

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.12
(1023 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
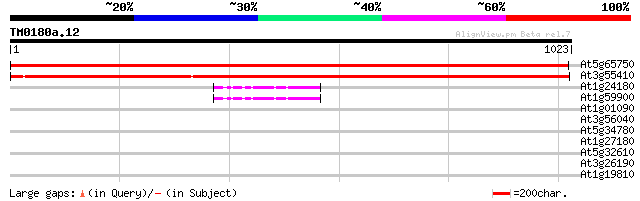
Score E
Sequences producing significant alignments: (bits) Value
At5g65750 2-oxoglutarate dehydrogenase, E1 component 1753 0.0
At3g55410 2-oxoglutarate dehydrogenase, E1 subunit - like protein 1744 0.0
At1g24180 pyruvate dehydrogenase E1 alpha subunit 45 2e-04
At1g59900 pyruvate dehydrogenase E1 alpha subunit, putative 44 5e-04
At1g01090 pyruvate dehydrogenase E1 alpha subunit 36 0.098
At3g56040 unknown protein 34 0.48
At5g34780 3-methyl-2-oxobutanoate dehydrogenase-like protein 31 3.1
At1g27180 disease resistance protein, putative 30 5.4
At5g32610 putative protein 30 7.0
At3g26190 cytochrome P450, putative 30 7.0
At1g19810 AAA ATPase, putative 30 9.1
>At5g65750 2-oxoglutarate dehydrogenase, E1 component
Length = 1025
Score = 1753 bits (4540), Expect = 0.0
Identities = 841/1021 (82%), Positives = 934/1021 (91%), Gaps = 3/1021 (0%)
Query: 1 MAWFRAAALSIAKHAIRRNLFRGGSSYYVTRTTNLPSTSRKLHTTIFKSEAH-AAPVPRP 59
M WFR + S+AK AIRR L + Y TRT LP +R H+TI KS+A AAPVPRP
Sbjct: 1 MVWFRIGS-SVAKLAIRRTLSQSRCGSYATRTRVLPCQTRCFHSTILKSKAESAAPVPRP 59
Query: 60 VPLSRLTDNFLDGTSSAYLEELQRAWEADPSSVDESWDNFFRNFVGQASTSPGISGQTIQ 119
VPLS+LTD+FLDGTSS YLEELQRAWEADP+SVDESWDNFFRNFVGQASTSPGISGQTIQ
Sbjct: 60 VPLSKLTDSFLDGTSSVYLEELQRAWEADPNSVDESWDNFFRNFVGQASTSPGISGQTIQ 119
Query: 120 ESMRLLLLVRAYQVNGHMKAKLDPLGLKERNIPDELDPGLYGFTEADLDREFFLGVWNMS 179
ESMRLLLLVRAYQVNGHMKAKLDPLGL++R IP++L PGLYGFTEADLDREFFLGVW MS
Sbjct: 120 ESMRLLLLVRAYQVNGHMKAKLDPLGLEKREIPEDLTPGLYGFTEADLDREFFLGVWRMS 179
Query: 180 GFLSENRPVQTLRSILTRLEQAYCGSIGYEYMHIPDRDKCNWLRDKIETPSPTQFSRERR 239
GFLSENRPVQTLRSIL+RLEQAYCG+IGYEYMHI DRDKCNWLRDKIETP+P Q++ ERR
Sbjct: 180 GFLSENRPVQTLRSILSRLEQAYCGTIGYEYMHIADRDKCNWLRDKIETPTPRQYNSERR 239
Query: 240 EVIFDRLAWSTLFENFLATKWTSAKRFGLEGGETLIPGMKEMFDRASDLGVENIVMGMAH 299
VI+DRL WST FENFLATKWT+AKRFGLEG E+LIPGMKEMFDR++DLGVENIV+GM H
Sbjct: 240 MVIYDRLTWSTQFENFLATKWTTAKRFGLEGAESLIPGMKEMFDRSADLGVENIVIGMPH 299
Query: 300 RGRLNVLGNVVRKPLRQIFCEFSGGL-PQDEVGLYTGTGDVKYHLGTSYDRPTRGGRRIH 358
RGRLNVLGNVVRKPLRQIF EFSGG P DEVGLYTGTGDVKYHLGTSYDRPTRGG+ +H
Sbjct: 300 RGRLNVLGNVVRKPLRQIFSEFSGGTRPVDEVGLYTGTGDVKYHLGTSYDRPTRGGKHLH 359
Query: 359 LSLMANPSHLEAVNPLVVGKTRAKQYYSNDAERMKNMGVLIHGDGSFAGQGVVYETLHLS 418
LSL+ANPSHLEAV+P+V+GKTRAKQYY+ D R KNMG+LIHGDGSFAGQGVVYETLHLS
Sbjct: 360 LSLVANPSHLEAVDPVVIGKTRAKQYYTKDENRTKNMGILIHGDGSFAGQGVVYETLHLS 419
Query: 419 ALPNYTTGGTIHIVFNNQVAFTTDPESGRSSQYSTDVAKALNAPIFHVNGDDVESVVHVC 478
ALPNY TGGT+HIV NNQVAFTTDP GRSSQY TDVAKAL+APIFHVN DD+E+VVH C
Sbjct: 420 ALPNYCTGGTVHIVVNNQVAFTTDPREGRSSQYCTDVAKALSAPIFHVNADDIEAVVHAC 479
Query: 479 ELAAEWRQTFHSDVVVDLVCYRRFGHNEIDEPSFTQPKMYKVIRNHPSALEIYQKKLLEL 538
ELAAEWRQTFHSDVVVDLVCYRRFGHNEIDEPSFTQPKMYKVIR+HPS+L+IYQ+KLL+
Sbjct: 480 ELAAEWRQTFHSDVVVDLVCYRRFGHNEIDEPSFTQPKMYKVIRSHPSSLQIYQEKLLQS 539
Query: 539 GELTQEDIDKIHKKVTSILNDEFLASKDYIPKRRDWLSAYWSGFKSPEQLSRIRNTGVKP 598
G++TQEDIDKI KKV+SILN+E+ ASKDYIP++RDWL+++W+GFKSPEQ+SRIRNTGVKP
Sbjct: 540 GQVTQEDIDKIQKKVSSILNEEYEASKDYIPQKRDWLASHWTGFKSPEQISRIRNTGVKP 599
Query: 599 DILKNVGKAITALPESLNPHRAVKKVYEQRAQMVETGEDIDWGFAEALAFATLIVEGNHV 658
+ILKNVGKAI+ PE+ PHR VK+VYEQRAQM+E+GE IDWG EALAFATL+VEGNHV
Sbjct: 600 EILKNVGKAISTFPENFKPHRGVKRVYEQRAQMIESGEGIDWGLGEALAFATLVVEGNHV 659
Query: 659 RLSGQDVERGTFSHRHAVVHDQATGEKYCPLDHVIMNQNEEMFTVSNSSLSEFGVLGFEL 718
RLSGQDVERGTFSHRH+V+HDQ TGE+YCPLDH+I NQ+ EMFTVSNSSLSEFGVLGFEL
Sbjct: 660 RLSGQDVERGTFSHRHSVLHDQETGEEYCPLDHLIKNQDPEMFTVSNSSLSEFGVLGFEL 719
Query: 719 GYSMENPNSLVIWEAQFGDFANGAQVIFDNFLSSGESKWLRQTGLVVLLPHGYDGQGPEH 778
GYSMENPNSLVIWEAQFGDFANGAQV+FD F+SSGE+KWLRQTGLVVLLPHGYDGQGPEH
Sbjct: 720 GYSMENPNSLVIWEAQFGDFANGAQVMFDQFISSGEAKWLRQTGLVVLLPHGYDGQGPEH 779
Query: 779 SSGRLERYLQMADDHPYIIPEMDPTLRKQIQECNLQIVNVTTPANFFHVLRRQIHREFRK 838
SSGRLER+LQM+DD+PY+IPEMDPTLRKQIQECN Q+VNVTTPAN+FHVLRRQIHR+FRK
Sbjct: 780 SSGRLERFLQMSDDNPYVIPEMDPTLRKQIQECNWQVVNVTTPANYFHVLRRQIHRDFRK 839
Query: 839 PLIVMSPKNLLRSKVCRSNLSEFDDVQGHPGFDKQGTRFKRLIKDQNDHSNVEEGIRRLI 898
PLIVM+PKNLLR K C SNLSEFDDV+GHPGFDKQGTRFKRLIKDQ+ HS++EEGIRRL+
Sbjct: 840 PLIVMAPKNLLRHKQCVSNLSEFDDVKGHPGFDKQGTRFKRLIKDQSGHSDLEEGIRRLV 899
Query: 899 LCSGKVYYELDEQRTKDDAKDVAICRVEQLCPFPYDLVQRELKRYPNAEVVWCQEEPMNM 958
LCSGKVYYELDE+R K + KDVAICRVEQLCPFPYDL+QRELKRYPNAE+VWCQEEPMNM
Sbjct: 900 LCSGKVYYELDEERKKSETKDVAICRVEQLCPFPYDLIQRELKRYPNAEIVWCQEEPMNM 959
Query: 959 GGYSYILPRLITSMKALGRGGYEDVKYVGRAPSAATATGFLKVHQREQAELVHKAMQHKP 1018
GGY YI RL T+MKAL RG + D+KYVGR PSAATATGF ++H +EQ +LV KA+Q P
Sbjct: 960 GGYQYIALRLCTAMKALQRGNFNDIKYVGRLPSAATATGFYQLHVKEQTDLVKKALQPDP 1019
Query: 1019 I 1019
I
Sbjct: 1020 I 1020
>At3g55410 2-oxoglutarate dehydrogenase, E1 subunit - like protein
Length = 1017
Score = 1744 bits (4517), Expect = 0.0
Identities = 836/1021 (81%), Positives = 940/1021 (91%), Gaps = 5/1021 (0%)
Query: 1 MAWFRAAALSIAKHAIRRNLFRGGSSYYVTRTTNLPSTSRKLHTTIFKSEAHAAPVPRPV 60
M WFRA + S+ K A+RR L +G S Y TRT ++PS +R H+TI + +A +APVPR V
Sbjct: 1 MVWFRAGS-SVTKLAVRRILNQGAS--YATRTRSIPSQTRSFHSTICRPKAQSAPVPRAV 57
Query: 61 PLSRLTDNFLDGTSSAYLEELQRAWEADPSSVDESWDNFFRNFVGQASTSPGISGQTIQE 120
PLS+LTD+FLDGTSS YLEELQRAWEADP+SVDESWDNFFRNFVGQA+TSPGISGQTIQE
Sbjct: 58 PLSKLTDSFLDGTSSVYLEELQRAWEADPNSVDESWDNFFRNFVGQAATSPGISGQTIQE 117
Query: 121 SMRLLLLVRAYQVNGHMKAKLDPLGLKERNIPDELDPGLYGFTEADLDREFFLGVWNMSG 180
SMRLLLLVRAYQVNGHMKAKLDPLGL++R IP++LD LYGFTEADLDREFFLGVW MSG
Sbjct: 118 SMRLLLLVRAYQVNGHMKAKLDPLGLEQREIPEDLDLALYGFTEADLDREFFLGVWQMSG 177
Query: 181 FLSENRPVQTLRSILTRLEQAYCGSIGYEYMHIPDRDKCNWLRDKIETPSPTQFSRERRE 240
F+SENRPVQTLRSILTRLEQAYCG+IG+EYMHI DRDKCNWLR+KIETP+P +++RERRE
Sbjct: 178 FMSENRPVQTLRSILTRLEQAYCGNIGFEYMHIADRDKCNWLREKIETPTPWRYNRERRE 237
Query: 241 VIFDRLAWSTLFENFLATKWTSAKRFGLEGGETLIPGMKEMFDRASDLGVENIVMGMAHR 300
VI DRLAWST FENFLATKWT+AKRFGLEGGE+LIPGMKEMFDRA+DLGVE+IV+GM+HR
Sbjct: 238 VILDRLAWSTQFENFLATKWTTAKRFGLEGGESLIPGMKEMFDRAADLGVESIVIGMSHR 297
Query: 301 GRLNVLGNVVRKPLRQIFCEFSGGL-PQDEVGLYTGTGDVKYHLGTSYDRPTRGGRRIHL 359
GRLNVLGNVVRKPLRQIF EFSGG+ P DEVG YTGTGDVKYHLGTSYDRPTRGG++IHL
Sbjct: 298 GRLNVLGNVVRKPLRQIFSEFSGGIRPVDEVG-YTGTGDVKYHLGTSYDRPTRGGKKIHL 356
Query: 360 SLMANPSHLEAVNPLVVGKTRAKQYYSNDAERMKNMGVLIHGDGSFAGQGVVYETLHLSA 419
SL+ANPSHLEA + +VVGKTRAKQYYSND +R KN+G+LIHGDGSFAGQGVVYETLHLSA
Sbjct: 357 SLVANPSHLEAADSVVVGKTRAKQYYSNDLDRTKNLGILIHGDGSFAGQGVVYETLHLSA 416
Query: 420 LPNYTTGGTIHIVFNNQVAFTTDPESGRSSQYSTDVAKALNAPIFHVNGDDVESVVHVCE 479
LPNYTTGGTIHIV NNQVAFTTDP +GRSSQY TDVAKAL+APIFHVNGDDVE+VVH CE
Sbjct: 417 LPNYTTGGTIHIVVNNQVAFTTDPRAGRSSQYCTDVAKALSAPIFHVNGDDVEAVVHACE 476
Query: 480 LAAEWRQTFHSDVVVDLVCYRRFGHNEIDEPSFTQPKMYKVIRNHPSALEIYQKKLLELG 539
LAAEWRQTFHSDVVVDLVCYRRFGHNEIDEPSFTQPKMYKVI+NHPS L+IY KKLLE G
Sbjct: 477 LAAEWRQTFHSDVVVDLVCYRRFGHNEIDEPSFTQPKMYKVIKNHPSTLQIYHKKLLECG 536
Query: 540 ELTQEDIDKIHKKVTSILNDEFLASKDYIPKRRDWLSAYWSGFKSPEQLSRIRNTGVKPD 599
E++Q+DID+I +KV +ILN+EF+ASKDY+PK+RDWLS W+GFKSPEQ+SR+RNTGVKP+
Sbjct: 537 EVSQQDIDRIQEKVNTILNEEFVASKDYLPKKRDWLSTNWAGFKSPEQISRVRNTGVKPE 596
Query: 600 ILKNVGKAITALPESLNPHRAVKKVYEQRAQMVETGEDIDWGFAEALAFATLIVEGNHVR 659
ILK VGKAI++LPE+ PHRAVKKVYEQRAQM+E+GE +DW AEALAFATL+VEGNHVR
Sbjct: 597 ILKTVGKAISSLPENFKPHRAVKKVYEQRAQMIESGEGVDWALAEALAFATLVVEGNHVR 656
Query: 660 LSGQDVERGTFSHRHAVVHDQATGEKYCPLDHVIMNQNEEMFTVSNSSLSEFGVLGFELG 719
LSGQDVERGTFSHRH+V+HDQ TGE+YCPLDH+IMNQ+ EMFTVSNSSLSEFGVLGFELG
Sbjct: 657 LSGQDVERGTFSHRHSVLHDQETGEEYCPLDHLIMNQDPEMFTVSNSSLSEFGVLGFELG 716
Query: 720 YSMENPNSLVIWEAQFGDFANGAQVIFDNFLSSGESKWLRQTGLVVLLPHGYDGQGPEHS 779
YSME+PNSLV+WEAQFGDFANGAQVIFD F+SSGE+KWLRQTGLV+LLPHGYDGQGPEHS
Sbjct: 717 YSMESPNSLVLWEAQFGDFANGAQVIFDQFISSGEAKWLRQTGLVMLLPHGYDGQGPEHS 776
Query: 780 SGRLERYLQMADDHPYIIPEMDPTLRKQIQECNLQIVNVTTPANFFHVLRRQIHREFRKP 839
S RLERYLQM+DD+PY+IP+M+PT+RKQIQECN QIVN TTPAN+FHVLRRQIHR+FRKP
Sbjct: 777 SARLERYLQMSDDNPYVIPDMEPTMRKQIQECNWQIVNATTPANYFHVLRRQIHRDFRKP 836
Query: 840 LIVMSPKNLLRSKVCRSNLSEFDDVQGHPGFDKQGTRFKRLIKDQNDHSNVEEGIRRLIL 899
LIVM+PKNLLR K C+SNLSEFDDVQGHPGFDKQGTRFKRLIKDQNDHS++EEGIRRL+L
Sbjct: 837 LIVMAPKNLLRHKDCKSNLSEFDDVQGHPGFDKQGTRFKRLIKDQNDHSDLEEGIRRLVL 896
Query: 900 CSGKVYYELDEQRTKDDAKDVAICRVEQLCPFPYDLVQRELKRYPNAEVVWCQEEPMNMG 959
CSGKVYYELD++R K A DVAICRVEQLCPFPYDL+QRELKRYPNAE+VWCQEE MNMG
Sbjct: 897 CSGKVYYELDDERKKVGATDVAICRVEQLCPFPYDLIQRELKRYPNAEIVWCQEEAMNMG 956
Query: 960 GYSYILPRLITSMKALGRGGYEDVKYVGRAPSAATATGFLKVHQREQAELVHKAMQHKPI 1019
+SYI PRL T+M+++ RG ED+KYVGR PSAATATGF H +EQA LV KA+ +PI
Sbjct: 957 AFSYISPRLWTAMRSVNRGDMEDIKYVGRGPSAATATGFYTFHVKEQAGLVQKAIGKEPI 1016
Query: 1020 N 1020
N
Sbjct: 1017 N 1017
>At1g24180 pyruvate dehydrogenase E1 alpha subunit
Length = 393
Score = 45.4 bits (106), Expect = 2e-04
Identities = 46/196 (23%), Positives = 92/196 (46%), Gaps = 15/196 (7%)
Query: 373 PLVVGKTRAKQYYSNDAERMKNMGVLIHGDGSFAGQGVVYETLHLSALPNYTTGGTIHIV 432
PL G A++Y ++A + ++GDG+ A QG ++E L++SAL + I +
Sbjct: 176 PLGCGLAFAQKYNKDEA-----VTFALYGDGA-ANQGQLFEALNISALWDLP---AILVC 226
Query: 433 FNNQVAFTTDPESGRSSQYSTDVAKALNAPIFHVNGDDVESVVHVCELAAEWRQTFHSDV 492
NN T + RS++ + P V+G D +V C+ A E + +
Sbjct: 227 ENNHYGMGT--ATWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKE-HALKNGPI 283
Query: 493 VVDLVCYRRFGHNEIDEPSFTQPKMYKV--IRNHPSALEIYQKKLLELGELTQEDIDKIH 550
++++ YR GH+ + +P T ++ +R +E +K LL T++++ +
Sbjct: 284 ILEMDTYRYHGHS-MSDPGSTYRTRDEISGVRQVRDPIERVRKLLLTHDIATEKELKDME 342
Query: 551 KKVTSILNDEFLASKD 566
K++ ++D +K+
Sbjct: 343 KEIRKEVDDAVAQAKE 358
>At1g59900 pyruvate dehydrogenase E1 alpha subunit, putative
Length = 389
Score = 43.9 bits (102), Expect = 5e-04
Identities = 44/196 (22%), Positives = 90/196 (45%), Gaps = 15/196 (7%)
Query: 373 PLVVGKTRAKQYYSNDAERMKNMGVLIHGDGSFAGQGVVYETLHLSALPNYTTGGTIHIV 432
PL G A++Y +A + ++GDG+ A QG ++E L++SAL + I +
Sbjct: 172 PLGCGIAFAQKYNKEEA-----VTFALYGDGA-ANQGQLFEALNISALWDLP---AILVC 222
Query: 433 FNNQVAFTTDPESGRSSQYSTDVAKALNAPIFHVNGDDVESVVHVCELAAEWRQTFHSDV 492
NN T R+++ + + P V+G D +V C+ A + +
Sbjct: 223 ENNHYGMGT--AEWRAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQ-HALEKGPI 279
Query: 493 VVDLVCYRRFGHNEIDEPSFTQPKMYKV--IRNHPSALEIYQKKLLELGELTQEDIDKIH 550
++++ YR GH+ + +P T ++ +R +E +K +L T++++ +
Sbjct: 280 ILEMDTYRYHGHS-MSDPGSTYRTRDEISGVRQERDPIERIKKLVLSHDLATEKELKDME 338
Query: 551 KKVTSILNDEFLASKD 566
K++ ++D +KD
Sbjct: 339 KEIRKEVDDAIAKAKD 354
>At1g01090 pyruvate dehydrogenase E1 alpha subunit
Length = 428
Score = 36.2 bits (82), Expect = 0.098
Identities = 45/172 (26%), Positives = 71/172 (41%), Gaps = 23/172 (13%)
Query: 397 VLIHGDGSFAGQGVVYETLHLSALPNYTTGGTIHIVFNNQVAF-------TTDPESGRSS 449
V GDG+ G +E L+++AL I +V NN A T+DPE +
Sbjct: 222 VAFFGDGT-CNNGQFFECLNMAALYKLPI---IFVVENNLWAIGMSHLRATSDPEIWKKG 277
Query: 450 QYSTDVAKALNAPIFHVNGDDVESVVHVCELAAEWRQTFHSDVVVDLVCYRRFGHN--EI 507
A P HV+G DV V V + A + +V+ YR GH+ +
Sbjct: 278 P-------AFGMPGVHVDGMDVLKVREVAKEAVTRARRGEGPTLVECETYRFRGHSLADP 330
Query: 508 DEPSFTQPKMYKVIRNHPSALEIYQKKLLELGELTQEDIDKIHKKVTSILND 559
DE K R+ +AL+ Y L+E + ++ I KK+ ++ +
Sbjct: 331 DELRDAAEKAKYAARDPIAALKKY---LIENKLAKEAELKSIEKKIDELVEE 379
>At3g56040 unknown protein
Length = 883
Score = 33.9 bits (76), Expect = 0.48
Identities = 13/56 (23%), Positives = 29/56 (51%)
Query: 367 HLEAVNPLVVGKTRAKQYYSNDAERMKNMGVLIHGDGSFAGQGVVYETLHLSALPN 422
++ VN + +++ Y+ ND R++ +++HG+ F V E H+ +P+
Sbjct: 777 NVNVVNRGIDWNSKSNVYWRNDVNRLETCKIILHGNAEFEASNVTIEGHHVFEVPD 832
>At5g34780 3-methyl-2-oxobutanoate dehydrogenase-like protein
Length = 365
Score = 31.2 bits (69), Expect = 3.1
Identities = 41/197 (20%), Positives = 81/197 (40%), Gaps = 16/197 (8%)
Query: 375 VVGKTRAKQYYSNDAERMKNMGVLIHGDGSFAGQGVVYETLHLSALPNYTTGGTIHIVFN 434
V+G ++AK + +A V GDG +G + L+ +A+ + I N
Sbjct: 14 VIGLSKAKDCWEKNA-----CAVTFIGDGG-TSEGDFHAGLNFAAVME---APVVFICRN 64
Query: 435 NQVAFTTDPESGRSSQYSTDVAKALNAPIFHVNGDDVESVVHVCELAAEWRQTFHSDVVV 494
N A +T S +A V+G+D +V A E T V++
Sbjct: 65 NGWAISTHISEQFRSDGIVVKGQAYGIRSIRVDGNDALAVYSAVCSAREMAVTEQRPVLI 124
Query: 495 DLVCYRRFGHNEIDEP----SFTQPKMYKVIRNHPSALEIYQKKLLELGELTQEDIDKIH 550
+++ YR H+ D+ + + + +K+ RN ++ ++K + + G ++ED K+
Sbjct: 125 EMMIYRVGHHSTSDDSTKYRAADEIQYWKMSRN---SVNRFRKSVEDNGWWSEEDESKLR 181
Query: 551 KKVTSILNDEFLASKDY 567
L A++ +
Sbjct: 182 SNARKQLLQAIQAAEKW 198
>At1g27180 disease resistance protein, putative
Length = 1556
Score = 30.4 bits (67), Expect = 5.4
Identities = 31/137 (22%), Positives = 54/137 (38%), Gaps = 32/137 (23%)
Query: 68 NFLDGTSSAYLEELQRAWEADPSSVDESWDNFFRNFVGQASTSPGISGQTIQESMRLLLL 127
NF D A ++E R W D VD D+ R + +A I++S+ +++
Sbjct: 30 NFTDRLYEALVKEELRVWNDDLERVDHDHDHELRPSLVEA----------IEDSVAFVVV 79
Query: 128 VRAYQVNGHMKA------------------KLDPLGLKERNIPDELD----PGLYGFTEA 165
+ N H++ K++P +KE+N P E D +G +
Sbjct: 80 LSPNYANSHLRLEELAKLCDLKCLMVPIFYKVEPREVKEQNGPFEKDFEEHSKRFGEEKI 139
Query: 166 DLDREFFLGVWNMSGFL 182
+ V N+SGF+
Sbjct: 140 QRWKGAMTTVGNISGFI 156
>At5g32610 putative protein
Length = 662
Score = 30.0 bits (66), Expect = 7.0
Identities = 18/76 (23%), Positives = 36/76 (46%), Gaps = 7/76 (9%)
Query: 872 KQGTRFKRLIKDQNDHSNVEEGIRRLILCSGKVYYE-------LDEQRTKDDAKDVAICR 924
+QG + +K+ +D S + +RR+ + S + Y E LD++ ++D+ +C
Sbjct: 560 RQGKVEENQVKNIDDQSKIWASLRRVTMDSSRKYVELLDRILDLDKKSKEEDSSPYVLCD 619
Query: 925 VEQLCPFPYDLVQREL 940
P DL + E+
Sbjct: 620 AVTTYQEPRDLKEGEI 635
>At3g26190 cytochrome P450, putative
Length = 499
Score = 30.0 bits (66), Expect = 7.0
Identities = 15/53 (28%), Positives = 28/53 (52%)
Query: 664 DVERGTFSHRHAVVHDQATGEKYCPLDHVIMNQNEEMFTVSNSSLSEFGVLGF 716
D+ + FS+ ++V A G+ + D V M++ EE+ S ++L F + F
Sbjct: 166 DLRKALFSYTASIVCRLAFGQNFHECDFVDMDKVEELVLESETNLGSFAFIDF 218
>At1g19810 AAA ATPase, putative
Length = 592
Score = 29.6 bits (65), Expect = 9.1
Identities = 18/53 (33%), Positives = 28/53 (51%), Gaps = 1/53 (1%)
Query: 309 VVRKPLRQIFCEFSGGLPQDEVGLYTGTGDVKYHLGTSYDRPTRGGRRIHLSL 361
VV +PL Q+ E SGG +D V + T + + + RP R G+ I++ L
Sbjct: 462 VVERPLTQLLNEMSGGKERDGVFVIGATNRPEM-MDPAITRPGRFGKHIYIPL 513
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,469,399
Number of Sequences: 26719
Number of extensions: 1124113
Number of successful extensions: 2492
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 2483
Number of HSP's gapped (non-prelim): 13
length of query: 1023
length of database: 11,318,596
effective HSP length: 109
effective length of query: 914
effective length of database: 8,406,225
effective search space: 7683289650
effective search space used: 7683289650
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0180a.12


