

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.7
(199 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
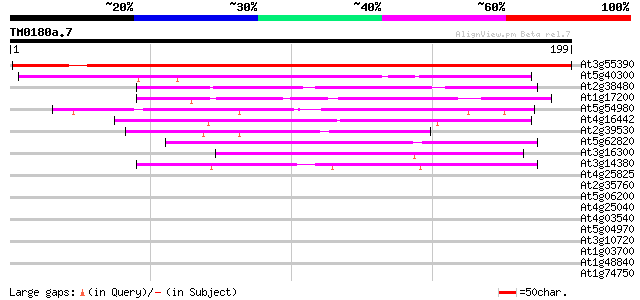
Score E
Sequences producing significant alignments: (bits) Value
At3g55390 unknown protein 256 7e-69
At5g40300 unknown protein 73 9e-14
At2g38480 unknown protein 57 8e-09
At1g17200 unknown protein 53 1e-07
At5g54980 unknown protein (At5g54980) 52 2e-07
At4g16442 unknown protein 51 5e-07
At2g39530 unknown protein 48 3e-06
At5g62820 unknown protein 48 4e-06
At3g16300 hypothetical protein 47 7e-06
At3g14380 unknown protein 43 1e-04
At4g25825 unknown protein 39 0.002
At2g35760 unknown protein 39 0.002
At5g06200 putative protein 37 0.005
At4g25040 unknown protein 34 0.058
At4g03540 hypothetical protein 33 0.100
At5g04970 pectinesterase 31 0.49
At3g10720 pectinesterase like protein 30 0.65
At1g03700 putative protein 28 2.5
At1g48840 unknown protein 28 3.2
At1g74750 28 4.2
>At3g55390 unknown protein
Length = 194
Score = 256 bits (653), Expect = 7e-69
Identities = 123/198 (62%), Positives = 149/198 (75%), Gaps = 6/198 (3%)
Query: 2 MRSPQPLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVF 61
MRSP RNG ++ H FHST+ KLRRFNSLIL+ RL SFSFSLAS+VF
Sbjct: 1 MRSPHAFRNGESPTLRDHTH------FHSTVTAQKLRRFNSLILLLRLASFSFSLASAVF 54
Query: 62 MLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFD 121
MLTN+ GS SPHWY +D FRFV ANAIVA+YS+FEM VWE SR T++PE QVWFD
Sbjct: 55 MLTNSRGSASPHWYDFDAFRFVFVANAIVALYSVFEMGTCVWEFSRETTLWPEAFQVWFD 114
Query: 122 FGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSL 181
FGHDQVF+YLLL+A +A A+ RT++ DTCTA+ AFC+QSD+A+ LG+A F+FL F+S
Sbjct: 115 FGHDQVFSYLLLSAGSAAAALARTMRGGDTCTANKAFCLQSDVAIGLGFAAFLFLAFSSC 174
Query: 182 LTGFRVVCFIINGSRFHL 199
+GFRV CF+I GSRFHL
Sbjct: 175 FSGFRVACFLITGSRFHL 192
>At5g40300 unknown protein
Length = 270
Score = 73.2 bits (178), Expect = 9e-14
Identities = 61/187 (32%), Positives = 95/187 (50%), Gaps = 8/187 (4%)
Query: 4 SPQPLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLI----LVFRLISFSFSLAS- 58
+P + NG + + + R P F S+ + + ++ SLI L FR+I+F L S
Sbjct: 79 TPFRVTNGEEEKKVSESRRQLRPSFSSSSSTPRESKWASLIRKALLGFRVIAFVSCLVSF 138
Query: 59 SVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQV 118
SV + G +Y+Y FRF LAAN I VYS F + V+ +S L+
Sbjct: 139 SVMVSDRDKGWAHDSFYNYKEFRFCLAANVIGFVYSGFMICDLVYLLSTSIRRSRHNLRH 198
Query: 119 WFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGF 178
+ +FG DQ+ AYLL ASA+ +A +R + + + ++ F + +VAL Y FV F
Sbjct: 199 FLEFGLDQMLAYLL--ASASTSASIR-VDDWQSNWGADKFPDLARASVALSYVSFVAFAF 255
Query: 179 TSLLTGF 185
SL +G+
Sbjct: 256 CSLASGY 262
>At2g38480 unknown protein
Length = 188
Score = 56.6 bits (135), Expect = 8e-09
Identities = 35/142 (24%), Positives = 72/142 (50%), Gaps = 9/142 (6%)
Query: 46 VFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEI 105
+ R I FSL + + M++N HG ++ Y+ +R+VLA + I +Y+ ++ A +
Sbjct: 50 ITRGICLLFSLIAFLIMVSNKHGYGR-NFNDYEEYRYVLAISIISTLYTAWQTFAHFSK- 107
Query: 106 SRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIA 165
+F + DF DQ+ AYLL++A+++ + +E N F + A
Sbjct: 108 ---REIFDRRTSILVDFSGDQIVAYLLISAASSAIPLTNIFREGQ----DNIFTDSAASA 160
Query: 166 VALGYAGFVFLGFTSLLTGFRV 187
+++ F+ L ++L +G+++
Sbjct: 161 ISMAIFAFIALALSALFSGYKL 182
>At1g17200 unknown protein
Length = 204
Score = 52.8 bits (125), Expect = 1e-07
Identities = 46/151 (30%), Positives = 68/151 (44%), Gaps = 19/151 (12%)
Query: 46 VFRLISFSFSLASSVFML----TNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVAS 101
+ RL +A+ V ML TN GS S + + FR+++ AN I A YSL A+
Sbjct: 36 MLRLAPVGLCVAALVVMLKDSETNEFGSIS--YSNLTAFRYLVHANGICAGYSLLS--AA 91
Query: 102 VWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQ 161
+ + R ++ P +VW F DQ+ YL+LAA A ++ D+ + C
Sbjct: 92 IAAMPRSSSTMP---RVWTFFCLDQLLTYLVLAAGAVSAEVLYLAYNGDSAITWSDAC-- 146
Query: 162 SDIAVALGYAGFVFLGFTSLLTGFRVVCFII 192
Y GF S++ F VVCF I
Sbjct: 147 ------SSYGGFCHRATASVIITFFVVCFYI 171
>At5g54980 unknown protein (At5g54980)
Length = 194
Score = 52.4 bits (124), Expect = 2e-07
Identities = 52/186 (27%), Positives = 84/186 (44%), Gaps = 26/186 (13%)
Query: 16 DNNNNH-----RSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVFMLTNTHGSD 70
DNNNN+ RSSS + A L+ +S + RL S+A+ +TN +
Sbjct: 3 DNNNNNTREEERSSSSKQQQPQAPMSLKIIDSCL---RLSVVPLSVATIWLTVTNHESNP 59
Query: 71 SPHWYHYDTF---RFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQV 127
Y++ ++++ +AI A+Y+L V+S W V V + W F DQV
Sbjct: 60 DYGNLEYNSIMGLKYMVGVSAISAIYALLSTVSS-W-------VTCLVSKAWLFFIPDQV 111
Query: 128 FAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQ-----SDIAVALGYAGFV--FLGFTS 180
AY++ + A T +V L + D + C S + +ALG FV F F S
Sbjct: 112 LAYVMTTSVAGATEIVYLLNKGDKIVTWSEMCSSYPHYCSKLTIALGLHVFVLFFFLFLS 171
Query: 181 LLTGFR 186
+++ +R
Sbjct: 172 VISAYR 177
>At4g16442 unknown protein
Length = 182
Score = 50.8 bits (120), Expect = 5e-07
Identities = 44/161 (27%), Positives = 67/161 (41%), Gaps = 14/161 (8%)
Query: 38 RRFNSLILVFRLISFSFSLASSVFMLTNTHGS------DSPHWYHYDTFRFVLAANAIVA 91
RR L+ R F+L + + ++T+T + + F++ AN I A
Sbjct: 6 RRMRLTELLLRCSISVFALLALILVVTDTEVKLIFTIKKTAKYTDMKAVVFLVVANGIAA 65
Query: 92 VYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETD- 150
VYSL + V V +G +F + L W F DQ AYL +AA AA +E +
Sbjct: 66 VYSLLQSVRCVVGTMKGKVLFSKPL-AWAFFSGDQAMAYLNVAAIAATAESGVIAREGEE 124
Query: 151 ------TCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGF 185
CT FC Q I V+ + + F S ++ F
Sbjct: 125 DLQWMRVCTMYGKFCNQMAIGVSSALLASIAMVFVSCISAF 165
>At2g39530 unknown protein
Length = 178
Score = 48.1 bits (113), Expect = 3e-06
Identities = 35/114 (30%), Positives = 62/114 (53%), Gaps = 9/114 (7%)
Query: 42 SLILVFRLISFSFSLASSVFMLTNTH----GSDSPHWYHYDTF--RFVLAANAIVAVYSL 95
+++L+ R+++ +F L + V + TNT S S D + R++L+A I VY++
Sbjct: 15 TVLLLLRVLTAAFLLITVVLISTNTVTLEISSTSIKLPFNDVYAYRYMLSAAVIGLVYAV 74
Query: 96 FEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKET 149
++ ++ + + G T L FDF D+V +YLL SAAG + + LK+T
Sbjct: 75 VQLFLTISQFATGKT---HPLTYQFDFYGDKVISYLLATGSAAGFGVSKDLKDT 125
>At5g62820 unknown protein
Length = 297
Score = 47.8 bits (112), Expect = 4e-06
Identities = 31/132 (23%), Positives = 60/132 (44%), Gaps = 3/132 (2%)
Query: 56 LASSVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEV 115
++ S+ T G + Y +R+ LA N I VYS FE + I++ + +
Sbjct: 163 ISFSIMAADKTQGWSGDSYDRYKEYRYCLAVNVIAFVYSAFEACDAACYIAKESYMINCG 222
Query: 116 LQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVF 175
F F DQ+ AYLL++AS+ V + + + F + ++A+ + F
Sbjct: 223 FHDLFVFSMDQLLAYLLMSASSCAATRV---DDWVSNWGKDEFTQMATASIAVSFLAFGA 279
Query: 176 LGFTSLLTGFRV 187
++L++ +R+
Sbjct: 280 FAVSALISSYRL 291
>At3g16300 hypothetical protein
Length = 212
Score = 47.0 bits (110), Expect = 7e-06
Identities = 26/116 (22%), Positives = 52/116 (44%), Gaps = 7/116 (6%)
Query: 74 WYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLL 133
W D+ +++ ++ +YSL +++ S + R + V P Q WF F DQ+ Y ++
Sbjct: 88 WSLSDSLIYLVVVSSATVLYSLIQLIISGTRLMRKSPVIPTRTQAWFCFVADQIIGYAMV 147
Query: 134 AASAAGTAM-------VRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLL 182
+ +A + +R + + C + FC ++ F+ L +SLL
Sbjct: 148 SGGSAALGVTNMNRTGIRHMPLPNFCKSLGFFCDHLAGSIVFALFAFLLLAASSLL 203
>At3g14380 unknown protein
Length = 178
Score = 42.7 bits (99), Expect = 1e-04
Identities = 38/152 (25%), Positives = 69/152 (45%), Gaps = 16/152 (10%)
Query: 46 VFRLISFSFSLASSVFMLTNTHGSD--SPHWYHYDTFRFVLAANAIVAVYSLFEMVASVW 103
V R+ S + S+ V M+ N+ ++ S + + F ++++AN + A YSL +A +
Sbjct: 26 VLRVASMALSITGLVIMIKNSISNEFGSVSYSNIGAFMYLVSANGVCAAYSLLSALAIL- 84
Query: 104 EISRGATVFP-EVLQVWFDFGHDQVFAYLLLAASAAGTAMVR-------TLKETDTCTAS 155
A P +QV F DQV Y++LAA A V + + C +
Sbjct: 85 -----ALPCPISKVQVRTLFLLDQVVTYVVLAAGAVSAETVYLAYYGNIPITWSSACDSY 139
Query: 156 NAFCVQSDIAVALGYAGFVFLGFTSLLTGFRV 187
+FC + I+V + + SL++ +R+
Sbjct: 140 GSFCHNALISVVFTFVVSLLYMLLSLISSYRL 171
>At4g25825 unknown protein
Length = 175
Score = 38.9 bits (89), Expect = 0.002
Identities = 35/156 (22%), Positives = 66/156 (41%), Gaps = 18/156 (11%)
Query: 45 LVFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLAANA------IVAVYSLFEM 98
++ RL F L +S + ++ + + + +FR++LA A +VA Y+L ++
Sbjct: 9 VILRLCIVFFLLLTSCLIGLDSQTKEIAYIHKNVSFRYLLALEAELYIDVVVAAYNLVQL 68
Query: 99 VASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAG-------TAMVRTLKETDT 151
+ + + T P+ WF + DQ AY++ A ++A R L+
Sbjct: 69 GLGWYNVEQ-KTSNPK----WFSYLLDQTAAYVVFAGTSAAAQHSLLVVTGSRELQWMKW 123
Query: 152 CTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRV 187
C FC Q A+ L Y + S ++ F +
Sbjct: 124 CYKFTRFCFQMGSAIILNYIAAALMVLLSSISAFNL 159
>At2g35760 unknown protein
Length = 201
Score = 38.5 bits (88), Expect = 0.002
Identities = 33/112 (29%), Positives = 49/112 (43%), Gaps = 8/112 (7%)
Query: 83 VLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLA--ASAAGT 140
++ N I A YSL + V V + +G +F + L W F DQ AYL +A A+AA +
Sbjct: 76 LVVVNGIAAGYSLVQAVRCVVGLMKGRVLFSKPL-AWAIFFGDQAVAYLCVAGVAAAAQS 134
Query: 141 AMVRTLKETD-----TCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRV 187
A L E + C FC Q +A + + S ++ F V
Sbjct: 135 AAFAKLGEPELQWMKICNMYGKFCNQVGEGIASALFACIGMVLISCISAFGV 186
>At5g06200 putative protein
Length = 202
Score = 37.4 bits (85), Expect = 0.005
Identities = 33/113 (29%), Positives = 49/113 (43%), Gaps = 11/113 (9%)
Query: 79 TFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAA 138
TF+F + A AIVA Y + + SV I R V P +L + D L AA++A
Sbjct: 88 TFQFFVVAIAIVAGYLVLSLPFSVVTIVRPLAVAPRLLLLVL----DTAALALDTAAASA 143
Query: 139 GTAMVRTLKETDT-------CTASNAFCVQSDIAVALGYAGFVFLGFTSLLTG 184
A+V +T C FC ++ AV +A FL +++G
Sbjct: 144 AAAIVYLAHNGNTNTNWLPICQQFGDFCQKTSGAVVSAFASVTFLAILVVISG 196
>At4g25040 unknown protein
Length = 170
Score = 33.9 bits (76), Expect = 0.058
Identities = 35/145 (24%), Positives = 62/145 (42%), Gaps = 12/145 (8%)
Query: 48 RLISFSFSLASSVFMLTN-----THGSDSPHWYHYDT-FRFVLAANAIVAVYSLFEMVAS 101
R+++ ++AS M+TN +G Y Y + FR+++ A V +LF +V +
Sbjct: 22 RVLTIGAAMASMWVMITNREVASVYGIAFEAKYSYSSAFRYLVYAQIAVCAATLFTLVWA 81
Query: 102 VWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKET---DTCTASNAF 158
+ R VF +FD AA A G K+ C + +
Sbjct: 82 CLAVRRRGLVF---ALFFFDLLTTLTAISAFSAAFAEGYVGKYGNKQAGWLPICGYVHGY 138
Query: 159 CVQSDIAVALGYAGFVFLGFTSLLT 183
C + I++A+ +A F+ L ++LT
Sbjct: 139 CSRVTISLAMSFASFILLFILTVLT 163
>At4g03540 hypothetical protein
Length = 164
Score = 33.1 bits (74), Expect = 0.100
Identities = 34/144 (23%), Positives = 59/144 (40%), Gaps = 26/144 (18%)
Query: 45 LVFRLISFSFSLASSVFMLTNTHGSD------SPHWYHYDTFRFVLAANAIVAVYSLFEM 98
LV RL +F +LA+ + M+T+ + + F++ + ANA+V+VYS +
Sbjct: 11 LVLRLAAFGAALAALIVMITSRERASFLAISLEAKYTDMAAFKYFVIANAVVSVYSFLVL 70
Query: 99 VASVWEISRGATVFPEVLQVW-FDFGHDQVFAYLLLAASAAGTAMVRTLKETDT------ 151
P+ +W F D V LL ++ +A A+ + K+ +
Sbjct: 71 ------------FLPKESLLWKFVVVLDLVMTMLLTSSLSAALAVAQVGKKGNANAGWLP 118
Query: 152 -CTASNAFCVQSDIAVALGYAGFV 174
C FC Q A+ G+ V
Sbjct: 119 ICGQVPKFCDQITGALIAGFVALV 142
>At5g04970 pectinesterase
Length = 624
Score = 30.8 bits (68), Expect = 0.49
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Query: 8 LRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVFMLTNTH 67
+RN ADG + +R S + T+ H LR+F ++ I F F A+++F N +
Sbjct: 419 VRNNADG---STFYRCSFEGYQDTLYVHSLRQFYRECDIYGTIDFIFGNAAAIFQNCNIY 475
>At3g10720 pectinesterase like protein
Length = 619
Score = 30.4 bits (67), Expect = 0.65
Identities = 17/60 (28%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query: 8 LRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVFMLTNTH 67
LRN A+G ++ +R S + T+ H LR+F ++ + F F A+++F N +
Sbjct: 414 LRNNAEG---SSFYRCSFEGYQDTLYVHSLRQFYRECDIYGTVDFIFGNAAAIFQNCNIY 470
>At1g03700 putative protein
Length = 164
Score = 28.5 bits (62), Expect = 2.5
Identities = 36/158 (22%), Positives = 59/158 (36%), Gaps = 26/158 (16%)
Query: 45 LVFRLISFSFSLASSVFMLTNTHGSDS------PHWYHYDTFRFVLAANAIVAVYSLFEM 98
LV R +F +L + + M+T+ S + F++ + ANAIV VYS +
Sbjct: 11 LVLRFAAFCAALGAVIAMITSRERSSFFVISLVAKYSDLAAFKYFVIANAIVTVYSFLVL 70
Query: 99 VASVWEISRGATVFPEVLQVW-FDFGHDQVFAYLLLAASAAGTAMVRTLKETDT------ 151
P+ +W F D + LL ++ +A A+ + K +
Sbjct: 71 ------------FLPKESLLWKFVVVLDLMVTMLLTSSLSAAVAVAQVGKRGNANAGWLP 118
Query: 152 -CTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVV 188
C FC Q A+ G V F + + VV
Sbjct: 119 ICGQVPRFCDQITGALIAGLVALVLYVFLLIFSIHHVV 156
>At1g48840 unknown protein
Length = 691
Score = 28.1 bits (61), Expect = 3.2
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 1/62 (1%)
Query: 127 VFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFR 186
V LLL A+ G ++ + D S FC+ S + G+ G+VF T + GF
Sbjct: 493 VMKELLLPATEIGNWLLSLVYWEDPLK-SFVFCLLSTFIIYRGWIGYVFAIATLFIAGFM 551
Query: 187 VV 188
V+
Sbjct: 552 VL 553
>At1g74750
Length = 855
Score = 27.7 bits (60), Expect = 4.2
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 9/51 (17%)
Query: 95 LFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLL--LAASAAGTAMV 143
L E SVWE++ G V+P+ L+ ++ ++Y L L + GTA++
Sbjct: 724 LKEEAGSVWEVAAGKNVYPDALR-------EKSYSYWLINLHVMSEGTAVI 767
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,208,562
Number of Sequences: 26719
Number of extensions: 150733
Number of successful extensions: 584
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 560
Number of HSP's gapped (non-prelim): 24
length of query: 199
length of database: 11,318,596
effective HSP length: 94
effective length of query: 105
effective length of database: 8,807,010
effective search space: 924736050
effective search space used: 924736050
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0180a.7


