

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0177.9
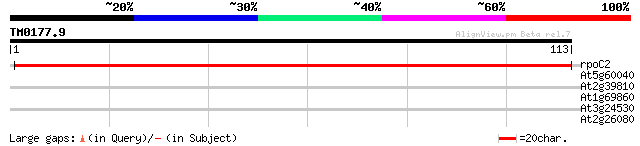
(113 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
rpoC2 -chloroplast genome- RNA polymerase beta' subunit-2 193 1e-50
At5g60040 DNA-directed RNA polymerase - like protein 30 0.20
At2g39810 unknown protein 26 2.8
At1g69860 putative peptide transporter 26 2.8
At3g24530 unknown protein 25 4.8
At2g26080 putative glycine dehydrogenase 25 6.3
>rpoC2 -chloroplast genome- RNA polymerase beta' subunit-2
Length = 1376
Score = 193 bits (491), Expect = 1e-50
Identities = 100/112 (89%), Positives = 107/112 (95%)
Query: 2 QSRISLVNKIQRVYRSQGVHIHNRHIEIIVRQITSKVLVSEDGMSNVFSLGELIGLLRAQ 61
QSRISLVNKIQ+VYRSQGV IHNRHIEIIVRQITSKVLVSE+GMSNVF GELIGLLRA+
Sbjct: 1191 QSRISLVNKIQKVYRSQGVQIHNRHIEIIVRQITSKVLVSEEGMSNVFLPGELIGLLRAE 1250
Query: 62 RTGRALEESICYRTILLGITKTSMNTQSFISEASFQETARVLAKAALRGRID 113
RTGRALEE+ICYR +LLGIT+ S+NTQSFISEASFQETARVLAKAALRGRID
Sbjct: 1251 RTGRALEEAICYRAVLLGITRASLNTQSFISEASFQETARVLAKAALRGRID 1302
>At5g60040 DNA-directed RNA polymerase - like protein
Length = 1328
Score = 30.0 bits (66), Expect = 0.20
Identities = 17/49 (34%), Positives = 28/49 (56%), Gaps = 2/49 (4%)
Query: 67 LEESICYRTILLGITKTSMNT--QSFISEASFQETARVLAKAALRGRID 113
L + + YR +LGI +T + +S + +ASF+ T L AA G++D
Sbjct: 1237 LADVMTYRGEVLGIQRTGIQKMDKSVLMQASFERTGDHLFSAAASGKVD 1285
>At2g39810 unknown protein
Length = 927
Score = 26.2 bits (56), Expect = 2.8
Identities = 14/44 (31%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query: 43 DGMSNVFSLGELIGLLRAQRTGRALEESICY-RTILLGITKTSM 85
DG+S + S+GE + LR + L E+ Y RT+ L + + ++
Sbjct: 492 DGVSELVSIGEAVTALRVRVECGLLSEAFTYQRTLCLKVKENNL 535
>At1g69860 putative peptide transporter
Length = 555
Score = 26.2 bits (56), Expect = 2.8
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query: 25 RHIEIIVRQITSKVLVSEDGMSNVFSLGELI--GLLRAQRTGRAL 67
RHIE I +Q L+ + G+ N+FS+ ++ G++ +R +L
Sbjct: 390 RHIENITKQNGGISLLQKVGIGNIFSISTMLISGIVERKRRDLSL 434
>At3g24530 unknown protein
Length = 481
Score = 25.4 bits (54), Expect = 4.8
Identities = 23/72 (31%), Positives = 33/72 (44%), Gaps = 4/72 (5%)
Query: 36 SKVLVSEDGMSNVFSLGELIGLLRAQRTGRALEESICYRTILLGITKTSMNTQSFISEAS 95
+K+ + ED +SN+ L EL LR G L+E R + L I +F+
Sbjct: 198 AKMELLEDELSNIVGLSELKTQLRKWAKGMLLDER--RRALGLNIGTRRPPHMAFLGNPG 255
Query: 96 FQET--ARVLAK 105
+T ARVL K
Sbjct: 256 TGKTMVARVLGK 267
>At2g26080 putative glycine dehydrogenase
Length = 1044
Score = 25.0 bits (53), Expect = 6.3
Identities = 11/45 (24%), Positives = 22/45 (48%)
Query: 67 LEESICYRTILLGITKTSMNTQSFISEASFQETARVLAKAALRGR 111
LE + Y+T++ +T M+ S + E + A + L+G+
Sbjct: 197 LESLLNYQTVITDLTGLPMSNASLLDEGTAAAEAMAMCNNILKGK 241
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.134 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,803,582
Number of Sequences: 26719
Number of extensions: 56910
Number of successful extensions: 127
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 123
Number of HSP's gapped (non-prelim): 6
length of query: 113
length of database: 11,318,596
effective HSP length: 89
effective length of query: 24
effective length of database: 8,940,605
effective search space: 214574520
effective search space used: 214574520
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0177.9


