142
Query: 158 NCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGL 217
+ F + K + I M++ +V +VLSQ+P F L ++ + ++S Y+ + +G
Sbjct: 143 SLFPQGTLK----LYEFIAMVT--VVMMVLSQLPSFHSLRHINCASLLLSLGYTFLVVGA 196
Query: 218 GFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPP 277
+ A + + + KV+ ++ IA + + IL EIQ T+ +PP
Sbjct: 197 CINLGLSKNAPKREYS----LEHSDSGKVFSAFTSISIIAAIFG-NGILPEIQATL-APP 250
Query: 278 SESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGF-----GFYNPFWLIDI 332
+ K +K V+ FY GY FG++S N+L P +I +
Sbjct: 251 ATGKMLKGLLLCYSVIFFTFYS-AAISGYWVFGNNSSSNILKNLMPDEGPTLAPIVVIGL 309
Query: 333 ANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWR 392
A +++ L VYSQ + +E +A+ +K +P RL+ R
Sbjct: 310 AVIFVLLQLFAIGLVYSQVAYEIMEKKSAD--TTKGIFSKRNLVP----------RLILR 357
Query: 393 TIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIKWICFQLF 452
T+++ ++ +LPFF DI ++GA GF PL P+ +Y + + + S +
Sbjct: 358 TLYMAFCGFMAAMLPFFGDINAVVGAFGFIPLDFVLPMLLYNMTYKPTRRSFTYWINMTI 417
Query: 453 SLACLIVTVAAAAGSVAGIFLDLQVYKPFKA 483
+ + A S+ + LD +K F +
Sbjct: 418 MVVFTCAGLMGAFSSIRKLVLDANKFKLFSS 448
>At3g55740 proline transporter 2
Length = 439
Score = 105 bits (263), Expect = 4e-23
Identities = 109/427 (25%), Positives = 186/427 (43%), Gaps = 47/427 (11%)
Query: 46 WTASAHIITAVIGSG-VLSLAWAI-AQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDP 103
W A ++T I S VL + + LGW+ G +IL + ++ Y + L+ + G
Sbjct: 33 WFQVAFVLTTGINSAYVLGYSGTVMVPLGWIGGVVGLILATAISLYANTLIAKLHEFG-- 90
Query: 104 VNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKS 163
GKR+ Y D+ G ++ +QY+NLF + G+ I + ++ A+
Sbjct: 91 --GKRHIRYRDLAGFIYGKKMYRVTWGLQYVNLFMINCGFIILAGSALKAVYVLF----- 143
Query: 164 EGKDPCHMNGNIYMISFGLVEIVLS-QIPDFDQLW-WLSILAAVMSFTYSTIGLGLGFGK 221
+D M ++ G+V + + IP L WL + + ++S Y + + L
Sbjct: 144 --RDDSLMKLPHFIAIAGVVCAIFAIGIPHLSALGIWLGV-STILSIIYIIVAIVLSAKD 200
Query: 222 VIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESK 281
+ ++ G ++ K++ A ++ FA++ M L EIQ TVK P K
Sbjct: 201 GVNKPERDYNIQGSSIN------KLFTITGAAANLVFAFNTGM-LPEIQATVKQPVV--K 251
Query: 282 TMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHL 341
M KA + V L GY A+G S+ LL P W+ +AN + +
Sbjct: 252 NMMKALYFQFTVGVLPMYAVTFIGYWAYGSSTSTYLLNSVS--GPVWVKALANISAFLQS 309
Query: 342 VGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTL 401
V + +++ P + ++ D+ + K + + N LFR V R ++ ++TL
Sbjct: 310 VISLHIFASPTYEYM---------DTKYGVKGSPLAMKNL----LFRTVARGSYIAVSTL 356
Query: 402 ISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKR-----IPK-WSIKWIC-FQLFSL 454
+S LLPF D + L GA+ +PLT MY++ + K W +C F L SL
Sbjct: 357 LSALLPFLGDFMSLTGAISTFPLTFILANHMYLVAMNDELSLVQKLWHWLNVCFFGLMSL 416
Query: 455 ACLIVTV 461
A I V
Sbjct: 417 AAAIAAV 423
>At2g39890 proline transporter 1
Length = 442
Score = 102 bits (254), Expect = 5e-22
Identities = 109/444 (24%), Positives = 194/444 (43%), Gaps = 48/444 (10%)
Query: 46 WTASAHIITAVIGSG-VLSLAWAI-AQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDP 103
W A ++T I S VL + I LGW+ G +++ + ++ Y + L+ + G
Sbjct: 36 WFQVAFVLTTGINSAYVLGYSGTIMVPLGWIGGVVGLLIATAISLYANTLIAKLHEFG-- 93
Query: 104 VNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKS 163
G+R+ Y D+ G L +QY+NLF + G+ I + ++ A+
Sbjct: 94 --GRRHIRYRDLAGFIYGRKAYHLTWGLQYVNLFMINCGFIILAGSALKAVYVLF----- 146
Query: 164 EGKDPCHMNGNIYMISFGLVEIVLS-QIPDFDQLW-WLSILAAVMSFTYSTIGLGLGFGK 221
+D M ++ GL+ + + IP L WL + + +S Y + + L
Sbjct: 147 --RDDHTMKLPHFIAIAGLICAIFAIGIPHLSALGVWLGV-STFLSLIYIVVAIVLSVRD 203
Query: 222 VIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESK 281
++ + + G ++ K++ A ++ FA++ M L EIQ TV+ P K
Sbjct: 204 GVKTPSRDYEIQGSSLS------KLFTITGAAANLVFAFNTGM-LPEIQATVRQPVV--K 254
Query: 282 TMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHL 341
M KA + L GY A+G S+ LL P W+ +AN + ++
Sbjct: 255 NMMKALYFQFTAGVLPMYAVTFIGYWAYGSSTSTYLLNSVN--GPLWVKALANVSAILQS 312
Query: 342 VGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTL 401
V + +++ P + +++ K + F K + LFR++ R ++ ++TL
Sbjct: 313 VISLHIFASPTYEYMDTKYGIK--GNPFAIKNL-----------LFRIMARGGYIAVSTL 359
Query: 402 ISMLLPFFNDIVGLLGALGFWPLTVYFPVEMYIIQKRIPKWSIK----WICFQLFSLACL 457
IS LLPF D + L GA+ +PLT MY K +++ W+ FSL
Sbjct: 360 ISALLPFLGDFMSLTGAVSTFPLTFILANHMYYKAKNNKLNAMQKLWHWLNVVFFSL--- 416
Query: 458 IVTVAAAAGSVAGIFLDLQVYKPF 481
++VAAA +V I +D + + F
Sbjct: 417 -MSVAAAIAAVRLIAVDSKNFHVF 439
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,726,549
Number of Sequences: 26719
Number of extensions: 449165
Number of successful extensions: 1587
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 1432
Number of HSP's gapped (non-prelim): 79
length of query: 485
length of database: 11,318,596
effective HSP length: 103
effective length of query: 382
effective length of database: 8,566,539
effective search space: 3272417898
effective search space used: 3272417898
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0181.18


