

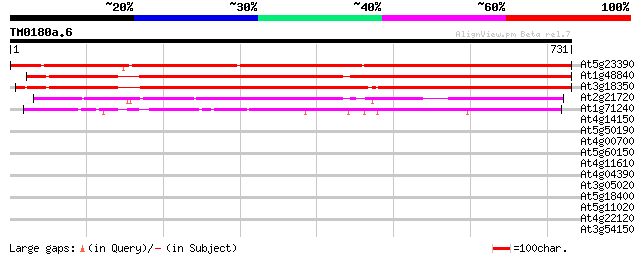
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.6
(731 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g23390 unknown protein 847 0.0
At1g48840 unknown protein 548 e-156
At3g18350 unknown protein 543 e-154
At2g21720 hypothetical protein 316 3e-86
At1g71240 hypothetical protein 258 1e-68
At4g14150 kinesin like protein 33 0.57
At5g50190 putative protein 32 0.97
At4g00700 putative phosphoribosylanthranilate transferase 32 0.97
At5g60150 putative protein 32 1.3
At4g11610 phosphoribosylanthranilate transferase like protein 31 2.2
At4g04390 hypothetical protein 31 2.2
At3g05020 acyl carrier protein 1 precursor (ACP) 31 2.8
At5g18400 unknown protein 30 6.3
At5g11020 ser/thr specific protein kinase-like protein 30 6.3
At4g22120 unknown protein 30 6.3
At3g54150 embryonic abundant protein -like 29 8.2
>At5g23390 unknown protein
Length = 730
Score = 847 bits (2189), Expect = 0.0
Identities = 443/735 (60%), Positives = 554/735 (75%), Gaps = 13/735 (1%)
Query: 2 EENGSENKSGGMWENFLRNHQNSLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSPL 61
E+ G GM ENF++ HQ+SLKSLF R SSS D A S + SPKPIP LS L
Sbjct: 3 EDGGGGGAKVGMLENFMKTHQSSLKSLFQRKKSSSGRDGD--ASPSPIASPKPIPQLSLL 60
Query: 62 ANSVVVRSSKILGISTEELQHAFDSELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYL 121
ANSVV R SKIL I TE+LQH FD ELP VK+LLTYARN LEFCS++ALH++ K DYL
Sbjct: 61 ANSVVSRCSKILNIQTEDLQHHFDVELPESVKQLLTYARNFLEFCSFQALHQVMKKPDYL 120
Query: 122 ADKDFRRLTYDMMLAWETPSVHTDE----TPPPSSKDEHGGDDDEASLFYSSSTNMAVQV 177
+D++FR+L +DMMLAWETPSV +++ PS +D D+D SLFYSS TNMA+QV
Sbjct: 121 SDQEFRQLMFDMMLAWETPSVTSEQENKDAASPSKQDSE--DEDGWSLFYSSPTNMAMQV 178
Query: 178 DDQKTVGLEAFSRIAPVCAPIADVITVHNLFDALTSSSGRRLHFLVYDKYIRFLDKVIKN 237
D++K+VG EAF+RIAPVC IAD ITVHNLFDALTSSSG RLH++VYDKY+R LDK+ K
Sbjct: 179 DEKKSVGQEAFARIAPVCPAIADAITVHNLFDALTSSSGHRLHYIVYDKYLRTLDKIFKA 238
Query: 238 SKNVLASSVGSLQLAEEEIVLDVDGTIPTQPVLQHIGISAWPGRLTLTNYALYFESLGVG 297
+K+ L S +LQLA+ EIVLD+DG P PVL+H+GISAWPG+LTLTN ALYF+S+G G
Sbjct: 239 AKSTLGPSAANLQLAKGEIVLDMDGANPVLPVLKHVGISAWPGKLTLTNCALYFDSMGGG 298
Query: 298 VHEKPVRYDLGTDLKQVIKPDLTGPLGARLFDKAVMYKSTSVAEPVYFEFPEFKANVRRD 357
EKP+RYDL D KQVIKP+LTGPLGAR+FDKA+MYKS +V EPV+FEF EFK N RRD
Sbjct: 299 --EKPMRYDLTEDTKQVIKPELTGPLGARIFDKAIMYKSITVPEPVFFEFTEFKGNARRD 356
Query: 358 YWLDISLEILRVHKFIRKYYLKETQKAEVLARAILGIYRYRAVREAFKYFSSHYKTLLSF 417
YWL I LEILRV FIR+Y K Q++E+LARAILGI+RYRA+REAF+ FSS YKTLL F
Sbjct: 357 YWLGICLEILRVQWFIRRYNFKGIQRSEILARAILGIFRYRAIREAFQVFSSQYKTLLIF 416
Query: 418 NLAEALPRGDMILETLSNSLTNLTV-ASSKRSSPRNVDTKKQLVVSPASAVALFRLGFKS 476
NLAE+LP GDM+LE LS+ ++ +T +S S + + L SP S L G
Sbjct: 417 NLAESLPGGDMVLEALSSRVSRITTNVASDIGSVQYMKWPSNL--SPVSLKLLEHFGLNL 474
Query: 477 DKFADIYEETTVVGDIRVGEINLLEMAVKKSLRDTGKAEAAQATVDQVKVEGIDTNVAVM 536
+ ++ EE T+VGD VGE + LE+A+K+S+ DT +AEAAQATV+QVKVEGIDTNVAVM
Sbjct: 475 ETGTNMGEELTIVGDFCVGETSPLEIALKQSILDTDRAEAAQATVEQVKVEGIDTNVAVM 534
Query: 537 KELLFPVIESANRLERLASWKDFYKSTAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWR 596
KELL P I+ + RLA W+D YKST F++L SYMI+ GWI ++LPSI ++VAI+M+WR
Sbjct: 535 KELLLPFIKLGLHINRLAYWQDPYKSTVFMILVSYMIISGWIGFILPSILLLVAIVMMWR 594
Query: 597 RHFRKGRSLEAFIVTPPPNRNPVEQLLTLQEAITQFESLIQAGNIVLLKVRALLLALLPQ 656
+ F KG+ + V PP++N VEQLL LQ+AI+QFESLIQA N+ LLK+RA+ LA+LPQ
Sbjct: 595 KQFNKGKEPKTVRVKAPPSKNAVEQLLVLQDAISQFESLIQAVNVGLLKIRAITLAILPQ 654
Query: 657 ATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMREWWIRIPA 716
AT+ A+ LV +A + A VP KY+I V F E++TRE+ RK SSDR RR+REWW R+PA
Sbjct: 655 ATDTTAISLVVVAVILAVVPVKYLITVAFVEWFTREVGWRKASSDRLERRIREWWFRVPA 714
Query: 717 APVELIKPDECKKRK 731
APV+LI+ ++ KK+K
Sbjct: 715 APVQLIRAEDSKKKK 729
>At1g48840 unknown protein
Length = 691
Score = 548 bits (1413), Expect = e-156
Identities = 324/713 (45%), Positives = 437/713 (60%), Gaps = 41/713 (5%)
Query: 23 NSLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSPLANSVVVRSSKILGISTEELQH 82
+SLK L + SS + + SA IP LSP+AN V+ R SKILG++ ELQ
Sbjct: 16 SSLKWLLGKQSSFDEEIEEIENSPSA--GANWIPELSPVANVVIRRCSKILGVAVSELQD 73
Query: 83 AFDSELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYLADKDFRRLTYDMMLAWETPSV 142
+F E VK+ + RN LE+C ++AL + +L+DK FRRLT+DMM+AWE PS
Sbjct: 74 SFKQEASESVKQPSMFPRNFLEYCCFRALALSVGVTGHLSDKSFRRLTFDMMVAWEVPS- 132
Query: 143 HTDETPPPSSKDEHGGDDDEASLFYSSSTNMAVQVDDQKTVGLEAFSRIAPVCAPIADVI 202
+++ + VD+ TVGLEAFSRIAP IADVI
Sbjct: 133 --------------------------AASQTLLSVDEDPTVGLEAFSRIAPAVPIIADVI 166
Query: 203 TVHNLFDALTS-SSGRRLHFLVYDKYIRFLDKVIKNSKNVLASSVGSLQLAEEEIVLDVD 261
NLF LTS S+ RL F VYDKY+ L++ IK K+ SS+ S ++ E +L++D
Sbjct: 167 ICENLFGILTSVSNSVRLQFYVYDKYLYGLERAIKKMKSQSESSLLSGVRSKGEKILELD 226
Query: 262 GTIPTQPVLQHIGISAWPGRLTLTNYALYFESLGVGVHEKPVRYDLGTDLKQVIKPDLTG 321
GT+ TQPVL+HIGIS WPGRL LT+++LYFE++ V + P RY L DLKQVIKP+LTG
Sbjct: 227 GTVTTQPVLEHIGISTWPGRLILTDHSLYFEAIKVVSFDTPKRYSLSDDLKQVIKPELTG 286
Query: 322 PLGARLFDKAVMYKSTSVAEPVYFEFPEFKANVRRDYWLDISLEILRVHKFIRKYYLKET 381
P G RLFDKAV YKS S+ EPV EFPE K + RRDYWL I LE+L VH++I+K+ +
Sbjct: 287 PWGTRLFDKAVSYKSISLPEPVVMEFPELKGHTRRDYWLAIILEVLYVHRYIKKFKINSV 346
Query: 382 QKAEVLARAILGIYRYRAVREAFKYFSSHYKTLLSFNLAEALPRGDMILETLSNSLTNLT 441
K E +++A+LGI R +A++E Y+ LL FNL + LP GD ILETL+
Sbjct: 347 AKDEAISKAVLGILRVQAIQEVGLTNPVRYENLLPFNLCDQLPGGDRILETLAE------ 400
Query: 442 VASSKRSSPRNVDTKKQLVVSPASAVALFRLGFKSDKFADIYEETTVVGDIRVGEINLLE 501
SS R R K+ + S +++ + +LG + + VVG++ VG++N LE
Sbjct: 401 -MSSSRVLDRTAKAKEGTLHSISASDMVSQLGLVFGATSPKSRSSLVVGEVMVGDVNPLE 459
Query: 502 MAVKKSLRDTGKAEAAQATVDQVKVEGIDTNVAVMKELLFPVIESANRLERLASWKDFYK 561
AVK+S ++ K AQ TV+ VKV+GIDTNVAVMKELL P E N L L W+D K
Sbjct: 460 KAVKQSRKNYEKVVLAQETVNGVKVDGIDTNVAVMKELLLPATEIGNWLLSLVYWEDPLK 519
Query: 562 STAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWRRHF--RKGRSLEAFIVTPPPNRNPV 619
S F LL++++I RGWI Y+ + +A M+ R+F R+ +E ++ PPP N +
Sbjct: 520 SFVFCLLSTFIIYRGWIGYVFAIATLFIAGFMVLTRYFSNREKVMIELKVMAPPP-MNTM 578
Query: 620 EQLLTLQEAITQFESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIAAVFAFVPPKY 679
EQLL +Q AI+Q E LIQ NIVLLK RALLL+L PQA+EK A+ +V A + A VP
Sbjct: 579 EQLLAVQNAISQLEQLIQDANIVLLKFRALLLSLFPQASEKFAVAIVIAATMMALVPWNN 638
Query: 680 IIFVIFFEYYTREMPCRKESSDRWVRRMREWWIRIPAAPVELIK-PDECKKRK 731
+I V+F E +TR P R+ S++R +RR++EWW IPAAPV L + D+ KK K
Sbjct: 639 LILVVFLELFTRYSPPRRASTERLMRRLKEWWFSIPAAPVLLEQSKDDNKKTK 691
>At3g18350 unknown protein
Length = 692
Score = 543 bits (1399), Expect = e-154
Identities = 312/725 (43%), Positives = 441/725 (60%), Gaps = 37/725 (5%)
Query: 8 NKSGGMWENFLRNHQNSLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSPLANSVVV 67
+K+ M E +R+ S K L + SS + + SA IP LSP+AN VV
Sbjct: 3 SKTRNMLEGLVRD--TSFKWLLGKQSSFDEEIEEMGRSPSA--GTNWIPELSPIANVVVR 58
Query: 68 RSSKILGISTEELQHAFDSELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYLADKDFR 127
R SKILG+S EL+ +F E +K+ + RN LE+C ++AL + +LADK FR
Sbjct: 59 RCSKILGVSANELRDSFKQEAFESLKQPSLFPRNFLEYCCFRALSLSVGVTGHLADKKFR 118
Query: 128 RLTYDMMLAWETPSVHTDETPPPSSKDEHGGDDDEASLFYSSSTNMAVQVDDQKTVGLEA 187
RLT+DMM+ WE P+V ++ + V++ TV LEA
Sbjct: 119 RLTFDMMVVWEVPAV---------------------------ASQALLSVEEDATVSLEA 151
Query: 188 FSRIAPVCAPIADVITVHNLFDALTSSSGRRLHFLVYDKYIRFLDKVIKNSKNVLASSVG 247
FSRIAP IADVI NLF LTSS+G RL F VYDKY+ L++ IK + SS+
Sbjct: 152 FSRIAPAVPIIADVIICDNLFQMLTSSTGGRLQFSVYDKYLHGLERAIKKMRTQSESSLL 211
Query: 248 SLQLAEEEIVLDVDGTIPTQPVLQHIGISAWPGRLTLTNYALYFESLGVGVHEKPVRYDL 307
S ++ E +L++DGT+ TQPVL+H+GIS WPGRL LT+++LYFE+L V ++ P RY L
Sbjct: 212 SGVRSKREKILEIDGTVTTQPVLEHVGISTWPGRLILTDHSLYFEALKVVSYDTPKRYHL 271
Query: 308 GTDLKQVIKPDLTGPLGARLFDKAVMYKSTSVAEPVYFEFPEFKANVRRDYWLDISLEIL 367
DLKQ+IKP+LTGP G RLFDKAV Y+S S++EPV EFPE K + RRDYWL I E+L
Sbjct: 272 SEDLKQIIKPELTGPWGTRLFDKAVSYQSISLSEPVVMEFPELKGHTRRDYWLTIIQEVL 331
Query: 368 RVHKFIRKYYLKETQKAEVLARAILGIYRYRAVREAFKYFSSHYKTLLSFNLAEALPRGD 427
VH++I KY + + E L++A+LG+ R +A++E + Y+ LL FNL + LP GD
Sbjct: 332 YVHRYINKYKITGLARDEALSKAVLGVMRVQALQELNLTNAMRYENLLPFNLCDQLPGGD 391
Query: 428 MILETLSNSLTNLTV-ASSKRSSPRNVDTKKQLVVSPASAVALFRLGFKSDKFADIYEET 486
+ILETL+ T+ + S+K + + +VS +V G S + + +
Sbjct: 392 LILETLAEMSTSRELHRSNKSKDTGTLHSSASDMVSQLGSV----FGGSSPR-SRRETSS 446
Query: 487 TVVGDIRVGEINLLEMAVKKSLRDTGKAEAAQATVDQVKVEGIDTNVAVMKELLFPVIES 546
VVG++ VG++N LE AVK+S + K AQ T++ VK+ GIDTN+AVMKEL+ P++E+
Sbjct: 447 LVVGEVVVGDVNPLERAVKESRKKYEKVVLAQETINGVKMGGIDTNLAVMKELMLPIMET 506
Query: 547 ANRLERLASWKDFYKSTAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWRRHFRKGRSLE 606
N + + W D KS+ F LLT+++I RGW+ Y+ + AI M+ R F + + +
Sbjct: 507 WNLILSVVYWDDPTKSSVFCLLTTFIIWRGWLVYVFALASLFSAIFMVLTRCFSREKLMI 566
Query: 607 AFIVTPPPNRNPVEQLLTLQEAITQFESLIQAGNIVLLKVRALLLALLPQATEKVALLLV 666
VT PP N +EQLL +Q I++ E IQ NIVLLK RALL +L PQA++K A+ +V
Sbjct: 567 ELKVTAPPPMNTMEQLLAVQNGISELEQNIQDANIVLLKFRALLFSLFPQASQKFAIAIV 626
Query: 667 FIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMREWWIRIPAAPVELIKPDE 726
A + AFVP +Y++ V+F E +TR P R+ S++R +RR+REWW IPAAPV L+
Sbjct: 627 VAATMMAFVPGRYLLSVVFVELFTRYSPPRRASTERLIRRLREWWFSIPAAPVVLLHDKN 686
Query: 727 CKKRK 731
KK+K
Sbjct: 687 NKKKK 691
>At2g21720 hypothetical protein
Length = 703
Score = 316 bits (810), Expect = 3e-86
Identities = 224/711 (31%), Positives = 365/711 (50%), Gaps = 73/711 (10%)
Query: 31 RNSSSSKNKTDTTADDSAVNSPKPIPLLSPLANSVVVRSSKILGISTEELQHAFDSELPL 90
+NS S K K D + + + + LS +AN VV R S+ L + ++L F+ +
Sbjct: 40 KNSKSKKLKQRILRDHESKKTMRKMKHLSSIANDVVQRCSQELETTIDDLVKEFECQWKP 99
Query: 91 GVKELLTYARNLLEFCSYKALHKLSKSS-DYLADKDFRRLTYDMMLAWETPSVHTDETPP 149
G TY++ +EFC+ K ++ ++ + + D F RLT+DMMLAW+ P +E+
Sbjct: 100 GSTGG-TYSKKFVEFCNSKVTSRVCENILERIKDGSFTRLTFDMMLAWQQPDADDNESYK 158
Query: 150 PS----SKDEH-----GGDDDEASLFYSSSTNMAVQVDDQKTVGLEAFSRIAPVCAPIAD 200
+ S+D+ + D+ SLFYS M + VD + +VG +AF + + D
Sbjct: 159 EAVGKESEDKRIQATLSPEQDDISLFYSDM--MPLLVDHEPSVGEDAFVYLGSIIPLPVD 216
Query: 201 VITVHNLFDALTSSSGRRLHFLVYDKYIRFLDKVIKNSKNVLASSVGSLQLAEEEIVLDV 260
+I F+ LT+ +G +LHF YD +++ + K +K+ + S+ ++LA++EI+L V
Sbjct: 217 IINGRYTFETLTAPTGHQLHFPAYDMFVKEIHKCMKHLQK--QSTPKGIELADDEIILHV 274
Query: 261 DGTIPTQPVLQHIGISAWPGRLTLTNYALYFESLGVGVHEKPVRYDLGTDLKQVIKPDLT 320
+GT+ +Q V++HI ++WPGRLTLTNYALYFE+ G+ +E ++ DL D ++ KP T
Sbjct: 275 EGTMASQRVIRHIKETSWPGRLTLTNYALYFEAAGIINYEDAIKIDLSKDNEKSTKPMST 334
Query: 321 GPLGARLFDKAVMYKSTSVAEPVYFEFPEFKANVRRDYWLDISLEILRVHKFIRKYYLKE 380
GPLGA LFDKA++Y+S E + EFPE ++ RRD+WL + EI +HKF+RK+ ++
Sbjct: 335 GPLGAPLFDKAIVYESPDFEEGIVIEFPEMTSSTRRDHWLMLVKEITLMHKFLRKFNVES 394
Query: 381 T-QKAEVLARAILGIYRYRAVREAFKYFSSHYKTLLSFNLAEALPRGDMILETLSNSLTN 439
Q E+ +R ILGI R A RE + K L F+L E +P+GD +LE L+
Sbjct: 395 PLQSWEIHSRTILGIIRLHAAREMLRISPPDPKNFLIFSLFEEVPKGDYVLEELAE---- 450
Query: 440 LTVASSKRSSPRNVDTKKQLVVSPASAVALFR------LG-FKSDKFADIYEETTVVGDI 492
S K + RN P SA ++ R LG ++ DI +E V
Sbjct: 451 ---ISLKIGTTRN----------PCSASSILRNMNMDQLGDMIKEEGEDICKEKVVKVTD 497
Query: 493 RVGEINLLEMAVKKSLRDTGKAEAAQATVDQVKVEGIDTNVAVMKELLFPVIESANRLER 552
+ + LE AV +S + E A+AT +++ EGI +VAV+ +L
Sbjct: 498 KEEMLASLESAVNQSREEGKVIEKARATTAELEEEGISESVAVLMKL------------- 544
Query: 553 LASWKDFYKSTAFLLLTSYMILRGWIHYLLPSIFV-VVAILMLWRRHFRKGRSLEAFIVT 611
+ R W+ + + + VVA + R +S +A V+
Sbjct: 545 ------------------NLCYREWVGKAIAACLIWVVAKMAQARNKMVHTKSEDAVTVS 586
Query: 612 PPPNRNPVEQLLTLQEAITQFESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIAAV 671
++ E +++ Q + + L+Q N+ +LK+R+L + + V L++ +A+
Sbjct: 587 TESDQTVTESIVSAQYGLIRLHQLMQHVNVTILKLRSLYTSKASKHASMVMALMLVLASF 646
Query: 672 FAFVPPK-YIIFVIFFEYYTREMPCRKESSDRWVRRMREWWIRIPAAPVEL 721
FA VP K +IIF I + + S+D+ RRM+EWW IP PV +
Sbjct: 647 FAVVPFKLFIIFGIVYCFVMTSSVGTYMSNDQSNRRMKEWWDSIPIVPVRV 697
>At1g71240 hypothetical protein
Length = 823
Score = 258 bits (658), Expect = 1e-68
Identities = 205/739 (27%), Positives = 352/739 (46%), Gaps = 69/739 (9%)
Query: 18 LRNHQNSLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSPLANSVVVRSSKILGIST 77
L+ SLK + L + ++ + + V S P LS A + + S++ GI+
Sbjct: 89 LKKKSQSLKKIDLEDQQDFEDLEELLTVEQTVRSDTPKGFLSFDAIISIEQFSRMNGITG 148
Query: 78 EELQHAFDSELPLGVKELLTYARNLLEFCSYKALHKLSKSSDY---LADKDFRRLTYDMM 134
+++Q F++ + L T AR L+E+C ++ L + SS++ L + F+RL + M
Sbjct: 149 KKMQDIFETIVS---PALSTDARYLVEYCCFRFLSR--DSSEFHPCLKEPAFQRLIFITM 203
Query: 135 LAWETPSVHTDETPPPSSKDEHGGDDDEASLFYSSSTNMAVQVDDQKTVGLEAFSRIAPV 194
LAW P K+ + +D + + +G EAF RIAP
Sbjct: 204 LAWANPYC----------KERNARNDASGKPSFQG-----------RFIGEEAFIRIAPA 242
Query: 195 CAPIADVITVHNLFDALTSSSGRR-LHFLVYDKYIRFLDKVIKNSKNVLASSVGSLQLAE 253
+ +AD TVHNLF AL +++ ++ + ++ YI+ L K+ + K+ + QL+
Sbjct: 243 ISGLADRATVHNLFKALATATDQKGISLEIWLAYIQELVKIHEGRKSHQTTDFP--QLSS 300
Query: 254 EEIVLDVDGTIPTQPVLQHIGISAWPGRLTLTNYALYFESLGVGVHEKPVRYDLGTDLKQ 313
E ++ PVL+ AWPG+LTLT+ ALYFE + + + +R DL D K
Sbjct: 301 ERLLCMAANR--KGPVLKWENNVAWPGKLTLTDKALYFEPVDIKGSKGVLRLDLAGD-KS 357
Query: 314 VIKPDLTGPLGARLFDKAVMYKSTSVAEPVYFEFPEFKANVRRDYWLDISLEILRVHKFI 373
++ GPLG LFD AV S EF + +RRD W I E++ +H F+
Sbjct: 358 TVEKAKVGPLGFSLFDSAVSVSSGPGLATWVLEFVDLGGELRRDVWHAIISEVIALHTFL 417
Query: 374 RKYYLKETQKA------------EVLARAILGIYRYRAVREAFKYFSSHYKTLLSFNLAE 421
R++ E K+ + +A A I R +A++ K L+ F+ +
Sbjct: 418 REFGPGEGDKSLYQVFGAKKGKEKAIASASNCIARLQALQYMRNLPDDPIK-LVQFSFLQ 476
Query: 422 ALPRGDMILETLSNSLTN---LTVASSKRSSPRNVDTKKQLV---VSPASAVALFRLGFK 475
+ GD++ +TL+ + LT S KR + VS + +
Sbjct: 477 QVAYGDIVCQTLAVNFWGGPLLTKVSDKRGDIARASRESYETFDNVSDLDGSVYLKRWMR 536
Query: 476 SD-----------KFADIYEETTVVGDIRVGEINLLEMAVKKSLRDTGKAEAAQATVDQV 524
S K + + + + + V ++ L+E A + + E QAT+D
Sbjct: 537 SPSWGSTASMNFWKNSSLRQGLVLSKHLAVADLTLVERATETCRQKYKVVEKTQATIDAA 596
Query: 525 KVEGIDTNVAVMKELLFPVIESANRLERLASWKDFYKSTAFLLLTSYMILRGWIHYLLPS 584
++GI +N+ + KEL+ P+ +A E+L W++ Y + +FL S +I R + Y+LP
Sbjct: 597 TIKGIPSNIDLFKELILPLSITATEFEKLRCWEEPYMTVSFLAFASTIIFRNLLQYVLPV 656
Query: 585 IFVVVAILMLW----RRHFRKGRSLEAFIVTPPPNRNPVEQLLTLQEAITQFESLIQAGN 640
+ +A ML RR R GR + P+ N +++++ +++A+ ES +Q N
Sbjct: 657 SLIFLATGMLTLKGLRRQGRLGRLFGIISIRDQPSSNTIQKIIAVKDAMQNLESYLQKVN 716
Query: 641 IVLLKVRALLLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESS 700
+VLLK+R ++L+ PQ T +VAL ++ IA V VP KY++ + ++ +TRE+ RKE
Sbjct: 717 VVLLKLRTIVLSGHPQITTEVALAMLSIATVLVIVPFKYVLAFVLYDQFTRELEFRKEMV 776
Query: 701 DRWVRRMREWWIRIPAAPV 719
++ +RE W +PAAPV
Sbjct: 777 KKFNAFLRERWEMVPAAPV 795
>At4g14150 kinesin like protein
Length = 1662
Score = 33.1 bits (74), Expect = 0.57
Identities = 21/84 (25%), Positives = 43/84 (51%), Gaps = 4/84 (4%)
Query: 18 LRNHQNSLKSLFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSPLANSVVVRSSKILGIST 77
+RN + SLK+ L +S ++ + ++A SP ++ ++++ + SK+ + T
Sbjct: 673 IRNSRKSLKTSELSTASQKDSEGENLVTEAADPSPATSKKMNNCSSALSTQKSKVFPVRT 732
Query: 78 EELQHAFDSELPLGVKELLTYARN 101
E L S L G+K L +Y ++
Sbjct: 733 ERLA----SSLHKGIKLLESYCQS 752
>At5g50190 putative protein
Length = 215
Score = 32.3 bits (72), Expect = 0.97
Identities = 30/113 (26%), Positives = 57/113 (49%), Gaps = 7/113 (6%)
Query: 500 LEMAVKKSLRDTGKAEAAQATVDQV--KVEGIDTNVAVMKELLFPVIE-SANRLERLASW 556
+EM K+SL +AE A+ V ++ K+E + V++ ++ ++ + N+ + W
Sbjct: 55 VEMQRKRSLEVETRAEIAEKKVAELGSKLENVRNMVSLGSDIDGKLLGLNLNKTQISQKW 114
Query: 557 KDFYKSTAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWRRHFRKGR-SLEAF 608
K ++ LT Y L+ I YL V ++ +W+ HF K + +LEA+
Sbjct: 115 IPSIKDV-YVTLTIY--LQPKIQYLTDKSIEVSRVMTIWKPHFEKIQVALEAY 164
>At4g00700 putative phosphoribosylanthranilate transferase
Length = 1006
Score = 32.3 bits (72), Expect = 0.97
Identities = 42/195 (21%), Positives = 80/195 (40%), Gaps = 40/195 (20%)
Query: 550 LERLASWKDFYKSTAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWRRHFR--------- 600
+E++ +WK T L+ Y +L + +LP++F+ +A++ +W F+
Sbjct: 821 MEQVCTWKT--PVTTALVHVLYTMLVTFPEMILPTVFLYMAVIGMWNYRFKPRFPPHMDA 878
Query: 601 --------KGRSLEAFIVTPPPNRNPVEQLLTLQEAITQFESLIQ--AGNIVLL--KVRA 648
L+ T P R P + + + + +Q AG+I +V+A
Sbjct: 879 KLSYADNVNSDELDEEFDTFPTVRAP-DIVKMRYDRLRSVAGKVQSVAGDIAAQGERVQA 937
Query: 649 LLLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMR 708
LL P+AT + + FI A+ ++ P ++ ++ Y+ R R
Sbjct: 938 LLSWRDPRAT-AIFVTFCFIIAMALYITPFKLVALLSGYYFMRHPKLRH----------- 985
Query: 709 EWWIRIPAAPVELIK 723
RIP+APV +
Sbjct: 986 ----RIPSAPVNFFR 996
>At5g60150 putative protein
Length = 1195
Score = 32.0 bits (71), Expect = 1.3
Identities = 23/60 (38%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Query: 18 LRNHQNSLKS-LFLRNSSSSKNKTDTTADDSAVNSPKPIPLLSPLANSVVVRSSKILGIS 76
L+N Q SL S F +N+SS+K+KT ++ A S P P L +V+ +SS+I +S
Sbjct: 237 LKNSQRSLGSESFSKNTSSTKSKTKSSL---ASKSSIPKPSLKQARRNVISKSSEIPTVS 293
>At4g11610 phosphoribosylanthranilate transferase like protein
Length = 1011
Score = 31.2 bits (69), Expect = 2.2
Identities = 45/192 (23%), Positives = 81/192 (41%), Gaps = 40/192 (20%)
Query: 553 LASWKDFYKSTAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWRRHFR-------KGRSL 605
+ SW++ T L+ +++L +LP++F+ + ++ LW FR +
Sbjct: 829 ICSWRN--PITTVLVHVLFLMLVCLPELILPTMFLYMFLIGLWNYRFRPRYPPHMNTKIS 886
Query: 606 EAFIVTP----------PPNRNPVEQLLTLQEAITQFESLIQA--GNIVLL--KVRALLL 651
+A V P P RNP + + + + IQ G++ + +ALL
Sbjct: 887 QAEAVHPDELDEEFDTFPTTRNP-DMVRLRYDRLRSVAGRIQTVIGDLATQGERFQALLS 945
Query: 652 ALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMREWW 711
P+AT + ++L FIAA+ F+ P I+ V ++T P R R
Sbjct: 946 WRDPRAT-AIFVILCFIAAIVFFITPIQIV-VALAGFFTMRHP-----------RFRH-- 990
Query: 712 IRIPAAPVELIK 723
R+P+ PV +
Sbjct: 991 -RLPSVPVNFFR 1001
>At4g04390 hypothetical protein
Length = 963
Score = 31.2 bits (69), Expect = 2.2
Identities = 20/62 (32%), Positives = 26/62 (41%)
Query: 407 FSSHYKTLLSFNLAEALPRGDMILETLSNSLTNLTVASSKRSSPRNVDTKKQLVVSPASA 466
F + LL F L E LPR + L L SSK +D K +L+V S
Sbjct: 674 FEPRVEDLLRFQLTEMLPRWQLSSLRLGRRSKRLRTRSSKLDGRFAIDKKTKLLVGHTSP 733
Query: 467 VA 468
+A
Sbjct: 734 IA 735
>At3g05020 acyl carrier protein 1 precursor (ACP)
Length = 137
Score = 30.8 bits (68), Expect = 2.8
Identities = 32/114 (28%), Positives = 52/114 (45%), Gaps = 19/114 (16%)
Query: 408 SSHYKTLLSFNLAEALPRGDMILETLSNSLTNLTVASSKRSSPRNVDTKKQLVVSPASAV 467
S+H KT LSFNL ++P S L+ A + + KKQL ++P V
Sbjct: 30 SNHGKTNLSFNLRRSIP---------SRRLSVSCAAKQETIEKVSAIVKKQLSLTPDKKV 80
Query: 468 ALFRLGFKSDKFADIYEETTVVGDIRVG---EINLLEMAVKKSLRDTGKAEAAQ 518
KFAD+ ++ +I +G E N ++MA +K+ + +AA+
Sbjct: 81 V------AETKFADLGADSLDTVEIVMGLEEEFN-IQMAEEKAQKIATVEQAAE 127
>At5g18400 unknown protein
Length = 272
Score = 29.6 bits (65), Expect = 6.3
Identities = 19/64 (29%), Positives = 35/64 (54%)
Query: 61 LANSVVVRSSKILGISTEELQHAFDSELPLGVKELLTYARNLLEFCSYKALHKLSKSSDY 120
+ + VV+ S +L I E + +S PL + + T + L+ S +A+ +SK+SD+
Sbjct: 13 VTDDVVLPVSSVLAIMKELGKEVIESFDPLIITQASTINQFPLDASSVEAVLAISKTSDF 72
Query: 121 LADK 124
+DK
Sbjct: 73 PSDK 76
>At5g11020 ser/thr specific protein kinase-like protein
Length = 402
Score = 29.6 bits (65), Expect = 6.3
Identities = 32/129 (24%), Positives = 49/129 (37%), Gaps = 27/129 (20%)
Query: 32 NSSSSKNKTDTTADDSAVNSPKPIPLLSPLANSVVVRSSKILGISTEELQHAFDSEL--- 88
N S++ K D +D+A + +LS L + ++ +LG ST + EL
Sbjct: 132 NISAAVKKLDCANEDAAKEFKSEVEILSKLQHPNII---SLLGYSTNDTARFIVYELMPN 188
Query: 89 ------------------PLGVKELLTYARNLL---EFCSYKALHKLSKSSDYLADKDFR 127
P+ +K L R L E C +H+ KSS+ L D +F
Sbjct: 189 VSLESHLHGSSQGSAITWPMRMKIALDVTRGLEYLHEHCHPAIIHRDLKSSNILLDSNFN 248
Query: 128 RLTYDMMLA 136
D LA
Sbjct: 249 AKISDFGLA 257
>At4g22120 unknown protein
Length = 771
Score = 29.6 bits (65), Expect = 6.3
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 4/88 (4%)
Query: 554 ASWKDFYKSTAFLLLTSYMILRGWI---HYLLPSIFVV-VAILMLWRRHFRKGRSLEAFI 609
A W D + L+ S ++L G + H L + F++ + +L + HF KGR AFI
Sbjct: 622 AFWPDVHGRVIAALVISQLLLMGLLGTKHAALAAPFLIALPVLTIGFHHFCKGRYEPAFI 681
Query: 610 VTPPPNRNPVEQLLTLQEAITQFESLIQ 637
P + L T +E + +Q
Sbjct: 682 RYPLQEAMMKDTLETAREPNLNLKGYLQ 709
>At3g54150 embryonic abundant protein -like
Length = 323
Score = 29.3 bits (64), Expect = 8.2
Identities = 26/114 (22%), Positives = 55/114 (47%), Gaps = 12/114 (10%)
Query: 198 IADVITVHNLFDALTSSSGRRLHFLVYDKYIRFLDKVIKNSKNVLASSVGSLQLAEEEI- 256
+ D++ N D + ++ +HF + + +V++ ++A V + + EI
Sbjct: 91 MVDLLGGENSVDLIVAAQA--VHFFDLNVFYNVAKRVLRKEGGLIAVWVYNDIIISHEID 148
Query: 257 -VLD--VDGTIPTQPVLQHIGISAWPGRLTLTNYALYFESLGVGVHEKPVRYDL 307
++ VD T+P + + ++ A+ G TLT FE++G+G KP+ D+
Sbjct: 149 PIMKRLVDSTLPFRTPIMNL---AFDGYKTLT---FPFETIGMGSEGKPITLDI 196
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,591,318
Number of Sequences: 26719
Number of extensions: 647575
Number of successful extensions: 2136
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 2106
Number of HSP's gapped (non-prelim): 17
length of query: 731
length of database: 11,318,596
effective HSP length: 107
effective length of query: 624
effective length of database: 8,459,663
effective search space: 5278829712
effective search space used: 5278829712
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0180a.6


