

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.4
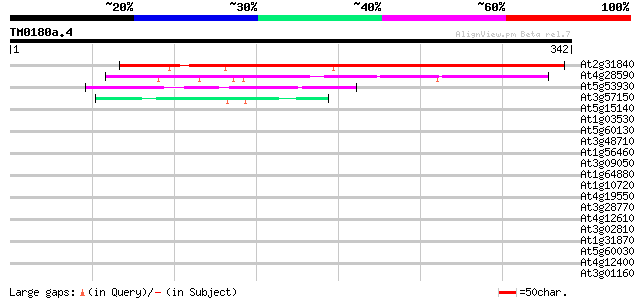
(342 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g31840 unknown protein 367 e-102
At4g28590 unknown protein 137 1e-32
At5g53930 unknown protein 45 4e-05
At3g57150 putative pseudouridine synthase (NAP57) 45 7e-05
At5g15140 putative aldose 1-epimerase - like protein 40 0.002
At1g03530 unknown protein 40 0.002
At5g60130 putative protein 39 0.005
At3g48710 unknown protein 39 0.005
At1g56460 unknown protein 37 0.020
At3g09050 unknown protein 36 0.035
At1g64880 unknown protein 36 0.035
At1g10720 unknown protein 36 0.035
At4g19550 hypothetical protein 35 0.045
At3g28770 hypothetical protein 35 0.045
At4g12610 putative protein 35 0.059
At3g02810 putative protein kinase 35 0.077
At1g31870 unknown protein 35 0.077
At5g60030 KED - like protein 34 0.10
At4g12400 stress-induced protein sti1 -like protein 34 0.10
At3g01160 unknown protein 34 0.10
>At2g31840 unknown protein
Length = 350
Score = 367 bits (942), Expect = e-102
Identities = 179/287 (62%), Positives = 226/287 (78%), Gaps = 21/287 (7%)
Query: 68 LKSDGAHRKSGKRE-ASSDSDSDSDDENDA---------PPPINDPYLMTLEERQEWRRK 117
+K+ G K GK+ + DSDS+DE D ++DP ER+EWR+
Sbjct: 62 VKAFGLVDKLGKKVWRKKEEDSDSEDEEDEVKEETFGGKEASLDDPV-----ERREWRKT 116
Query: 118 IRQVMDMKPNVQE--ESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFN 175
IR+V+D P+++E E D EK++KM+KL+ DYPLVV+EEDPNWPEDADGWGFS QFFN
Sbjct: 117 IREVIDKHPDIEEDEEIDMVEKRRKMQKLLADYPLVVNEEDPNWPEDADGWGFSFNQFFN 176
Query: 176 KITIKDNKKAKDDDDDDDEGN----KVVWQDDDYIRPIKDIKSSEWEETVFKDISPLIIL 231
KITIK+ KK ++DDD+D EG+ ++VWQDD+YIRPIKD+ ++EWEE VFKDISPL++L
Sbjct: 177 KITIKNEKKEEEDDDEDSEGDDSEKEIVWQDDNYIRPIKDLTTAEWEEAVFKDISPLMVL 236
Query: 232 VHNRYRRPKENERIRDELEKAVHIIWNCRLPSPRCVALDAVVETELVAALKVSVFPEMIF 291
VHNRY+RPKENE+ R+ELEKA+ +IWNC LPSPRCVA+DAVVET+LV+ALKVSVFPE+IF
Sbjct: 237 VHNRYKRPKENEKFREELEKAIQVIWNCGLPSPRCVAVDAVVETDLVSALKVSVFPEIIF 296
Query: 292 TKSGKILFRDKAIRNADELSKIMAYFYFGAAKPPCLNSFTDFQEDIP 338
TK+GKIL+R+K IR ADELSKIMA+FY+GAAKPPCLN + QE IP
Sbjct: 297 TKAGKILYREKGIRTADELSKIMAFFYYGAAKPPCLNGVVNSQEQIP 343
>At4g28590 unknown protein
Length = 331
Score = 137 bits (344), Expect = 1e-32
Identities = 91/293 (31%), Positives = 150/293 (51%), Gaps = 33/293 (11%)
Query: 59 SKAPRRVQALKSDGAHRKSGKREASSDSDSD------SDDENDAPPPINDPYLMTLEERQ 112
S++ +R A+ SD K + S + ++ EN+ P P R
Sbjct: 41 SRSVKRFAAVSSDSVLDPESKNQTRSRRKNKEAVTPIAETENNEKFPTKVPRKSKRGRRS 100
Query: 113 EW---RRKIRQVMDMKPNVQEESDPE--EKKKKM---------EKLMKDYPLVVDEEDPN 158
E +R ++ + +E +PE E K+K E+ ++ +VV+EEDP+
Sbjct: 101 EADAVEDYVRSSLERTFSTIKEQNPEVFENKEKANFIKDRGVDEEEEEEEEMVVEEEDPD 160
Query: 159 WPEDAD-GWGFSLGQFFNKITIKDNKKAKDDDDDDDEGNKVVWQDDDYIRPIKDIKSSEW 217
WP D D GWG ++F+ IK+ D+G+++ W+ + +K+I EW
Sbjct: 161 WPVDTDVGWGIKASEYFDTHPIKNVV--------GDDGSEIDWEGEIDDSWVKEINCLEW 212
Query: 218 EETVFKDISPLIILVHNRYRRPKENERIRDELEKAVHIIWNC--RLPSPRCVALDAVVET 275
E F SPL++LV RY+R +N + ELEKA+ + W+ RLP PR V +D +ET
Sbjct: 213 ESFAFHP-SPLVVLVFERYKRASDNWKTLKELEKAIKVYWDAKDRLP-PRAVKIDLNIET 270
Query: 276 ELVAALKVSVFPEMIFTKSGKILFRDKAIRNADELSKIMAYFYFGAAKPPCLN 328
+L ALK P+++F + +IL+R+K R ADEL ++A+FY+ A +P C++
Sbjct: 271 DLAYALKAKECPQILFLRGNRILYREKDFRTADELVHMIAHFYYKAKRPSCVD 323
>At5g53930 unknown protein
Length = 529
Score = 45.4 bits (106), Expect = 4e-05
Identities = 40/166 (24%), Positives = 73/166 (43%), Gaps = 20/166 (12%)
Query: 47 SLSSNLSLLVGDSKAPRRVQALKSDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLM 106
S S+ SL RR + +S + ++S KR +SS+SD DSDD+
Sbjct: 43 SSGSDSSLYSSSEDDYRRKKKRRSKLSKKRSRKRYSSSESDDDSDDDR------------ 90
Query: 107 TLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGW 166
L++++ +RK V K V K++K + + D + +D W
Sbjct: 91 LLKKKKRSKRKDENVGKKKKKV-----VSRKRRKRDLSSSSTSSEQSDNDGSESDDGKRW 145
Query: 167 GFSLGQFFNKITIKDNK-KAKDDDDDDDEGNKVVWQDDDYIRPIKD 211
G+ K +KD++ +++D+ + + E WQ +D + P K+
Sbjct: 146 SRDRGRRLGK--VKDSRSRSRDELEGESEEPDECWQGEDEVIPEKN 189
>At3g57150 putative pseudouridine synthase (NAP57)
Length = 565
Score = 44.7 bits (104), Expect = 7e-05
Identities = 43/169 (25%), Positives = 68/169 (39%), Gaps = 45/169 (26%)
Query: 53 SLLVGDSKAPRRVQALKSDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLE-ER 111
+++ G + AP ++A +G ++ KR+ DD +D+P P+ T E E
Sbjct: 415 AIIAGAAAAPEEIKADAENGEAGEARKRK--------HDDSSDSPAPVTTKKSKTKEVEG 466
Query: 112 QEWRRKIRQVMDMKPNVQEE-------SDPEEKKKKME-------------------KLM 145
+E K++ K +EE S+ +EKKKK + K
Sbjct: 467 EEAEEKVKSSKKKKKKDKEEEKEEEAGSEKKEKKKKKDKKEEVIEEVASPKSEKKKKKKS 526
Query: 146 KDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDE 194
KD VD ED + E ++ K KD KK D +DD+E
Sbjct: 527 KDTEAAVDAEDESAAEKSE----------KKKKKKDKKKKNKDSEDDEE 565
>At5g15140 putative aldose 1-epimerase - like protein
Length = 490
Score = 39.7 bits (91), Expect = 0.002
Identities = 28/161 (17%), Positives = 67/161 (41%), Gaps = 24/161 (14%)
Query: 44 LNLSLSSNLSLLVGDSKAPRRVQALKSDGAHRKSGKREASSDSDSDSDDENDAPPPINDP 103
++++ + N+ L D ++ ++ + + D H++ G + D D++ +D
Sbjct: 20 ISVTFADNVELKSKDLESSKKDKKVDHDSTHKEKGGEKVDGAGSDDEDNDKKEKKKEHDV 79
Query: 104 YLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDA 163
+ + + ++ +D K + + D +++KK +K ++ N +D+
Sbjct: 80 QKKDKQHENKDKDDEKKHVDKKKSGGHDKDDDDEKKHKDK---------KKDGHNDDDDS 130
Query: 164 DGWGFSLGQFFNKITIKDNKKAKDDDDDDDEGNKVVWQDDD 204
D D DDDDDDD+ ++V D++
Sbjct: 131 DD---------------DTDDDDDDDDDDDDDDEVDGDDNE 156
>At1g03530 unknown protein
Length = 801
Score = 39.7 bits (91), Expect = 0.002
Identities = 33/119 (27%), Positives = 53/119 (43%), Gaps = 7/119 (5%)
Query: 89 DSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDY 148
D D+++D D + E E D + +EESD +E K+ K + +
Sbjct: 230 DDDEKSDEAKGEMD----SAESESETSSSSASSSDSSSSEEEESDEDESDKEENKKEEKF 285
Query: 149 P-LVVDEEDPNWPEDADGWGFSLGQFF--NKITIKDNKKAKDDDDDDDEGNKVVWQDDD 204
+VV +ED E +G +L + + I +D+ DDDDDDD V W +D+
Sbjct: 286 EHMVVGKEDDLAGELEEGEIENLDEENGDDDIEDEDDDDDDDDDDDDDVNEMVAWSNDE 344
>At5g60130 putative protein
Length = 300
Score = 38.5 bits (88), Expect = 0.005
Identities = 25/101 (24%), Positives = 49/101 (47%), Gaps = 14/101 (13%)
Query: 110 ERQEWRRKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEEDP----NWPEDADG 165
E + +KIR+ D ++ ESD +E+ + + ++ +DE+D N ED D
Sbjct: 82 EHRTMCKKIRRSSDQSEEIKVESDSDEQNQASDDVLS-----LDEDDDDSDYNCGEDNDS 136
Query: 166 WGFSLGQFFNKITIKDNKKAKDDDDDDDEGNKVVWQDDDYI 206
+ ++ ++ + DD+D D+ + V +DDDY+
Sbjct: 137 -----DDYADEAAVEKDDNDADDEDVDNVADDVPVEDDDYV 172
Score = 28.9 bits (63), Expect = 4.2
Identities = 26/118 (22%), Positives = 48/118 (40%), Gaps = 18/118 (15%)
Query: 75 RKSGKREASSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDP 134
R+S + +SDSD++N A + ++L+E + D N E++D
Sbjct: 91 RRSSDQSEEIKVESDSDEQNQASDDV-----LSLDEDDD---------DSDYNCGEDNDS 136
Query: 135 EEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDD 192
++ + D D +D + AD + +D+ KA DDD+D+
Sbjct: 137 DDYADEAAVEKDDN----DADDEDVDNVADDVPVEDDDYVEAFDSRDHAKADDDDEDE 190
>At3g48710 unknown protein
Length = 462
Score = 38.5 bits (88), Expect = 0.005
Identities = 38/159 (23%), Positives = 68/159 (41%), Gaps = 19/159 (11%)
Query: 38 KPRKQFLNLSLSSNLSLLVGDSKAPRRVQALKSDGAHRKSGKREAS--SDSDSDSDDEND 95
K RK+ + +++S S V A RR QA K + G E+ S+ +DS+ E+D
Sbjct: 215 KKRKKSTSKNVTSGESSHV---PAKRRRQAKKQEQPTETEGNGESDVGSEGTNDSNGEDD 271
Query: 96 APPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEE 155
P E + + + + D K +E++ +KK+ ++ K+ P +E+
Sbjct: 272 VAP-----------EEENNKSEDTETEDEKDKAKEKTKSTDKKRLSKRTKKEKPAAEEEK 320
Query: 156 DPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDE 194
+ F + +K T +KK K D DD +
Sbjct: 321 SIKGSAKSSRKSF---RQVDKSTTSSSKKQKVDKDDSSK 356
>At1g56460 unknown protein
Length = 502
Score = 36.6 bits (83), Expect = 0.020
Identities = 33/126 (26%), Positives = 57/126 (45%), Gaps = 30/126 (23%)
Query: 73 AHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLEERQEWRR-KIRQVMDMKPNVQEE 131
++R S +R + DS DD+ EE Q RR K+ +V+ ++ +V ++
Sbjct: 201 SNRISKRRVLDEELDSLDDDD---------------EEIQFLRRMKMAKVVAVEEDVDDD 245
Query: 132 SDPEEKKKKMEKLMK---DYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDD 188
D K KK+ K+MK ++P V + + +D G F D KDD
Sbjct: 246 EDRTRKHKKLSKVMKQNVEFPRGVGTSEKSAKKDKMGKAFD-----------DADYVKDD 294
Query: 189 DDDDDE 194
D++++E
Sbjct: 295 DEEEEE 300
>At3g09050 unknown protein
Length = 258
Score = 35.8 bits (81), Expect = 0.035
Identities = 39/170 (22%), Positives = 72/170 (41%), Gaps = 25/170 (14%)
Query: 83 SSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKKME 142
S D SDSD + D P P D RQE +I + K + + D E+ + +
Sbjct: 38 SGDGTSDSDSDPDPPKPEGDT------RRQELLARIAMIQTSKVRLTDFLD--ERSEYLT 89
Query: 143 KLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDEG-NKVVWQ 201
K ++ D+ + +D D I +N ++K ++ G N++ +
Sbjct: 90 KFAEEANAEFDKVGEDAMKDLDE---------ASTRILENIESKMQAFEESAGLNRLEIE 140
Query: 202 DDDYIRPIKDIKSSEWEETVFKDISPLIILVHNRYRRPKENERIRDELEK 251
++D K +E+EE + D + + R ++P + E+ R+E EK
Sbjct: 141 ENDN-------KLAEFEEKIQVDRNEGLFFKSLRDKKPVDREQAREETEK 183
>At1g64880 unknown protein
Length = 515
Score = 35.8 bits (81), Expect = 0.035
Identities = 51/204 (25%), Positives = 81/204 (39%), Gaps = 52/204 (25%)
Query: 66 QALKSDGAHRKSGKREASSDSDS-----DSDDENDAPPPINDPYLM------------TL 108
Q LK DG + +RE SSDSDS D D+E N + TL
Sbjct: 129 QNLKDDG---ELNRREESSDSDSEYDEIDLDEERKKEDIFNKKFQRHKELLQTLTKSETL 185
Query: 109 EERQEWRRKIRQVMDMKPNVQEE-------------SDPEEKKKKMEKLMKDYPLVVDEE 155
+E W K+ + + ++ E ++ +EK +KL + LV +E
Sbjct: 186 DEAFRWMNKLDKFEEKHFKLRPEYRVIGELMNRLKVAEGKEKFVLQQKLNRAMRLVEWKE 245
Query: 156 --DPNWP--------EDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDEGNKVVWQDDDY 205
DPN P ++ G G + ++ K DDDDD++E + + +DD
Sbjct: 246 AFDPNNPANYGVIERDNVQGGGEEREE---RLVADGPKDDNDDDDDEEEFDDMKERDDIL 302
Query: 206 IRPIK------DIKSSEWEETVFK 223
+ + +IK SE + T K
Sbjct: 303 LEKLNAIDKKLEIKLSELDHTFGK 326
>At1g10720 unknown protein
Length = 429
Score = 35.8 bits (81), Expect = 0.035
Identities = 27/118 (22%), Positives = 48/118 (39%), Gaps = 8/118 (6%)
Query: 81 EASSDSDSDSDDENDAPPPINDPYLMTLEER----QEWRRKIRQVMDMKPNVQEESDPEE 136
E + S S+ N APP P + E +++ + V EE ++
Sbjct: 296 EEEDITPSTSNYYNHAPPEFLSPRIYAFEPPSIMYRDFEHGFENAQFIDKAVIEEKPIQK 355
Query: 137 KKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDE 194
K L + VVD++D +WPE+ D S + T+ ++ + D + DD+
Sbjct: 356 NDKNSASLSQTSKDVVDDDDDDWPEEED----SANSWAPMFTVNEDDVSFSDLEGDDD 409
>At4g19550 hypothetical protein
Length = 251
Score = 35.4 bits (80), Expect = 0.045
Identities = 31/136 (22%), Positives = 64/136 (46%), Gaps = 7/136 (5%)
Query: 69 KSDGAHRKS-GKREASSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPN 127
K D A++ + K+ +++ ++S+++ EN P + Y + L++ + ++ KP
Sbjct: 119 KKDPANKIAPSKKTSATKTESNAETENKKRPSLEINYNV-LDKLFDPENSPKRAKLDKPV 177
Query: 128 VQEESDPEEKKKKMEKLMKDYPLVVDEED---PNWPEDADGWGFSLGQFFNKITIKDNKK 184
V + K+ E L+K +EE+ P+W ED G + +N +
Sbjct: 178 VVGDQIEYSKQNSEESLLKSQYSEEEEEEAEEPDWNEDYSNEDAYRGN--EECYGNENLE 235
Query: 185 AKDDDDDDDEGNKVVW 200
+D DDD+++ V+W
Sbjct: 236 FEDLDDDEEDDEGVIW 251
>At3g28770 hypothetical protein
Length = 2081
Score = 35.4 bits (80), Expect = 0.045
Identities = 49/221 (22%), Positives = 94/221 (42%), Gaps = 35/221 (15%)
Query: 36 KDKPRKQFLNLSLSSNLSLLVGDSKAPRRVQALKSDGAHRKSGKREASSDSDS-DSDDEN 94
K++ +K+++N L + + + KS+ + K ++ +S DS +N
Sbjct: 959 KEEDKKEYVNNELKK---------QEDNKKETTKSENSKLKEENKDNKEKKESEDSASKN 1009
Query: 95 DAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEEKKKKMEK-LMKDYPLVVD 153
+ T EE ++ ++K + D K +EE D EE+K K EK +D
Sbjct: 1010 REKKEYEEKKSKTKEEAKKEKKKSQ---DKK---REEKDSEERKSKKEKEESRDLKAKKK 1063
Query: 154 EEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDDDDDDDEGNKVVWQDDDYIRPIKDIK 213
EE+ ++++ ++K K +D + E NK + +++D K +
Sbjct: 1064 EEETKEKKESE----------------NHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEE 1107
Query: 214 S-SEWEETVFKDISPLIILVHNRYRRPKENERIRDELEKAV 253
S S +E KD+ L N+ + K NE+ + + K V
Sbjct: 1108 SKSRKKEEDKKDMEKLEDQNSNKKKEDK-NEKKKSQHVKLV 1147
Score = 30.8 bits (68), Expect = 1.1
Identities = 29/135 (21%), Positives = 55/135 (40%), Gaps = 15/135 (11%)
Query: 69 KSDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNV 128
K++ + + ++ SDS +DSD+ + D T +E R+K
Sbjct: 1388 KNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKK----------- 1436
Query: 129 QEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDD 188
+ S E KK+K K K+ P ++ N E + G S+ + + +A
Sbjct: 1437 -QTSVAENKKQKETKEEKNKP---KDDKKNTTEQSGGKKESMESESKEAENQQKSQATTQ 1492
Query: 189 DDDDDEGNKVVWQDD 203
+ D+ N+++ Q D
Sbjct: 1493 GESDESKNEILMQAD 1507
Score = 30.4 bits (67), Expect = 1.5
Identities = 29/135 (21%), Positives = 55/135 (40%), Gaps = 15/135 (11%)
Query: 69 KSDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMKPNV 128
K++ + + ++ SDS +DSD+ + D T +E R+K
Sbjct: 1277 KNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKK----------- 1325
Query: 129 QEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDD 188
+ S E KK+K K K+ P ++ N + + G S+ + + +A
Sbjct: 1326 -QTSVAENKKQKETKEEKNKP---KDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQ 1381
Query: 189 DDDDDEGNKVVWQDD 203
D D+ N+++ Q D
Sbjct: 1382 ADSDESKNEILMQAD 1396
Score = 30.4 bits (67), Expect = 1.5
Identities = 29/140 (20%), Positives = 59/140 (41%), Gaps = 7/140 (5%)
Query: 57 GDSKAPRRVQALKSDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLE-ERQEWR 115
G+S + V A +D H+K K E ++ EN N+ + E +
Sbjct: 398 GESTNDKMVNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEESMKGENLENKVGN 457
Query: 116 RKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFF- 174
+++ ++ ES EEK+++ ++ + Y ++ E+ + G S+G
Sbjct: 458 EELKGNASVEAKTNNESSKEEKREESQRSNEVYM----NKETTKGENVNIQGESIGDSTK 513
Query: 175 -NKITIKDNKKAKDDDDDDD 193
N + K++ K K D ++ D
Sbjct: 514 DNSLENKEDVKPKVDANESD 533
Score = 28.1 bits (61), Expect = 7.2
Identities = 33/161 (20%), Positives = 65/161 (39%), Gaps = 18/161 (11%)
Query: 58 DSKAPRRVQALKS--DGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLEERQEWR 115
D KA ++ + K + + KS K+E D ++N + D E + R
Sbjct: 1057 DLKAKKKEEETKEKKESENHKSKKKE-----DKKEHEDNKSMKKEEDKKEKKKHEESKSR 1111
Query: 116 RKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFN 175
+K DM+ + S+ +++ K +K + LV E D ++ +
Sbjct: 1112 KKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENE----------E 1161
Query: 176 KITIKDNKKAKDDDDDDDEGNKVVWQDDDYIRPIKDIKSSE 216
K K+ + +K ++ D+ K +D + K++K SE
Sbjct: 1162 KSETKEIESSKSQKNEVDKKEKKSSKDQQK-KKEKEMKESE 1201
>At4g12610 putative protein
Length = 649
Score = 35.0 bits (79), Expect = 0.059
Identities = 30/131 (22%), Positives = 57/131 (42%), Gaps = 11/131 (8%)
Query: 69 KSDGAHRKSG---KREASSDSDSDSDDENDAPPPINDPYLMTLEERQEWRRKIRQVMDMK 125
+ + A RKS R+++ D D + D +D +E+ +W + D
Sbjct: 244 EEEEASRKSRLGLNRKSNDDDDEEGPRGGDLDMDDDD-----IEKGDDWEHEEIFTDD-- 296
Query: 126 PNVQEESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKA 185
+ +DPEE++ + + P + +ED E+ +G G+ K+ K N
Sbjct: 297 -DEAVGNDPEEREDLLAPEIPAPPEIKQDEDDEENEEEEGGLSKSGKELKKLLGKANGLD 355
Query: 186 KDDDDDDDEGN 196
+ D+DDDD+ +
Sbjct: 356 ESDEDDDDDSD 366
Score = 28.5 bits (62), Expect = 5.5
Identities = 16/44 (36%), Positives = 23/44 (51%), Gaps = 6/44 (13%)
Query: 182 NKKAKDDDDDDDEGNKVVWQDDDYIRPIKDIKSSEWE-ETVFKD 224
N+K+ DDDD++ + DDD I K +WE E +F D
Sbjct: 257 NRKSNDDDDEEGPRGGDLDMDDDDIE-----KGDDWEHEEIFTD 295
>At3g02810 putative protein kinase
Length = 558
Score = 34.7 bits (78), Expect = 0.077
Identities = 32/125 (25%), Positives = 51/125 (40%), Gaps = 19/125 (15%)
Query: 81 EASSDSDSDSDDENDAPP-PI---NDPYLMTLEERQEWRRKIRQVMDMKPNVQEESDPEE 136
E+ +SDS+S+ E D PP PI N + ++ R W D+ N + S
Sbjct: 424 ESDDESDSNSEHEKDQPPKPIDEKNQAQSLKIKYRYSWE-------DIDVNDERLSSKSS 476
Query: 137 KKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNKKAKDD-----DDD 191
+K E Y D+++ E+ + + I D+ K DD D+D
Sbjct: 477 QKSNDESTSSRYDSDRDQDEKGKEEEEEE---EAEEKHTHIEHIDSSKTDDDQSVYFDND 533
Query: 192 DDEGN 196
DD G+
Sbjct: 534 DDSGD 538
>At1g31870 unknown protein
Length = 561
Score = 34.7 bits (78), Expect = 0.077
Identities = 32/116 (27%), Positives = 49/116 (41%), Gaps = 31/116 (26%)
Query: 130 EESDPEEKKKKMEKLMKDYP------LVVDEEDPNWPEDADGWGFSLGQFFNKITIKDNK 183
E SD EKKKK +K K LVVDE DP W + D
Sbjct: 15 ESSDVVEKKKKKKKQKKPSKPEPRGVLVVDE-DPVWQKQVD------------------- 54
Query: 184 KAKDDDDDDDEGNKVVWQDDDY----IRPIKDIKSSEWEETVFKDISPLIILVHNR 235
++D+++DD + D+D +R +++IK+ + +D S + L NR
Sbjct: 55 -PEEDENEDDSAEETPLVDEDIEVKRMRRLEEIKARRAHNAIAEDGSGWVTLPLNR 109
>At5g60030 KED - like protein
Length = 292
Score = 34.3 bits (77), Expect = 0.10
Identities = 50/202 (24%), Positives = 82/202 (39%), Gaps = 27/202 (13%)
Query: 70 SDGAHRKSGKREASSDSDSDSD--DE---------NDAPPPINDPYLMTLEERQEWRRKI 118
S KS + + SSD+ S+ + DE N + Y +E++ + K
Sbjct: 66 SSDRETKSTETKQSSDAKSERNVIDEFDGRKIRYRNSEAVSVESVYGRERDEKKMKKSKD 125
Query: 119 RQVMDMKPNVQ-------EESDPEEKKKKMEKLMKDYPLVVDEEDPNWPEDADGWGFSLG 171
V+D K N + EE +K+KK +K KD VVDE+ ED
Sbjct: 126 ADVVDEKVNEKLEAEQRSEERRERKKEKKKKKNNKDED-VVDEKVKEKLEDE-------- 176
Query: 172 QFFNKITIKDNKKAKDDDDDDDEGNKVVWQDDDYIRPIKDIKSSEWEETVFKDISPLIIL 231
Q + KK+K ++D+D K +D+ IK+ K ++ E+ V + +
Sbjct: 177 QKSADRKERKKKKSKKNNDEDVVDEKEKLEDEQKSAEIKEKKKNKDEDVVDEKEKEKLED 236
Query: 232 VHNRYRRPKENERIRDELEKAV 253
R KE ++ R E+ V
Sbjct: 237 EQRSGERKKEKKKKRKSDEEIV 258
Score = 28.5 bits (62), Expect = 5.5
Identities = 18/87 (20%), Positives = 40/87 (45%), Gaps = 15/87 (17%)
Query: 56 VGDSKAPRRVQALKSDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLEERQEWR 115
V D K +++ + ++ K+++ ++D D DE +E+ E
Sbjct: 164 VVDEKVKEKLEDEQKSADRKERKKKKSKKNNDEDVVDE---------------KEKLEDE 208
Query: 116 RKIRQVMDMKPNVQEESDPEEKKKKME 142
+K ++ + K N E+ E++K+K+E
Sbjct: 209 QKSAEIKEKKKNKDEDVVDEKEKEKLE 235
>At4g12400 stress-induced protein sti1 -like protein
Length = 558
Score = 34.3 bits (77), Expect = 0.10
Identities = 28/109 (25%), Positives = 52/109 (47%), Gaps = 12/109 (11%)
Query: 50 SNLSLLVGDSKAPRRVQALKSDGAHRKSGKREASSDSDSDSDDENDAPPPINDPYLMTLE 109
+NL+L + D + + + L + SG+ D++ DE P P +P +T E
Sbjct: 165 NNLNLYMKDKRVMKALGVLLNVKFGGSSGE-----DTEMKEADERKEPEPEMEPMELTEE 219
Query: 110 ERQEWRRKIRQVMDMKPNVQEESDPEEKKKKMEKLMKDY--PLVVDEED 156
ERQ+ RK + + + + E + KKK + ++ Y + +D+ED
Sbjct: 220 ERQKKERKEKALKE-----KGEGNVAYKKKDFGRAVEHYTKAMELDDED 263
>At3g01160 unknown protein
Length = 713
Score = 34.3 bits (77), Expect = 0.10
Identities = 42/193 (21%), Positives = 82/193 (41%), Gaps = 14/193 (7%)
Query: 43 FLNLSLSSNLSLLVGDSKAPRRVQALKSDGAHRKSGK---REASSDSDSDSDDENDAPPP 99
F + +L + L D P R++ L + +E + +SDSD+E+D
Sbjct: 356 FQSRALQMSKVNLSWDEDEPHRIKTLNQKFNPEQLANLEMKEFLASDESDSDEEDDLGNE 415
Query: 100 INDPYLMTLEERQEWRRKIR-QVMDMKPNVQEESDPEEKKKKMEKLMKDYPLVVDEEDPN 158
+ + +++ ++R I + +D +++EE+D ++ + ++D + ++ N
Sbjct: 416 VINQSKKKDKKKDKYRALIEAEDVDSDKDLEEEND-QDMEVTFNTGLEDLSKEILKKKDN 474
Query: 159 WPEDADGWGFSLGQFFNKITIKDNKK-----AKDDDDDDDEGNKVVWQ--DDDYIRPIKD 211
E W L Q K + NK+ + DDDDD + K V DDD+
Sbjct: 475 QSESV--WETYLRQRREKKRARKNKQKDDDSSPDDDDDYNIDRKAVKDDGDDDFFMEEPP 532
Query: 212 IKSSEWEETVFKD 224
+K + E K+
Sbjct: 533 LKKKKKEGKTKKE 545
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,968,877
Number of Sequences: 26719
Number of extensions: 446051
Number of successful extensions: 3576
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 163
Number of HSP's that attempted gapping in prelim test: 2886
Number of HSP's gapped (non-prelim): 510
length of query: 342
length of database: 11,318,596
effective HSP length: 100
effective length of query: 242
effective length of database: 8,646,696
effective search space: 2092500432
effective search space used: 2092500432
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0180a.4


