

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0177.6
(1032 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
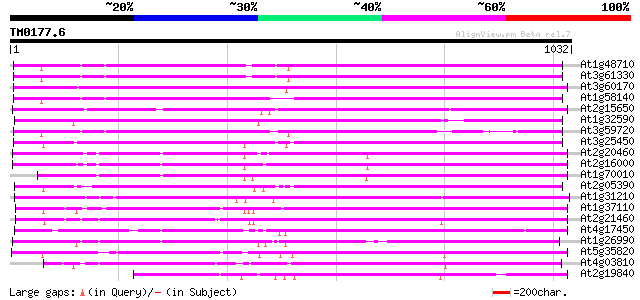
Score E
Sequences producing significant alignments: (bits) Value
At1g48710 hypothetical protein 751 0.0
At3g61330 copia-type polyprotein 751 0.0
At3g60170 putative protein 751 0.0
At1g58140 hypothetical protein 744 0.0
At2g15650 putative retroelement pol polyprotein 711 0.0
At1g32590 hypothetical protein, 5' partial 659 0.0
At3g59720 copia-type reverse transcriptase-like protein 643 0.0
At3g25450 hypothetical protein 634 0.0
At2g20460 putative retroelement pol polyprotein 630 e-180
At2g16000 putative retroelement pol polyprotein 622 e-178
At1g70010 hypothetical protein 580 e-165
At2g05390 putative retroelement pol polyprotein 574 e-163
At1g31210 putative reverse transcriptase 572 e-163
At1g37110 546 e-155
At2g21460 putative retroelement pol polyprotein 540 e-153
At4g17450 retrotransposon like protein 531 e-150
At1g26990 polyprotein, putative 530 e-150
At5g35820 copia-like retrotransposable element 526 e-149
At4g03810 putative retrotransposon protein 488 e-138
At2g19840 copia-like retroelement pol polyprotein 474 e-133
>At1g48710 hypothetical protein
Length = 1352
Score = 751 bits (1939), Expect = 0.0
Identities = 410/1019 (40%), Positives = 579/1019 (56%), Gaps = 26/1019 (2%)
Query: 7 KHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV----DSSPC 62
++ WYLDSG S HM G + MF EL G V G K ++ G G I +
Sbjct: 331 ENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQF 390
Query: 63 IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLS 122
I NV + + N+LS+ QL +KGYD+ + Q + KN ++ + +
Sbjct: 391 ISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLNIR 450
Query: 123 ELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQK 182
AQ +K + EE W+WH R GH + + LS+ +VRGLP + + +CE C
Sbjct: 451 NDIAQCLK--MCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQ-VCEGCLL 507
Query: 183 DKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTR 242
K K+ F ++ +SLEL+H D+ GP+K +S+G Y ++ +DD+SR TWV FL
Sbjct: 508 GKQFKMSFPKESSSRAQKSLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKE 567
Query: 243 KDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQ 302
K E +F F A V+ E I +RSD GG+F + +F + GI + PR+PQQ
Sbjct: 568 KSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQ 627
Query: 303 NGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNI 362
NGV ERKNRT+ EMAR+ML+ + K WAEAV A Y+ NR + + KTP E W
Sbjct: 628 NGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGR 687
Query: 363 KPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESIH 422
K +S+ FG + + ++ K D KS K + +GY SKG++ YN D K S +
Sbjct: 688 KSGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRN 747
Query: 423 VRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSRIT 482
+ FD++ + D + N D P E +E EP E PS T + T
Sbjct: 748 IVFDEEGEWDWNS----------NEEDYNFFPH-FEEDEPEPTREEPPSEEPTTPPTSPT 796
Query: 483 AAHPKELIMGNKDEPVRTRSAFRPSEETL----LSLKGLVSLIEPKSIDEALQDKDWILA 538
++ +E + + R RS E T L+L L + EP EA++ K W A
Sbjct: 797 SSQIEE---SSSERTPRFRSIQELYEVTENQENLTLFCLFAECEPMDFQEAIEKKTWRNA 853
Query: 539 MEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGI 598
M+EE+ KND W L P IG KWV++ K N KG+V R KARLVA+GY Q+ GI
Sbjct: 854 MDEEIKSIQKNDTWELTSLPNGHKTIGVKWVYKAKKNSKGEVERYKARLVAKGYIQRAGI 913
Query: 599 DYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQPPGFEDEKK 658
DY E FAPVARLE +RL+IS + + +HQ+DVKSAFLNG + EEVY+ QP G+ + +
Sbjct: 914 DYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGE 973
Query: 659 PDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKDDILIVQIYVD 718
D V +LKK+LYGLKQAPRAW R+ + E +F + + L+ K K+DILI +YVD
Sbjct: 974 EDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVD 1033
Query: 719 DIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKK 778
D+IF N S+ +EF + M EFEM+ +G + Y+LGI+V Q G +I Q Y KE+LKK
Sbjct: 1034 DLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKK 1093
Query: 779 FNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRPDILFSVHLCARF 838
F M +S TPM L K+++ V ++ +GSL YLT +RPDIL++V + +R+
Sbjct: 1094 FKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRY 1153
Query: 839 *SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQ 898
P TH A KRILRY+KGT N GL Y TS+YKL GY D+D+ GD +RKSTSG
Sbjct: 1154 MEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVF 1213
Query: 899 FLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQI-LESNIPIYCDN 957
++G +W SK+Q + LST EAEY++A C +W+++ L++ + E I+ DN
Sbjct: 1214 YIGDTAFTWMSKKQPIVVLSTCEAEYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDN 1273
Query: 958 TAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRF 1016
+AI+L+KNP+ H R+KHI+ +YH+IR+ V K + L++V T Q ADIFTKPL + F
Sbjct: 1274 KSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIFTKPLKREDF 1332
>At3g61330 copia-type polyprotein
Length = 1352
Score = 751 bits (1938), Expect = 0.0
Identities = 409/1019 (40%), Positives = 579/1019 (56%), Gaps = 26/1019 (2%)
Query: 7 KHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV----DSSPC 62
++ WYLDSG S HM G + MF EL G V G K ++ G G I +
Sbjct: 331 ENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQF 390
Query: 63 IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLS 122
I NV + + N+LS+ QL +KGYD+ + Q + KN ++ + +
Sbjct: 391 ISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLNIR 450
Query: 123 ELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQK 182
AQ +K + EE W+WH R GH + + LS+ +VRGLP + + +CE C
Sbjct: 451 NDIAQCLK--MCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQ-VCEGCLL 507
Query: 183 DKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTR 242
K K+ F ++ + LEL+H D+ GP+K +S+G Y ++ +DD+SR TWV FL
Sbjct: 508 GKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKE 567
Query: 243 KDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQ 302
K E +F F A V+ E I +RSD GG+F + +F + GI + PR+PQQ
Sbjct: 568 KSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQ 627
Query: 303 NGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNI 362
NGVVERKNRT+ EMAR+ML+ + K WAEAV A Y+ NR + + KTP E W
Sbjct: 628 NGVVERKNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGR 687
Query: 363 KPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESIH 422
KP +S+ FG + + ++ K D KS K + +GY SKG++ YN D K S +
Sbjct: 688 KPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRN 747
Query: 423 VRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSRIT 482
+ FD++ + D + N D P E +E EP E PS T + T
Sbjct: 748 IVFDEEGEWDWNS----------NEEDYNFFPH-FEEDEPEPTREEPPSEEPTTPPTSPT 796
Query: 483 AAHPKELIMGNKDEPVRTRSAFRPSEETL----LSLKGLVSLIEPKSIDEALQDKDWILA 538
++ +E + + R RS E T L+L L + EP +A++ K W A
Sbjct: 797 SSQIEE---SSSERTPRFRSIQELYEVTENQENLTLFCLFAECEPMDFQKAIEKKTWRNA 853
Query: 539 MEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGI 598
M+EE+ KND W L P IG KWV++ K N KG+V R KARLVA+GYSQ+ GI
Sbjct: 854 MDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRVGI 913
Query: 599 DYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQPPGFEDEKK 658
DY E FAPVARLE +RL+IS + + +HQ+DVKSAFLNG + EEVY+ QP G+ + +
Sbjct: 914 DYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGE 973
Query: 659 PDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKDDILIVQIYVD 718
D V +LKK LYGLKQAPRAW R+ + E +F + + L+ K K+DILI +YVD
Sbjct: 974 EDKVLRLKKVLYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVD 1033
Query: 719 DIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKK 778
D+IF N S+ +EF + M EFEM+ +G + Y+LGI+V Q G +I Q Y KE+LKK
Sbjct: 1034 DLIFTGNNPSIFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKK 1093
Query: 779 FNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRPDILFSVHLCARF 838
F + +S TPM L K+++ V ++ +GSL YLT +RPDIL++V + +R+
Sbjct: 1094 FKIDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRY 1153
Query: 839 *SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQ 898
P TH A KRILRY+KGT N GL Y TS+YKL GY D+D+ GD +RKSTSG
Sbjct: 1154 MEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVF 1213
Query: 899 FLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQI-LESNIPIYCDN 957
++G +W SK+Q + LST EAEY++A C +W+++ L++ + E I+ DN
Sbjct: 1214 YIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDN 1273
Query: 958 TAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRF 1016
+AI+L+KNP+ H R+KHI+ +YH+IR+ V K + L++V T Q AD FTKPL + F
Sbjct: 1274 KSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADFFTKPLKRENF 1332
>At3g60170 putative protein
Length = 1339
Score = 751 bits (1938), Expect = 0.0
Identities = 420/1039 (40%), Positives = 605/1039 (57%), Gaps = 22/1039 (2%)
Query: 7 KHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSS---PCI 63
+ + W+LDSGCS HMTG + F EL+ V G + + +VG G++ V + I
Sbjct: 296 RDEVWFLDSGCSNHMTGSKEWFSELEEGFNRTVKLGNDTRMSVVGKGSVKVKVNGVTQVI 355
Query: 64 DNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSE 123
V V L +NLLS+ QL ++G ++ +C+ G+++ + N ++ + S+
Sbjct: 356 PEVYYVPELRNNLLSLGQLQERGLAILIRDGTCKVYHPSKGAIMETNMSGNRMFFLLASK 415
Query: 124 LEAQNVKCLLS---VNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEAC 180
+ +N CL + +++E +WH R GH + + L+ +V GLP LK A+ +C C
Sbjct: 416 PQ-KNSLCLQTEEVMDKENHLWHCRFGHLNQEGLKLLAHKKMVIGLPILK-ATKEICAIC 473
Query: 181 QKDKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFL 240
K + K +S L+L+H D+ GP+ S GKRY + +DD++R TWV FL
Sbjct: 474 LTGKQHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKTWVYFL 533
Query: 241 TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTP 300
K E+ A F F A V+ E + +R+D GG+F +++F S+GI+ + TP
Sbjct: 534 HEKSEAFATFKIFKASVEKEIGAFLTCLRTDRGGEFTSNEFGEFCRSHGISRQLTAAFTP 593
Query: 301 QQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWK 360
QQNGV ERKNRT+ R+ML E + K FW+EA + +IQNR + TP E W
Sbjct: 594 QQNGVAERKNRTIMNAVRSMLSERQVPKMFWSEATKWSVHIQNRSPTAAVEGMTPEEAWS 653
Query: 361 NIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEES 420
KP + YF FGC+ YV + K D KS KC+ LG SE SK +R Y+ K I S
Sbjct: 654 GRKPVVEYFRVFGCIGYVHIPDQKRSKLDDKSKKCVFLGVSEESKAWRLYDPVMKKIVIS 713
Query: 421 IHVRFDD--KLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSN---SQT 475
V FD+ D DQ+ + K L D K E VEP G N S
Sbjct: 714 KDVVFDEDKSWDWDQADVEAKEVTLECGDEDDEKNSEVVEPIAVASPNHVGSDNNVSSSP 773
Query: 476 LKKSRITAAHPKELIMGNKDEPVRTRSAFRPSE----ETLLSLKGLVSLIE--PKSIDEA 529
+ A P + + P + + E E LS+ L+ + E P D+A
Sbjct: 774 ILAPSSPAPSPVAAKVTRERRPPGWMADYETGEGEEIEENLSVMLLMMMTEADPIQFDDA 833
Query: 530 LQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVA 589
++DK W AME E+ KN+ W L P+ IG KWV++ KLNE G+V + KARLVA
Sbjct: 834 VKDKIWREAMEHEIESIVKNNTWELTTLPKGFTPIGVKWVYKTKLNEDGEVDKYKARLVA 893
Query: 590 QGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQ 649
+GY+Q GIDYTE FAPVARL+ +R +++ S N + Q+DVKSAFL+G + EEVYV Q
Sbjct: 894 KGYAQCYGIDYTEVFAPVARLDTVRTILAISSQFNWEIFQLDVKSAFLHGELKEEVYVRQ 953
Query: 650 PPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKDD 709
P GF E + + V+KL+K+LYGLKQAPRAWY R+ ++ L+ EFER + TLF KT +
Sbjct: 954 PEGFIREGEEEKVYKLRKALYGLKQAPRAWYSRIEAYFLKEEFERCPSEHTLFTKTRVGN 1013
Query: 710 ILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 769
ILIV +YVDD+IF +++++C EF + M EFEMS +G++K+FLGI+V Q+ G +I Q
Sbjct: 1014 ILIVSLYVDDLIFTGSDKAMCDEFKKSMMLEFEMSDLGKMKHFLGIEVKQSDGGIFICQR 1073
Query: 770 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRPDIL 829
+Y +E+L +F M ES K P+ P L K++ KV + +++ +GSL+YLT +RPD++
Sbjct: 1074 RYAREVLARFGMDESNAVKNPIVPGTKLTKDENGEKVDETMFKQLVGSLMYLTVTRPDLM 1133
Query: 830 FSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMY--KKTSEYKLSGYCDADYAGDR 887
+ V L +RF S+PR +H A KRILRYLKGT LG+ Y +K KL + D+DYAGD
Sbjct: 1134 YGVCLISRFMSNPRMSHWLAAKRILRYLKGTVELGIFYRRRKNRSLKLMAFTDSDYAGDL 1193
Query: 888 TERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQIL 947
+R+STSG + S + WASK+Q +ALST EAEYI+AA C+ Q +W++ LE
Sbjct: 1194 NDRRSTSGFVFLMASGAICWASKKQPVVALSTTEAEYIAAAFCACQCVWLRKVLEKLGAE 1253
Query: 948 E-SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADI 1006
E S I CDN++ I LSK+P+LH ++KHIEV++H++RD V V+ L++ T+ Q ADI
Sbjct: 1254 EKSATVINCDNSSTIQLSKHPVLHGKSKHIEVRFHYLRDLVNGDVVKLEYCPTEDQVADI 1313
Query: 1007 FTKPLAEDRFNFILKNLNM 1025
FTKPL ++F + L M
Sbjct: 1314 FTKPLKLEQFEKLRALLGM 1332
>At1g58140 hypothetical protein
Length = 1320
Score = 744 bits (1921), Expect = 0.0
Identities = 400/1015 (39%), Positives = 566/1015 (55%), Gaps = 50/1015 (4%)
Query: 7 KHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV----DSSPC 62
++ WYLDSG S HM G + MF EL G V G K ++ G G I +
Sbjct: 331 ENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQF 390
Query: 63 IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLS 122
I NV + + N+LS+ QL +KGYD+ + Q + KN ++ + +
Sbjct: 391 ISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLNIR 450
Query: 123 ELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQK 182
AQ +K + EE W+WH R GH + + LS+ +VRGLP + + +CE C
Sbjct: 451 NDIAQCLK--MCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQ-VCEGCLL 507
Query: 183 DKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTR 242
K K+ F ++ + LEL+H D+ GP+K +S+G Y ++ +DD+SR TWV FL
Sbjct: 508 GKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKE 567
Query: 243 KDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQ 302
K E +F F A V+ E I +RSD GG+F + +F + GI + PR+PQQ
Sbjct: 568 KSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQ 627
Query: 303 NGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNI 362
NGV ERKNRT+ EMAR+ML+ + K WAEAV A Y+ NR + + KTP E W
Sbjct: 628 NGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGR 687
Query: 363 KPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESIH 422
KP +S+ FG + + ++ K D KS K + +GY SKG++ YN D K S +
Sbjct: 688 KPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRN 747
Query: 423 VRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSRIT 482
+ FD++ + D + E + DK + E P E+ P++SQ +K
Sbjct: 748 IVFDEEGEWDWNSNEEDYNFFPHFEEDKPEPTREEPPSEEPTTPPTSPTSSQIEEKC--- 804
Query: 483 AAHPKELIMGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEEE 542
EP EA++ K W AM+EE
Sbjct: 805 ---------------------------------------EPMDFQEAIEKKTWRNAMDEE 825
Query: 543 LNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTE 602
+ KND W L P IG KWV++ K N KG+V R KARLVA+GYSQ+ GIDY E
Sbjct: 826 IKSIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRAGIDYDE 885
Query: 603 TFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHV 662
FAPVARLE +RL+IS + + +HQ+DVKSAFLNG + EEVY+ QP G+ + + D V
Sbjct: 886 VFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKV 945
Query: 663 FKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKDDILIVQIYVDDIIF 722
+LKK+LYGLKQAPRAW R+ + E +F + + L+ K K+DILI +YVDD+IF
Sbjct: 946 LRLKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVDDLIF 1005
Query: 723 GSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNML 782
N S+ +EF + M EFEM+ +G + Y+LGI+V Q G +I Q Y KE+LKKF M
Sbjct: 1006 TGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMD 1065
Query: 783 ESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRPDILFSVHLCARF*SDP 842
+S TPM L K+++ V ++ +GSL YLT +RPDIL++V + +R+ P
Sbjct: 1066 DSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHP 1125
Query: 843 RETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGS 902
TH A KRILRY+KGT N GL Y TS+YKL GY D+D+ GD +RKSTSG ++G
Sbjct: 1126 TTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGD 1185
Query: 903 NLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQI-LESNIPIYCDNTAAI 961
+W SK+Q + LST EAEY++A C +W+++ L++ + E I+ DN +AI
Sbjct: 1186 TAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAI 1245
Query: 962 SLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRF 1016
+L+KNP+ H R+KHI+ +YH+IR+ V K + L++V T Q ADIFTKPL + F
Sbjct: 1246 ALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIFTKPLKREDF 1300
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 711 bits (1834), Expect = 0.0
Identities = 399/1041 (38%), Positives = 590/1041 (56%), Gaps = 40/1041 (3%)
Query: 6 LKHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV---DSSPC 62
L+ W +DSGC+ HMT E R F + + + G G I V
Sbjct: 321 LREDVWLVDSGCTNHMTKEERYFSNINKSIKVPIRVRNGDIVMTAGKGDITVMTRHGKRI 380
Query: 63 IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLS 122
I NV LV GL NLLS+ Q+ GY V F K C + +G + N + + +KI+LS
Sbjct: 381 IKNVFLVPGLEKNLLSVPQIISSGYWVRFQDKRC-IIQDANGKEIMNIEMTDKSFKIKLS 439
Query: 123 ELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQK 182
+E + + + E WH+RLGH S +++ Q+ LV GLP K + C+AC
Sbjct: 440 SVEEEAMTANVQTEE---TWHKRLGHVSNKRLQQMQDKELVNGLPRFKVTKET-CKACNL 495
Query: 183 DKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTR 242
K ++ F ++ T LE++H D+ GP++ +SI G RY ++ +DDY+ WV FL +
Sbjct: 496 GKQSRKSFPKESQTKTREKLEIVHTDVCGPMQHQSIDGSRYYVLFLDDYTHMCWVYFLKQ 555
Query: 243 KDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQ 302
K E+ A F F A V+ + C I +R E + GI + P +PQQ
Sbjct: 556 KSETFATFKKFKALVEKQSNCSIKTLRP----------MEVFCEDEGINRQVTLPYSPQQ 605
Query: 303 NGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNK-TPYELWKN 361
NG ERKNR+L EMAR+ML E + WAEAV T+ Y+QNR+ + I + TP E W
Sbjct: 606 NGAAERKNRSLVEMARSMLVEQDLPLKLWAEAVYTSAYLQNRLPSKAIEDDVTPMEKWCG 665
Query: 362 IKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESI 421
KPN+S+ FG +CYV + K DAK+ +L+GYS ++KG+R + + + +E S
Sbjct: 666 HKPNVSHLRIFGSICYVHIPDQKRRKLDAKAKCGILIGYSNQTKGYRVFLLEDEKVEVSR 725
Query: 422 HVRF--DDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEE-----DEPEEEAGPSNSQ 474
V F D K D D+ + V+K +SIN + + +E + D G ++S
Sbjct: 726 DVVFQEDKKWDWDKQEEVKKTFVMSINDIQESRDQQETSSHDLSQIDDHANNGEGETSSH 785
Query: 475 TL-----KKSRITAAHPKELIMGNKDEPVRTRSAFRPSEETLLSLKG-LVSLIEPKSIDE 528
L ++ R T+ PK+ + + ++ ++E ++ LV+ EP++ DE
Sbjct: 786 VLSQVNDQEERETSESPKKY---KSMKEILEKAPRMENDEAAQGIEACLVANEEPQTYDE 842
Query: 529 ALQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLV 588
A DK+W AM EE+ KN W LV KPE +VI KW+++ K + G+ V++KARLV
Sbjct: 843 ARGDKEWEEAMNEEIKVIEKNRTWKLVDKPEKKNVISVKWIYKIKTDASGNHVKHKARLV 902
Query: 589 AQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVH 648
A+G+SQ+ GIDY ETFAPV+R + IR L++++ L+Q+DVKSAFLNG + EEVYV
Sbjct: 903 ARGFSQEYGIDYLETFAPVSRYDTIRALLAYAAQMKWRLYQMDVKSAFLNGELEEEVYVT 962
Query: 649 QPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKD 708
QPPGF E K + V +L K+LYGLKQAPRAWYER+ S+ ++N F R D L+ K +
Sbjct: 963 QPPGFVIEGKEEKVLRLYKALYGLKQAPRAWYERIDSYFIQNGFARSMNDAALYSKKKGE 1022
Query: 709 DILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 768
D+LIV +YVDD+I N L F + M+ EFEM+ +G L YFLG++V+Q G ++ Q
Sbjct: 1023 DVLIVSLYVDDLIITGNNTHLINTFKKNMKDEFEMTDLGLLNYFLGMEVNQDDSGIFLSQ 1082
Query: 769 SKYTKELLKKFNMLESTVAKTPMHPTCI---LEKEDKSGKVCQKLYRGTIGSLLYLTASR 825
KY +L+ KF M ES TP+ P +E +DK K YR +G LLYL ASR
Sbjct: 1083 EKYANKLIDKFGMKESKSVSTPLTPQGKRKGVEGDDKEFADPTK-YRRIVGGLLYLCASR 1141
Query: 826 PDILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAG 885
PD++++ +R+ S P H KR+LRY+KGT+N G+++ +L GY D+D+ G
Sbjct: 1142 PDVMYASSYLSRYMSSPSIQHYQEAKRVLRYVKGTSNFGVLFTSKETPRLVGYSDSDWGG 1201
Query: 886 DRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQ 945
++KST+G LG + W S +Q T+A STAEAEYI+ + Q +W++ ED+
Sbjct: 1202 SLEDKKSTTGYVFTLGLAMFCWQSCKQQTVAQSTAEAEYIAVCAATNQAIWLQRLFEDFG 1261
Query: 946 I-LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWA 1004
+ + IPI CDN +AI++ +NP+ H R KHIE+KYHF+R+ KG++ L++ + Q A
Sbjct: 1262 LKFKEGIPILCDNKSAIAIGRNPVQHRRTKHIEIKYHFVREAEHKGLIQLEYCKGEDQLA 1321
Query: 1005 DIFTKPLAEDRFNFILKNLNM 1025
D+ TK L+ RF + + L +
Sbjct: 1322 DVLTKALSVSRFEGLRRKLGV 1342
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 659 bits (1700), Expect = 0.0
Identities = 368/1026 (35%), Positives = 558/1026 (53%), Gaps = 50/1026 (4%)
Query: 9 QSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSS---PCIDN 65
Q W+LDSGCS HM G R F EL V G + + + G G + ++ I +
Sbjct: 256 QIWFLDSGCSNHMCGTREWFLELDSGFKQNVRLGDDRRMAVEGKGKLRLEVDGRIQVISD 315
Query: 66 VLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNN----IYKIRL 121
V V GL +NL S+ QL KG I C + + ++ +S N ++
Sbjct: 316 VYFVPGLKNNLFSVGQLQQKGLRFIIEGDVCEVWHKTEKRMVMHSTMTKNRMFVVFAAVK 375
Query: 122 SELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASD-ALCEAC 180
E + +CL + + +WH+R GH + + + L++ +V+GLP + A+C+ C
Sbjct: 376 KSKETEETRCLQVIGKANNMWHKRFGHLNHQGLRSLAEKEMVKGLPKFDLGEEEAVCDIC 435
Query: 181 QKDKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFL 240
K K + ++ +++ L+L+H D+ GP+ S GKRY + +DD+SR W L
Sbjct: 436 LKGKQIRESIPKESAWKSTQVLQLVHTDICGPINPASTSGKRYILNFIDDFSRKCWTYLL 495
Query: 241 TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTP 300
+ K E+ F F A+V+ E ++V +RSD GG++ + +F+ +GI + TP
Sbjct: 496 SEKSETFQFFKEFKAEVERESGKKLVCLRSDRGGEYNSREFDEYCKEFGIKRQLTAAYTP 555
Query: 301 QQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWK 360
QQNGV ERKNR++ M R ML E + + FW EAV A YI NR + + + TP E W
Sbjct: 556 QQNGVAERKNRSVMNMTRCMLMEMSVPRKFWPEAVQYAVYILNRSPSKALNDITPEEKWS 615
Query: 361 NIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEES 420
+ KP++ + FG + Y L + K D KS KC++ G S+ SK +R Y+ I S
Sbjct: 616 SWKPSVEHLRIFGSLAYALVPYQKRIKLDEKSIKCVMFGVSKESKAYRLYDPATGKILIS 675
Query: 421 IHVRFDDKLD---SDQSKLVEKFADLSINVSDKGKAPE------EVEPEEDEPEEEAGPS 471
V+FD++ D+S E D S + + PE + + E +E EE +
Sbjct: 676 RDVQFDEERGWEWEDKSLEEELVWDNSDHEPAGEEGPEINHNGQQDQEETEEEEETVAET 735
Query: 472 NSQTLKKSRITAAHPKELIMGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQ 531
Q L ++ + KD V +E L + +P +EA Q
Sbjct: 736 VHQNLPAVGTGGVRQRQQPVWMKDYVVGNARVLITQDEEDEVLALFIGPDDPVCFEEAAQ 795
Query: 532 DKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQG 591
+ W AME E+ +N+ W LV+ PE VIG KW+F+ K NEKG+V + KARLVA+G
Sbjct: 796 LEVWRKAMEAEITSIEENNTWELVELPEEAKVIGLKWIFKTKFNEKGEVDKFKARLVAKG 855
Query: 592 YSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQPP 651
Y Q+ G+D+ E FAPVA+ + IRL++ + + Q+DVKSAFL+G + E+V+V QP
Sbjct: 856 YHQRYGVDFYEVFAPVAKWDTIRLILGLAAEKGWSVFQLDVKSAFLHGDLKEDVFVEQPK 915
Query: 652 GFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKDDIL 711
GFE E++ V+KLKK+LYGLKQAPRAWY R+ F + FE+ + TLF K + D L
Sbjct: 916 GFEVEEESSKVYKLKKALYGLKQAPRAWYSRIEEFFGKEGFEKCYCEHTLFVKKERSDFL 975
Query: 712 IVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKY 771
+V +YVDD+I+ ++ + + F M EF M+ +G++KYFLG++V Q G +I+Q KY
Sbjct: 976 VVSVYVDDLIYTGSSMEMIEGFKNSMMEEFAMTDLGKMKYFLGVEVIQDERGIFINQRKY 1035
Query: 772 TKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRPDILFS 831
E++KK+ M K P+ P +K K+G V
Sbjct: 1036 AAEIIKKYGMEGCNSVKNPIVPG---QKLTKAGAV------------------------- 1067
Query: 832 VHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERK 891
+R+ P E HL AVKRILRY++GT +LG+ Y++ +L G+ D+DYAGD +RK
Sbjct: 1068 ----SRYMESPNEQHLLAVKRILRYVQGTLDLGIQYERGGATELVGFVDSDYAGDVDDRK 1123
Query: 892 STSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQI-LESN 950
STSG LG ++WASK+Q + LST EAE++SA+ + Q +W+++ LE+ E
Sbjct: 1124 STSGYVFMLGGGAIAWASKKQPIVTLSTTEAEFVSASYGACQAVWLRNVLEEIGCRQEGG 1183
Query: 951 IPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKP 1010
++CDN++ I LSKNP+LH R+KHI V+YHF+R+ V++G + L + T Q ADI TK
Sbjct: 1184 TLVFCDNSSTIKLSKNPVLHGRSKHIHVRYHFLRELVKEGTIRLDYCTTTDQVADIMTKA 1243
Query: 1011 LAEDRF 1016
+ + F
Sbjct: 1244 VKREVF 1249
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 643 bits (1658), Expect = 0.0
Identities = 374/1019 (36%), Positives = 530/1019 (51%), Gaps = 106/1019 (10%)
Query: 7 KHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV----DSSPC 62
++ WYLDSG S HM G + MF EL G V G K ++ G G I +
Sbjct: 331 ENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDHQF 390
Query: 63 IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLS 122
I NV + + N+LS+ QL +KGYD+ + + + KN ++ + +
Sbjct: 391 ISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDKESNLITKVPMSKNRMFVLNIR 450
Query: 123 ELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQK 182
AQ +K + EE W+WH R GH + + LS+ +VRGLP + + +CE C
Sbjct: 451 NDIAQCLK--MCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQ-VCEGCLL 507
Query: 183 DKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTR 242
K+ F ++ + LEL+H D+ GP+K +S+G Y ++ +DD+SR TWV FL
Sbjct: 508 GNQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKE 567
Query: 243 KDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQ 302
K E +F F A V+ E I +RSD GG+F + +F + GI + PR+PQQ
Sbjct: 568 KSEVFEIFKKFKAHVEKESGLVIKTMRSDSGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQ 627
Query: 303 NGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNI 362
NGV ERKNRT+ EMAR+ML+ + K WAEAV A Y+ NR + + KTP E W
Sbjct: 628 NGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGR 687
Query: 363 KPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESIH 422
KP +S+ FG + + ++ +K D KS K + +GY SKG++ YN D K S +
Sbjct: 688 KPGVSHLRVFGSIAHAHVPDEKRNKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRN 747
Query: 423 VRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSRIT 482
+ FD++ + D + E + DK + E P E+ P++SQ + S
Sbjct: 748 IVFDEEGEWDWNSNEEDYNFFPHFEEDKPEPTREEPPSEEPTTPPTSPTSSQIEESS--- 804
Query: 483 AAHPKELIMGNKDEPVRTRSAFRPSEETL----LSLKGLVSLIEPKSIDEALQDKDWILA 538
+ R RS E T L+L L + EP EA++ K W A
Sbjct: 805 -----------SERTPRFRSIQELYEVTENQENLTLFCLFAECEPMDFQEAIEKKTWRNA 853
Query: 539 MEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGI 598
M+EE+ KND W L P IG KWV++ K N KG+V R KARLVA+GYSQ+ GI
Sbjct: 854 MDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRAGI 913
Query: 599 DYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQPPGFEDEKK 658
DY E FAPVARLE +RL+IS + + +HQ+DVKSAFLNG + EEVY+ QP G+ + +
Sbjct: 914 DYDEIFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGE 973
Query: 659 PDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKDDILIVQIYVD 718
D V +LKK LYGLKQAPRAW R+ + E +F + + L+ K K+DILI +YVD
Sbjct: 974 EDKVLRLKKVLYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVD 1033
Query: 719 DIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKK 778
D+IF N S+ +EF + M EFEM+ +G + Y+LGI+V Q G +I Q Y KE+LKK
Sbjct: 1034 DLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKK 1093
Query: 779 FNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRPDILFSVHLCARF 838
F M +S + +GSL YLT +RPDIL++V + +R+
Sbjct: 1094 FKMDDSNPS--------------------------LVGSLRYLTCTRPDILYAVGVVSRY 1127
Query: 839 *SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQ 898
P TH A KRILRY+KGT N GL Y TS DY
Sbjct: 1128 MEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTS----------DY--------------- 1162
Query: 899 FLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQI-LESNIPIYCDN 957
+C +W+++ L++ + E I+ DN
Sbjct: 1163 ---------------------------KLVVCHA--IWLRNLLKELSLPQEEPTKIFVDN 1193
Query: 958 TAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRF 1016
+AI+L+KNP+ H R+KHI+ +YH+IR+ V K + L++V T Q ADIFTKPL + F
Sbjct: 1194 KSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIFTKPLKREDF 1252
>At3g25450 hypothetical protein
Length = 1343
Score = 634 bits (1635), Expect = 0.0
Identities = 368/1038 (35%), Positives = 568/1038 (54%), Gaps = 41/1038 (3%)
Query: 8 HQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSS----PCI 63
+ +WYLD+G S HMTG R F +L G+V FG + I G G+I S +
Sbjct: 289 NNAWYLDNGASNHMTGNRAWFCKLDEMITGKVRFGDDSCINIKGKGSIPFISKGGERKIL 348
Query: 64 DNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKR-KNNIYKIRLS 122
+V + L N+LS+ Q + G D+ + + +G++L ++R +N +YK+
Sbjct: 349 FDVYYIPDLKSNILSLGQATESGCDIRMREDYL-TLHDREGNLLIKAQRSRNRLYKV--- 404
Query: 123 ELEAQNVKCL-LSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQ 181
LE +N KCL L+ E +WH RLGH S I + K LV G+ + C +C
Sbjct: 405 SLEVENSKCLQLTTTNESTIWHARLGHISFETIKAMIKKELVIGISSSVPQEKETCGSCL 464
Query: 182 KDKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLT 241
K + F ++ LEL+H DL GP+ + KRY V++DD+SR+ W L
Sbjct: 465 FGKQARHSFPKATSYRAAQVLELIHGDLCGPISPSTAAKKRYVFVLIDDHSRYMWSILLK 524
Query: 242 RKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQ 301
K E+ F F A V+ E I R+D GG+F + +F+ GI + P TPQ
Sbjct: 525 EKSEAFGKFKEFKALVEQECGAIIKTFRTDRGGEFLSHEFQEFCAKEGINRHLTAPYTPQ 584
Query: 302 QNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKN 361
QNGVVER+NRTL M R++L+ M + W EAV + Y+ NR+ R + N+TPYE++K+
Sbjct: 585 QNGVVERRNRTLLGMTRSILKHMNMPNYLWGEAVRHSTYLINRVGTRSLSNQTPYEVFKH 644
Query: 362 IKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESI 421
KPN+ + FGCV Y L K D +S + LG SK +R + + I S
Sbjct: 645 KKPNVEHLRVFGCVSYAKVEVPNLKKLDDRSRMLVYLGTEPGSKAYRLLDPTKRRIFVSR 704
Query: 422 HVRFDDKLD----SDQSKLVEKFADLSINVSD---KGKAPEEVEPEEDEPEEEAGPSNSQ 474
V FD+ S+ ++ +I +S+ G ++ E +E EE +
Sbjct: 705 DVVFDENRSWMWQESSSETDKESGTFTITLSEFGNNGVTENDISTEPEETEEAEINGEDE 764
Query: 475 TLKKSRITAAHPKELIMGNKDEPVRT--RSAFRPS-------------EETLLSLKGLVS 519
+ + T H + + +PVR R RP+ E LL++
Sbjct: 765 NIIEEAETEEHDQSQ---EEPQPVRRSQRQVIRPNYLKDYVLCAEIEAEHLLLAVND--- 818
Query: 520 LIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGD 579
EP EA + K+W A +EE+ KN WSLV P IG KWVF+ K N G
Sbjct: 819 --EPWDFKEANKSKEWRDACKEEIQSIEKNRTWSLVDLPVGSKAIGVKWVFKLKHNSDGS 876
Query: 580 VVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNG 639
+ + KARLVA+GY Q+ G+D+ E FAPVAR+E +RL+I+ + ++ +H +DVK+AFL+G
Sbjct: 877 INKYKARLVAKGYVQRHGVDFEEVFAPVARIETVRLIIALAASNGWEIHHLDVKTAFLHG 936
Query: 640 YISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDT 699
+ E+VYV QP GF +++ + V+KL K+LYGL+QAPRAW +L+ L E +FE+ +
Sbjct: 937 ELREDVYVSQPEGFTNKESKEKVYKLHKALYGLRQAPRAWNTKLNEILKELKFEKCHKEP 996
Query: 700 TLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQ 759
+L+ K ++IL+V +YVDD++ +N + F + M +FEMS +G+L Y+LGI+V Q
Sbjct: 997 SLYRKQEGENILVVAVYVDDLLVTGSNLDIILNFKKGMVGKFEMSDLGKLTYYLGIEVLQ 1056
Query: 760 TPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLL 819
+ +G + Q +Y K++L++ M + TPM + L K ++ + YR IG L
Sbjct: 1057 SKDGITLKQERYAKKILEEAGMSKCNTVNTPMIASLELSKAQDEKRIDETDYRRNIGCLR 1116
Query: 820 YLTASRPDILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYC 879
YL +RPD+ ++V + +R+ +PRE+H A+K+ILRYL+GTT+ GL +KK L GY
Sbjct: 1117 YLLHTRPDLSYNVGILSRYLQEPRESHGAALKQILRYLQGTTSHGLYFKKGENAGLIGYS 1176
Query: 880 DADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH 939
D+ + D + KST G+ +L ++W S++Q + LS+ EAE+++A + Q +W++
Sbjct: 1177 DSSHNVDLDDGKSTGGHIFYLNDCPITWCSQKQQVVTLSSCEAEFMAATEAAKQAIWLQE 1236
Query: 940 *LEDYQILE-SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVD 998
L + E + I DN +AI+L+KNP+ H R+KHI +YHFIR+ V+ G + ++ V
Sbjct: 1237 LLAEVIGTECEKVTIRVDNKSAIALTKNPVFHGRSKHIHRRYHFIRECVENGQIEVEHVP 1296
Query: 999 TDHQWADIFTKPLAEDRF 1016
Q ADI TK L + +F
Sbjct: 1297 GVRQKADILTKALGKIKF 1314
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 630 bits (1624), Expect = e-180
Identities = 365/1037 (35%), Positives = 567/1037 (54%), Gaps = 37/1037 (3%)
Query: 6 LKHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSSPCIDN 65
L +W +DSG + H++ +R++F+ L V +I G GT+ ++ + N
Sbjct: 437 LSSDTWVIDSGATHHVSHDRKLFQTLDTSIVSFVNLPTGPNVRISGVGTVLINKDIILQN 496
Query: 66 VLLVDGLTHNLLSISQLA-DKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSEL 124
VL + NL+SIS L D G VIF+ C+ G L KR N+Y + ++
Sbjct: 497 VLFIPEFRLNLISISSLTTDLGTRVIFDPSCCQIQDLTKGLTLGEGKRIGNLYVLD-TQS 555
Query: 125 EAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQKDK 184
A +V ++ V+ VWH+RLGH S ++ LS+ V G K A C C K
Sbjct: 556 PAISVNAVVDVS----VWHKRLGHPSFSRLDSLSE---VLGTTRHKNKKSAYCHVCHLAK 608
Query: 185 FTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKD 244
K+ F + N + S + ELLHID++GP E++ G +Y + IVDD+SR TW+ L K
Sbjct: 609 QKKLSFPSANNICNS-TFELLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWIYLLKSKS 667
Query: 245 ESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQNG 304
+ VF FI V+N+ R+ VRSD+ + F + + GI SCP TP+QN
Sbjct: 668 DVLTVFPAFIDLVENQYDTRVKSVRSDNAKELA---FTEFYKAKGIVSFHSCPETPEQNS 724
Query: 305 VVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKP 364
VVERK++ + +AR ++ ++ M+ +W + V TA ++ NR + NKTP+E+ P
Sbjct: 725 VVERKHQHILNVARALMFQSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGKLP 784
Query: 365 NISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESIHVR 424
+ S FGC+CY + + HKF +S C+ LGY KG++ + ++ + S +V
Sbjct: 785 DYSQLKTFGCLCYSSTSSKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVE 844
Query: 425 FDDKL----DSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSR 480
F ++L S QS ++ G + P + PS + K R
Sbjct: 845 FHEELFPLASSQQSATTASDVFTPMDPLSSGNSITS-----HLPSPQISPSTQ--ISKRR 897
Query: 481 ITA--AHPKEL--IMGNKDE--PVRTRSAFRP-SEETLLSLKGLVSLIEPKSIDEALQDK 533
IT AH ++ NKD+ P+ + ++ S +L + + + P+S EA K
Sbjct: 898 ITKFPAHLQDYHCYFVNKDDSHPISSSLSYSQISPSHMLYINNISKIPIPQSYHEAKDSK 957
Query: 534 DWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYS 593
+W A+++E+ + D W + P +G KWVF K + G + R KAR+VA+GY+
Sbjct: 958 EWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWVFTVKFHADGSLERFKARIVAKGYT 1017
Query: 594 QQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQPPGF 653
Q+EG+DYTETF+PVA++ ++LL+ S + L+Q+D+ +AFLNG + E +Y+ P G+
Sbjct: 1018 QKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQLDISNAFLNGDLEETIYMKLPDGY 1077
Query: 654 EDEK----KPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKDD 709
D K P+ V +LKKS+YGLKQA R W+ + S+ LL FE+ D TLF + +
Sbjct: 1078 ADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSNSLLALGFEKQHGDHTLFVRCIGSE 1137
Query: 710 ILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 769
+++ +YVDDI+ S + + E ++A F++ +G LKYFLG++V +T EG + Q
Sbjct: 1138 FIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRELGPLKYFLGLEVARTSEGISLSQR 1197
Query: 770 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRPDIL 829
KY ELL +ML+ + PM P L K D +++YR +G L+YLT +RPDI
Sbjct: 1198 KYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLLEDKEMYRRLVGKLMYLTITRPDIT 1257
Query: 830 FSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTE 889
F+V+ +F S PR HL AV ++L+Y+KGT GL Y + L GY DAD+
Sbjct: 1258 FAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGLFYSAEDDLTLKGYTDADWGTCPDS 1317
Query: 890 RKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQILES 949
R+ST+G F+GS+L+SW SK+Q T++ S+AEAEY + A+ S +M W+ L ++ S
Sbjct: 1318 RRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYRALALASCEMAWLSTLLLALRV-HS 1376
Query: 950 NIPI-YCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFT 1008
+PI Y D+TAA+ ++ NP+ H R KHIE+ H +R+ + G L L V T Q ADI T
Sbjct: 1377 GVPILYSDSTAAVYIATNPVFHERTKHIEIDCHTVREKLDNGQLKLLHVKTKDQVADILT 1436
Query: 1009 KPLAEDRFNFILKNLNM 1025
KPL +F +L +++
Sbjct: 1437 KPLFPYQFAHLLSKMSI 1453
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 622 bits (1604), Expect = e-178
Identities = 358/1037 (34%), Positives = 564/1037 (53%), Gaps = 33/1037 (3%)
Query: 6 LKHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSSPCIDN 65
L +W +DSG + H++ +R +F L V KI G GT+ ++ + N
Sbjct: 426 LSSATWVIDSGATHHVSHDRSLFSSLDTSVLSAVNLPTGPTVKISGVGTLKLNDDILLKN 485
Query: 66 VLLVDGLTHNLLSISQLADK-GYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSEL 124
VL + NL+SIS L D G VIF++ SC I G +L +R N+Y + + +
Sbjct: 486 VLFIPEFRLNLISISSLTDDIGSRVIFDKNSCEIQDLIKGRMLGQGRRVANLYLLDVGD- 544
Query: 125 EAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQKDK 184
++ +V ++ ++ +WHRRLGHAS++++ +S G K C C K
Sbjct: 545 QSISVNAVVDIS----MWHRRLGHASLQRLDAISDS---LGTTRHKNKGSDFCHVCHLAK 597
Query: 185 FTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKD 244
K+ F N V +LLHID++GP E++ G +Y + IVDD+SR TW+ L K
Sbjct: 598 QRKLSFPTSNKVC-KEIFDLLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWMYLLKTKS 656
Query: 245 ESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQNG 304
E VF FI QV+N+ ++ VRSD+ + KF S + GI SCP TP+QN
Sbjct: 657 EVLTVFPAFIQQVENQYKVKVKAVRSDNAPEL---KFTSFYAEKGIVSFHSCPETPEQNS 713
Query: 305 VVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKP 364
VVERK++ + +AR ++ ++ + W + V TA ++ NR + ++NKTPYE+ P
Sbjct: 714 VVERKHQHILNVARALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEILTGTAP 773
Query: 365 NISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESIHVR 424
FGC+CY + + HKF +S CL LGY KG++ + ++ T+ S +V+
Sbjct: 774 VYEQLRTFGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQ 833
Query: 425 FDDKL--------DSDQSKLVEKFADLSINV-SDKGKAPEEVEPE-EDEPEEEAGPSNSQ 474
F +++ KL +S + SD +P + + D P + +SQ
Sbjct: 834 FHEEVFPLAKNPGSESSLKLFTPMVPVSSGIISDTTHSPSSLPSQISDLPPQ----ISSQ 889
Query: 475 TLKK--SRITAAHPKELIMGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQD 532
++K + + H + +K T S + S + + + + P + EA
Sbjct: 890 RVRKPPAHLNDYHCNTMQSDHKYPISSTISYSKISPSHMCYINNITKIPIPTNYAEAQDT 949
Query: 533 KDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGY 592
K+W A++ E+ K + W + P+ +G KWVF K G++ R KARLVA+GY
Sbjct: 950 KEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGY 1009
Query: 593 SQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQPPG 652
+Q+EG+DYT+TF+PVA++ I+LL+ S + L Q+DV +AFLNG + EE+++ P G
Sbjct: 1010 TQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEG 1069
Query: 653 FEDEK----KPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYKD 708
+ + K + V +LK+S+YGLKQA R W+++ SS LL F++ D TLF K Y
Sbjct: 1070 YAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDG 1129
Query: 709 DILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 768
+ +IV +YVDDI+ S +++ + E + F++ +G+LKYFLG++V +T G I Q
Sbjct: 1130 EFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVARTTAGISICQ 1189
Query: 769 SKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRPDI 828
KY ELL+ ML PM P + K+D + YR +G L+YLT +RPDI
Sbjct: 1190 RKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDI 1249
Query: 829 LFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRT 888
F+V+ +F S PR THLTA R+L+Y+KGT GL Y +S+ L G+ D+D+A +
Sbjct: 1250 TFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQD 1309
Query: 889 ERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQILE 948
R+ST+ F+G +L+SW SK+Q T++ S+AEAEY + A+ + +M+W+ L Q
Sbjct: 1310 SRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQASP 1369
Query: 949 SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFT 1008
+Y D+TAAI ++ NP+ H R KHI++ H +R+ + G L L V T+ Q ADI T
Sbjct: 1370 PVPILYSDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGELKLLHVRTEDQVADILT 1429
Query: 1009 KPLAEDRFNFILKNLNM 1025
KPL +F + +++
Sbjct: 1430 KPLFPYQFEHLKSKMSI 1446
>At1g70010 hypothetical protein
Length = 1315
Score = 580 bits (1495), Expect = e-165
Identities = 345/1002 (34%), Positives = 530/1002 (52%), Gaps = 35/1002 (3%)
Query: 52 TGTICVDSSPCIDNVLLVDGLTHNLLSISQLADK-GYDVIFNQKSCRAVSQIDGSVLFNS 110
+G++ + +++VL + NLLS+S L G + F++ SC ++
Sbjct: 314 SGSVHLGRHLILNDVLFIPQFKFNLLSVSSLTKSMGCRIWFDETSCVLQDATRELMVGMG 373
Query: 111 KRKNNIYKIRLSELEAQNVKCLLSVNE--EQWVWHRRLGHASMRKISQLSKLNLVRGLPN 168
K+ N+Y + L L ++V +WH+RLGH S++K+ +S L P
Sbjct: 374 KQVANLYIVDLDSLSHPGTDSSITVASVTSHDLWHKRLGHPSVQKLQPMSSL---LSFPK 430
Query: 169 LKFASDALCEACQKDKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIV 228
K +D C C K +PF + N S SR +L+HID +GP ++ G RY + IV
Sbjct: 431 QKNNTDFHCRVCHISKQKHLPFVSHNNKS-SRPFDLIHIDTWGPFSVQTHDGYRYFLTIV 489
Query: 229 DDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSY 288
DDYSR TWV L K + V TF+ V+N+ I VRSD+ + F + S
Sbjct: 490 DDYSRATWVYLLRNKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPELN---FTQFYHSK 546
Query: 289 GIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVR 348
GI SCP TPQQN VVERK++ + +AR++ ++ + +W + + TA Y+ NR+
Sbjct: 547 GIVPYHSCPETPQQNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINRLPAP 606
Query: 349 PILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFR 408
+ +K P+E+ P + FGC+CY + HKF ++ C +GY KG++
Sbjct: 607 ILEDKCPFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYK 666
Query: 409 FYNTDAKTIEESIHVRFDDKL----DSDQSKLVEKF-ADLS------------INVSDKG 451
+ + +I S HV F ++L SD S+ + F DL+ +N SD
Sbjct: 667 LLDLETHSIIVSRHVVFHEELFPFLGSDLSQEEQNFFPDLNPTPPMQRQSSDHVNPSDSS 726
Query: 452 KAPE---EVEPEEDEPEEEAGPSNSQTLKKSRITAAHPKELIMGNKDEPVRTRSAFRPSE 508
+ E P + PE S+ + K + + + ++ E + S R ++
Sbjct: 727 SSVEILPSANPTNNVPEPSVQTSHRKAKKPAYLQDYYCHSVVSSTPHEIRKFLSYDRIND 786
Query: 509 ETLLSLKGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKW 568
L L L EP + EA + + W AM E + W + P IG +W
Sbjct: 787 PYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRW 846
Query: 569 VFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLH 628
+F+ K N G V R KARLVAQGY+Q+EGIDY ETF+PVA+L +++LL+ + + L
Sbjct: 847 IFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLT 906
Query: 629 QIDVKSAFLNGYISEEVYVHQPPGFE----DEKKPDHVFKLKKSLYGLKQAPRAWYERLS 684
Q+D+ +AFLNG + EE+Y+ P G+ D P+ V +LKKSLYGLKQA R WY + S
Sbjct: 907 QLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFS 966
Query: 685 SFLLENEFERGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMS 744
S LL F + D T F K L V +Y+DDII S N + M++ F++
Sbjct: 967 STLLGLGFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLR 1026
Query: 745 MMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSG 804
+GELKYFLG+++ ++ +G +I Q KY +LL + L + PM P+ + +
Sbjct: 1027 DLGELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGD 1086
Query: 805 KVCQKLYRGTIGSLLYLTASRPDILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLG 864
V YR IG L+YL +RPDI F+V+ A+F PR+ HL AV +IL+Y+KGT G
Sbjct: 1087 FVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQG 1146
Query: 865 LMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEY 924
L Y TSE +L Y +ADY R R+STSG C FLG +L+ W S++Q ++ S+AEAEY
Sbjct: 1147 LFYSATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEY 1206
Query: 925 ISAAICSTQMLWMKH*LEDYQI-LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFI 983
S ++ + +++W+ + L++ Q+ L ++CDN AAI ++ N + H R KHIE H +
Sbjct: 1207 RSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSV 1266
Query: 984 RDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1025
R+ + KG+ L ++T+ Q AD FTKPL F+ ++ + +
Sbjct: 1267 RERLLKGLFELYHINTELQIADPFTKPLYPSHFHRLISKMGL 1308
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 574 bits (1479), Expect = e-163
Identities = 353/1032 (34%), Positives = 546/1032 (52%), Gaps = 60/1032 (5%)
Query: 10 SWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSS----PCIDN 65
SWYLD+G S HMTG + F +L G+V FG + + I G G+I + + + +
Sbjct: 279 SWYLDNGASNHMTGNLQWFSKLNEMITGKVRFGDDSRIDIKGKGSIVLITKGGIRKTLTD 338
Query: 66 VLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKR-KNNIYKIRLSEL 124
V + L N++S+ Q + G DV + +G +L + R +N +YK+ +L
Sbjct: 339 VYFIPDLKSNIISLGQATEAGCDVRMKDDQL-TLHDREGCLLLRATRSRNRLYKV---DL 394
Query: 125 EAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQKDK 184
+NVKCL + + + + LV G+ N+ + C +C K
Sbjct: 395 NVENVKCL------------------QLEAATMVRKELVIGISNIPKEKET-CGSCLLGK 435
Query: 185 FTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKD 244
+ PF S+ LEL+H DL GP+ + KRY +V++DD++R+ W L K
Sbjct: 436 QARQPFPKATTYRASQVLELVHGDLCGPITQSTTAKKRYILVLIDDHTRYMWSMLLKEKS 495
Query: 245 ESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQNG 304
E+ F F +V+ E +I R+D GG+F + +F+ GI + P TPQQNG
Sbjct: 496 EAFEKFRDFKTKVEQESGVKIKTFRTDKGGEFVSQEFQDFCAKEGINRHLTAPYTPQQNG 555
Query: 305 VVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKP 364
VVER+NRTL M R++L+ M + W EAV + YI NR+ R + N+TPYE++K KP
Sbjct: 556 VVERRNRTLLGMTRSILKHMKMPNYLWGEAVRHSTYIINRVGTRSLQNQTPYEVFKQRKP 615
Query: 365 NISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYN-TDAKTIEESIHV 423
N+ + FGC+ Y L K D +S + LG SK +R + T+ K I+ +
Sbjct: 616 NVEHLRVFGCIGYAKIEGPHLRKLDDRSKMLVYLGTEPGSKAYRLLDPTNRKIIKWNNSD 675
Query: 424 RFDDKLDSDQSKLVEKFADLSINVSD------KGKAPEEVEPEEDEPE---------EEA 468
+ S + +F + I SD G+ E EE E E EE
Sbjct: 676 SETRDISGTFSLTLGEFGNNGIQESDDIETEKNGEESENSHEEEGENEHNEQEQIDAEET 735
Query: 469 GPSNSQ---TLKKSRITAAHPKELIMGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKS 525
PS++ TL++S P L D+ V E+ LL++ EP
Sbjct: 736 QPSHATPLPTLRRSTRQVGKPNYL-----DDYVLMAEI--EGEQVLLAIND-----EPWD 783
Query: 526 IDEALQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKA 585
EA + K+W A +EE+ KN WSL+ P VIG KWVF+ K N G + + KA
Sbjct: 784 FKEANKLKEWRDACKEEILSIEKNKTWSLIDLPVRRKVIGLKWVFKIKRNSDGSINKYKA 843
Query: 586 RLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEV 645
RLVA+GY Q+ GIDY E FA VAR+E IR++I+ + ++ +H +DVK+AFL+G + E+V
Sbjct: 844 RLVAKGYVQRHGIDYDEVFAHVARIETIRVIIALAASNGWEVHHLDVKTAFLHGELREDV 903
Query: 646 YVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKT 705
YV QP GF ++ V+KL K+LYGLKQAPRAW +L+ L E F + + +++ +
Sbjct: 904 YVTQPEGFTNKDNEGKVYKLHKALYGLKQAPRAWNTKLNKILQELNFVKCSKEPSVYRRQ 963
Query: 706 YKDDILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTY 765
+ +LIV IYVDD++ ++ L F + M +FEMS +G+L Y+LGI+V G
Sbjct: 964 EEKKLLIVAIYVDDLLVTGSSLDLILCFKKDMAGKFEMSDLGQLTYYLGIEVLHRKNGII 1023
Query: 766 IHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASR 825
+ Q +Y +++++ M PM L K + + ++ YR IG L Y+ +R
Sbjct: 1024 LRQERYAMKIIEEAGMSNCNPVLIPMAAGLELCKAQEEKCITERDYRRMIGCLRYIVHTR 1083
Query: 826 PDILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAG 885
PD+ + V + +R+ PRE+H A+K++LRYLKGT + GL K+ + L GY D+ ++
Sbjct: 1084 PDLSYCVGVLSRYLQQPRESHGNALKQVLRYLKGTMSHGLYLKRGFKSGLVGYSDSSHSA 1143
Query: 886 DRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*L-EDY 944
D + KST+G+ +L ++W S++Q +ALS+ EAE+++A + Q +W++ E
Sbjct: 1144 DLDDGKSTAGHIFYLHQCPITWCSQKQQVVALSSCEAEFMAATEAAKQAIWLQDLFAEVC 1203
Query: 945 QILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWA 1004
+ I DN +AI+L+KN + H R+KHI +YHFIR+ V+ ++ + V Q A
Sbjct: 1204 GTTSEKVMIRVDNKSAIALTKNLVFHGRSKHIHRRYHFIRECVENNLVEVDHVPGVEQRA 1263
Query: 1005 DIFTKPLAEDRF 1016
DI TKPL +F
Sbjct: 1264 DILTKPLGRIKF 1275
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 572 bits (1475), Expect = e-163
Identities = 349/1057 (33%), Positives = 554/1057 (52%), Gaps = 47/1057 (4%)
Query: 9 QSWYLDSGCSRHMTGERRMFRELKLKPGGE-VGFGGNEKGKIVGTGTICVDSSPC---ID 64
+ W+ DS + H+T + G + V G I TG+ + SS ++
Sbjct: 320 KEWHPDSAATAHVTSSTNGLQSATEYEGDDAVLVGDGTYLPITHTGSTTIKSSNGKIPLN 379
Query: 65 NVLLVDGLTHNLLSISQLADK-GYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSE 123
VL+V + +LLS+S+L D V F+ + V+ R+N +Y + E
Sbjct: 380 EVLVVPNIQKSLLSVSKLCDDYPCGVYFDANKVCIIDLQTQKVVTTGPRRNGLYVLENQE 439
Query: 124 LEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQKD 183
A + EE VWH RLGHA+ + + L ++ K + +CE CQ
Sbjct: 440 FVALYSNRQCAATEE--VWHHRLGHANSKALQHLQNSKAIQ---INKSRTSPVCEPCQMG 494
Query: 184 KFTKVPFKAKNVVSTSR---SLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFL 240
K +++PF ++S SR L+ +H DL+GP S G +Y + VDDYSR++W L
Sbjct: 495 KSSRLPF----LISDSRVLHPLDRIHCDLWGPSPVVSNQGLKYYAIFVDDYSRYSWFYPL 550
Query: 241 TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTP 300
K E +VF +F V+N+ +I +SD GG+F ++K ++ +GI H SCP TP
Sbjct: 551 HNKSEFLSVFISFQKLVENQLNTKIKVFQSDGGGEFVSNKLKTHLSEHGIHHRISCPYTP 610
Query: 301 QQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWK 360
QQNG+ ERK+R L E+ +ML + + FW E+ TA YI NR+ + N +PYE
Sbjct: 611 QQNGLAERKHRHLVELGLSMLFHSHTPQKFWVESFFTANYIINRLPSSVLKNLSPYEALF 670
Query: 361 NIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFR-FYNTDAK---- 415
KP+ S FG CY +KFD +S +C+ LGY+ + KG+R FY K
Sbjct: 671 GEKPDYSSLRVFGSACYPCLRPLAQNKFDPRSLQCVFLGYNSQYKGYRCFYPPTGKVYIS 730
Query: 416 --TIEESIHVRFDDKLDS----DQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAG 469
I + F +K S + L++ + I+ AP ++ + + AG
Sbjct: 731 RNVIFNESELPFKEKYQSLVPQYSTPLLQAWQHNKISEISVPAAPVQLFSKPIDLNTYAG 790
Query: 470 PSNSQTLKKSRITA----------------AHPKELIMGNKDEPVRTRSAFRPSEETLLS 513
++ L T+ A +E ++ + R+++ +
Sbjct: 791 SQVTEQLTDPEPTSNNEGSDEEVNPVAEEIAANQEQVINSHAMTTRSKAGIQKPNTRYAL 850
Query: 514 LKGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNK 573
+ ++ EPK++ A++ W A+ EE+N+ WSLV + ++++ +KWVF+ K
Sbjct: 851 ITSRMNTAEPKTLASAMKHPGWNEAVHEEINRVHMLHTWSLVPPTDDMNILSSKWVFKTK 910
Query: 574 LNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVK 633
L+ G + + KARLVA+G+ Q+EG+DY ETF+PV R IRL++ S + + Q+DV
Sbjct: 911 LHPDGSIDKLKARLVAKGFDQEEGVDYLETFSPVVRTATIRLVLDVSTSKGWPIKQLDVS 970
Query: 634 SAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFE 693
+AFL+G + E V+++QP GF D +KP HV +L K++YGLKQAPRAW++ S+FLL+ F
Sbjct: 971 NAFLHGELQEPVFMYQPSGFIDPQKPTHVCRLTKAIYGLKQAPRAWFDTFSNFLLDYGFV 1030
Query: 694 RGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFL 753
K D +LF IL + +YVDDI+ ++QSL ++ + ++ F M +G +YFL
Sbjct: 1031 CSKSDPSLFVCHQDGKILYLLLYVDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFL 1090
Query: 754 GIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRG 813
GIQ++ G ++HQ+ Y ++L++ M + TP+ L+ + +R
Sbjct: 1091 GIQIEDYANGLFLHQTAYATDILQQAGMSDCNPMPTPLPQQ--LDNLNSELFAEPTYFRS 1148
Query: 814 TIGSLLYLTASRPDILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEY 873
G L YLT +RPDI F+V+ + P + +KRILRY+KGT +GL K+ S
Sbjct: 1149 LAGKLQYLTITRPDIQFAVNFICQRMHSPTTSDFGLLKRILRYIKGTIGMGLPIKRNSTL 1208
Query: 874 KLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQ 933
LS Y D+D+AG + R+ST+G C LGSNL+SW++KRQ T++ S+ EAEY + + +
Sbjct: 1209 TLSAYSDSDHAGCKNTRRSTTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTYAARE 1268
Query: 934 MLWMKH*LEDYQILE-SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVL 992
+ W+ L D I + +YCDN +A+ LS NP LH+R+KH + YH+IR+ V G++
Sbjct: 1269 ITWISFLLRDLGIPQYLPTQVYCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVALGLI 1328
Query: 993 LLKFVDTDHQWADIFTKPLAEDRFNFILKNLNMDFCP 1029
+ + Q AD+FTK L F + L + P
Sbjct: 1329 ETQHISATFQLADVFTKSLPRRAFVDLRSKLGVSGSP 1365
>At1g37110
Length = 1356
Score = 546 bits (1408), Expect = e-155
Identities = 349/1068 (32%), Positives = 552/1068 (51%), Gaps = 76/1068 (7%)
Query: 11 WYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSS----PCIDNV 66
W LDSGC+ HMT R F + K + G + + G GTI +D+ ++NV
Sbjct: 308 WILDSGCTSHMTSRRDWFISFQEKGNTTILLGDDHSVESQGQGTIRIDTHGGTIKILENV 367
Query: 67 LLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIR----LS 122
V L NL+S L GY + R + N +Y + +S
Sbjct: 368 KYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFK--NNKTALRGSLSNGLYVLDGSTVMS 425
Query: 123 EL---EAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLV--RGLPNLKFASDALC 177
EL E VK L WH RLGH SM + L+ L+ + + L+F C
Sbjct: 426 ELCNAETDKVKTAL--------WHSRLGHMSMNNLKVLAGKGLIDRKEINELEF-----C 472
Query: 178 EACQKDKFTKVPFKAKNVVSTSRSLELLHIDLFG-PVKTESIGGKRYGMVIVDDYSRWTW 236
E C K KV F S +L +H DL+G P T SI GK+Y + I+DD +R W
Sbjct: 473 EHCVMGKSKKVSFNVGKHTSED-ALSYVHADLWGSPNVTPSISGKQYFLSIIDDKTRKVW 531
Query: 237 VKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSC 296
+ FL KDE+ F + + V+N+ ++ +R+D+G +F N +F+S +GI +C
Sbjct: 532 LYFLKSKDETFDKFCEWKSLVENQVNKKVKCLRTDNGLEFCNSRFDSYCKEHGIERHRTC 591
Query: 297 PRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPY 356
TPQQNGV ER NRT+ E R +L ++G+ + FWAEA TA Y+ NR I + P
Sbjct: 592 TYTPQQNGVAERMNRTIMEKVRCLLNKSGVEEVFWAEAAATAAYLINRSPASAINHNVPE 651
Query: 357 ELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKT 416
E+W N KP + FG + YV + +L ++ K LGY +KG++ + + +
Sbjct: 652 EMWLNRKPGYKHLRKFGSIAYVHQDQGKLKP---RALKGFFLGYPAGTKGYKVWLLEEEK 708
Query: 417 IEESIHVRFDDKL----------DSDQSKLVE---------KFADLSIN------VSDKG 451
S +V F + + D+D E KFA+ S + SD
Sbjct: 709 CVISRNVVFQESVVYRDLKVKEDDTDNLNQKETTSSEVEQNKFAEASGSGGVIQLQSDSE 768
Query: 452 KAPEEVEPEEDEPEEEAGPSNSQTLKKSRITAAH-PKELIMGNKDEPVRTRSAFRPSEET 510
E + + E E E +T K++ +T ++ + N + P R F
Sbjct: 769 PITEGEQSSDSEEEVEYSEKTQETPKRTGLTTYKLARDRVRRNINPPTR----FTEESSV 824
Query: 511 LLSLKGLVSLI--EPKSIDEALQDKD---WILAMEEELNQFSKNDVWSLVKKPESVHVIG 565
+L + + I EP+S EA++ +D W +A +E++ KN W LV KP+ +IG
Sbjct: 825 TFALVVVENCIVQEPQSYQEAMESQDCEKWDMATHDEMDSLMKNGTWDLVDKPKDRKIIG 884
Query: 566 TKWVFRNKLNEKG-DVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHN 624
+W+F+ K G + R KARLVA+GY+Q+EG+DY E FAPV + +IR+L+S V+ +
Sbjct: 885 CRWLFKLKSGIPGVEPTRFKARLVAKGYTQREGVDYQEIFAPVVKHVSIRILMSLVVDKD 944
Query: 625 IVLHQIDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLS 684
+ L Q+DVK+ FL+G + EE+Y+ QP GF + + V +LKKSLYGLKQ+PR W +R
Sbjct: 945 LELEQMDVKTTFLHGDLEEELYMEQPEGFVSDSSENKVCRLKKSLYGLKQSPRQWNKRFD 1004
Query: 685 SFLLENEFERGKVDTTLFCKTYKD-DILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEM 743
F+ +F R + D ++ K + D + + +YVDD++ A+++ E + EFEM
Sbjct: 1005 RFMSSQQFIRSEHDACVYVKHVSEHDFIYLLLYVDDMLIAGASKAEINRVKEQLSTEFEM 1064
Query: 744 SMMGELKYFLGIQVDQTPEGTYIHQSK--YTKELLKKFNMLESTVAKTPM---HPTCILE 798
MG LGI + + +G + S+ Y +++L +FNM + + P+ +
Sbjct: 1065 KDMGGASRILGIDIYRDRKGGVLKLSQEIYIRKVLDRFNMSGAKMTNAPVGAHFKLAAVR 1124
Query: 799 KEDKSGKVCQKLYRGTIGSLLY-LTASRPDILFSVHLCARF*SDPRETHLTAVKRILRYL 857
+ED+ Y +GS++Y + +RPD+ +++ L +R+ S P H AVK ++RYL
Sbjct: 1125 EEDECVDTDVVPYSSAVGSIMYAMLGTRPDLAYAICLISRYMSKPGSMHWEAVKWVMRYL 1184
Query: 858 KGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIAL 917
KG +L L++ K ++ ++GYCD++YA D R+S SG +G N VSW + Q +A+
Sbjct: 1185 KGAQDLNLVFTKEKDFTVTGYCDSNYAADLDRRRSISGYVFTIGGNTVSWKASLQPVVAM 1244
Query: 918 STAEAEYISAAICSTQMLWMKH*LEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIE 977
ST EAEYI+ A + + +W+K L+D + + + I+CD+ +AI LSKN + H R KHI+
Sbjct: 1245 STTEAEYIALAEAAKEAMWIKGLLQDMGMQQDKVKIWCDSQSAICLSKNSVYHERTKHID 1304
Query: 978 VKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1025
V++++IRD V+ G + + + T D TK + ++F L L +
Sbjct: 1305 VRFNYIRDVVESGDVDVLKIHTSRNPVDALTKCIPVNKFKSALGVLKL 1352
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 540 bits (1392), Expect = e-153
Identities = 345/1055 (32%), Positives = 551/1055 (51%), Gaps = 51/1055 (4%)
Query: 11 WYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSSPC----IDNV 66
W +D+GCS HMT +R F +L GG V G K+ G GTI V + + NV
Sbjct: 287 WVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRLTNV 346
Query: 67 LLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEA 126
+ + NLLS+ GY + ++ SVL +R +Y ++ +
Sbjct: 347 RYIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIA--GDSVLLTVRRCYTLYLLQWRPVTE 404
Query: 127 QNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQKDKFT 186
+++ ++ ++ +WHRRLGH S + + L K L L K + CE C K
Sbjct: 405 ESLS-VVKRQDDTILWHRRLGHMSQKNMDLLLKKGL---LDKKKVSKLETCEDCIYGKAK 460
Query: 187 KVPFKAKNVVSTSRSLELLHIDLFG-PVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDE 245
++ F T LE +H DL+G P S+G +Y + +DDY+R + FL KDE
Sbjct: 461 RIGFNLAQH-DTREKLEYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKTKDE 519
Query: 246 SHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQNGV 305
+ F + V+N+ RI +R+D+G +F N F+ GI +C TPQQNGV
Sbjct: 520 AFDKFVEWANLVENQTDKRIKTLRTDNGLEFCNRSFDEFCSQKGILWHRTCAYTPQQNGV 579
Query: 306 VERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKPN 365
ER NRTL E R+ML ++G+ K FWAEA +T + N+ + + P + W P
Sbjct: 580 AERMNRTLMEKVRSMLSDSGLPKKFWAEATHTTAILINKTPSSALNYEVPDKRWSGKSPI 639
Query: 366 ISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESIHVRF 425
SY FGC+ +V +T D K + ++ K +L+GY KG++ + + K S +V F
Sbjct: 640 YSYLRRFGCIAFV-HTDDG--KLNPRAKKGILVGYPIGVKGYKIWLLEEKKCVVSRNVIF 696
Query: 426 DDKL---DSDQSKLVEK---------FADLSIN----VSDKGKAP--EEVEPEEDEPEEE 467
+ D QSK EK + DL ++ ++ G P E P P
Sbjct: 697 QENASYKDMMQSKDAEKDENEAPPSSYLDLDLDHEEVITSGGDDPIVEAQSPFNPSPATT 756
Query: 468 AGPSNSQTLKKSRITAAHPKELIMGNKDEPVRTRSAFRPSE---ETLLSLKGLVSLIEPK 524
S + I + +L+ +R F + E L + + IEP
Sbjct: 757 QTYSEGVNSETDIIQSPLSYQLVRDRDRRTIRAPVRFDDEDYLAEALYTTEDSGE-IEPA 815
Query: 525 SIDEALQDKDWI---LAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKG-DV 580
EA + +W LAM EE+ KN W++VK+P+ VIG++W+++ KL G +
Sbjct: 816 DYSEAKRSMNWNKWKLAMNEEMESQIKNHTWTVVKRPQHQKVIGSRWIYKFKLGIPGVEE 875
Query: 581 VRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGY 640
R KARLVA+GY+Q++GIDY E FAPV + +IR+L+S ++ L Q+DVK+AFL+G
Sbjct: 876 GRFKARLVAKGYAQRKGIDYHEIFAPVVKHVSIRILMSIVAQEDLELEQLDVKTAFLHGE 935
Query: 641 ISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTT 700
+ E++Y+ P G+E+ K D V L KSLYGLKQAP+ W E+ ++++ E F R D+
Sbjct: 936 LKEKIYMVPPEGYEEMFKEDEVCLLNKSLYGLKQAPKQWNEKFNAYMSEIGFIRSLYDSC 995
Query: 701 LFCKTYKDDILI-VQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQ 759
+ K D + + +YVDD++ + N+ + E + F+M +G K LG+++ +
Sbjct: 996 AYIKELSDGSRVYLLLYVDDMLVAAKNKEDISQLKEELSQRFDMKDLGAAKRILGMEIIR 1055
Query: 760 TPEGT--YIHQSKYTKELLKKFNMLESTVAKTPMHP-----TCILEKEDKSGKVCQKL-Y 811
E ++ Q+ Y ++L+ +NM ES TP+ +EK+++ + + Y
Sbjct: 1056 NREENTLWLSQNGYLNKILETYNMAESKHVVTPLGAHLKMRAATVEKQEQDEDYMKSIPY 1115
Query: 812 RGTIGSLLY-LTASRPDILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKT 870
+GS++Y + +RPD+ + V + +R+ S P H VK +LRY+KG+ L YK++
Sbjct: 1116 SSAVGSIMYAMIGTRPDLAYPVGIISRYMSQPAREHWLGVKWVLRYIKGSLGTKLQYKRS 1175
Query: 871 SEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAIC 930
S++K+ GYCDAD+A + R+S +G LG + +SW S +Q +ALST EAEY+S
Sbjct: 1176 SDFKVVGYCDADHAACKDRRRSITGLVFTLGGSTISWKSGQQRVVALSTTEAEYMSLTEA 1235
Query: 931 STQMLWMKH*LEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKG 990
+ +WMK L+++ + ++ I+CD+ +AI+LSKN + H R KHI+V+Y +IRD + G
Sbjct: 1236 VKEAVWMKGLLKEFGYEQKSVEIFCDSQSAIALSKNNVHHERTKHIDVRYQYIRDIIANG 1295
Query: 991 VLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1025
+ +DT+ ADIFTK + ++F L L +
Sbjct: 1296 DGDVVKIDTEKNPADIFTKIVPVNKFQAALTLLQV 1330
>At4g17450 retrotransposon like protein
Length = 1433
Score = 531 bits (1367), Expect = e-150
Identities = 327/1042 (31%), Positives = 525/1042 (50%), Gaps = 72/1042 (6%)
Query: 9 QSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 68
+ W +DSG + H+T R ++ + V + KI G G I + + + NVL
Sbjct: 430 RGWVIDSGATHHVTHNRDLYLNFRSLENTFVRLPNDCTVKIAGIGFIQLSDAISLHNVLY 489
Query: 69 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 128
+ NL+S + + SQ+ + + N+ ++ +
Sbjct: 490 IPEFKFNLIS----------ELTKELMIGRGSQVGNLYVLDFNENNHTVSLKGTTSMCPE 539
Query: 129 VKCLLSVNEEQWVWHRRLGHASMRKISQLSK-LNL-VRGLPNLKFASDALCEACQKDKFT 186
SV + WH+RLGH + KI LS LNL V+ + +C C K
Sbjct: 540 FSVCSSVVVDSVTWHKRLGHPAYSKIDLLSDVLNLKVKKINKEHSPVCHVCHVCHLSKQK 599
Query: 187 KVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDES 246
+ F+++ + S + +L+HID +GP + TW+ L K +
Sbjct: 600 HLSFQSRQNMC-SAAFDLVHIDTWGPFSVPTNDA--------------TWIYLLKNKSDV 644
Query: 247 HAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCPRTPQQNGVV 306
VF FI V + ++ VRSD+ + KF LF ++GI SCP TP+QN VV
Sbjct: 645 LHVFPAFINMVHTQYQTKLKSVRSDNAHEL---KFTDLFAAHGIVAYHSCPETPEQNSVV 701
Query: 307 ERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKPNI 366
ERK++ + +AR +L ++ + FW + V TA ++ NR+ + NK+PYE KNI P
Sbjct: 702 ERKHQHILNVARALLFQSNIPLEFWGDCVLTAVFLINRLPTPVLNNKSPYEKLKNIPPAY 761
Query: 367 SYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTIEESIHVRFD 426
FGC+CY + + HKF+ ++ C+ LGY KG++ + + + S HV F
Sbjct: 762 ESLKTFGCLCYSSTSPKQRHKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISRHVIFH 821
Query: 427 DKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSRITAAHP 486
+ + F +S + D K + P P ++ L+++ I HP
Sbjct: 822 EDI----------FPFISSTIKDDIK---DFFPLLQFPAR----TDDLPLEQTSIIDTHP 864
Query: 487 KELIMGNKD----EPVRTRSAFRP------------SEETLLSLKGLVSLIEPKSIDEAL 530
+ + +K +P+ R P +E + + + + P+ EA
Sbjct: 865 HQDVSSSKALVPFDPLSKRQKKPPKHLQDFHCYNNTTEPFHAFINNITNAVIPQRYSEAK 924
Query: 531 QDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQ 590
K W AM+EE+ + + WS+V P + IG KWVF K N G + R KARLVA+
Sbjct: 925 DFKAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCKWVFTIKHNADGSIERYKARLVAK 984
Query: 591 GYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQP 650
GY+Q+EG+DY ETF+PVA+L ++R+++ + +HQ+D+ +AFLNG + EE+Y+ P
Sbjct: 985 GYTQEEGLDYEETFSPVAKLTSVRMMLLLAAKMKWSVHQLDISNAFLNGDLDEEIYMKIP 1044
Query: 651 PGFED---EKKPDH-VFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTY 706
PG+ D E P H + +L KS+YGLKQA R WY +LS+ L F++ D TLF K
Sbjct: 1045 PGYADLVGEALPPHAICRLHKSIYGLKQASRQWYLKLSNTLKGMGFQKSNADHTLFIKYA 1104
Query: 707 KDDILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYI 766
++ V +YVDDI+ S + +F +++ F++ +G KYFLGI++ ++ +G I
Sbjct: 1105 NGVLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKLRDLGAAKYFLGIEIARSEKGISI 1164
Query: 767 HQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGTIGSLLYLTASRP 826
Q KY ELL L S + P+ P+ L KED YR +G L+YL +RP
Sbjct: 1165 CQRKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGVPLTDSTSYRKLVGKLMYLQITRP 1224
Query: 827 DILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGD 886
DI ++V+ +F P HL+AV ++LRYLKGT GL Y ++ L GY D+D+
Sbjct: 1225 DIAYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQGLFYSADDKFDLRGYTDSDFGSC 1284
Query: 887 RTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQI 946
R+ + C F+G LVSW SK+Q T+++STAEAE+ + + + +M+W+ +D+++
Sbjct: 1285 TDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAEFRAMSQGTKEMIWLSRLFDDFKV 1344
Query: 947 LESNIP---IYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQW 1003
IP +YCDNTAA+ + N + H R K +E+ + R+ V+ G L FV+T Q
Sbjct: 1345 --PFIPPAYLYCDNTAALHIVNNSVFHERTKFVELDCYKTREAVESGFLKTMFVETGEQV 1402
Query: 1004 ADIFTKPLAEDRFNFILKNLNM 1025
AD TK + +F+ ++ + +
Sbjct: 1403 ADPLTKAIHPAQFHKLIGKMGV 1424
>At1g26990 polyprotein, putative
Length = 1436
Score = 530 bits (1364), Expect = e-150
Identities = 332/1038 (31%), Positives = 529/1038 (49%), Gaps = 71/1038 (6%)
Query: 6 LKHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSSPCIDN 65
L ++W +DSG S H+T ER ++ K V KI GTG I + + + N
Sbjct: 416 LSLRAWVIDSGASHHVTHERNLYHTYKALDRTFVRLPNGHTVKIEGTGFIQLTDALSLHN 475
Query: 66 VLLVDGLTHNLLSISQLADKGYD-VIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRL--- 121
VL + NLLS+S L V F C + +L + N+Y + L
Sbjct: 476 VLFIPEFKFNLLSVSVLTKTLQSKVSFTSDECMIQALTKELMLGKGSQVGNLYILNLDKS 535
Query: 122 ----SELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDAL- 176
S ++V SV E +WH+RLGH S KI LS + + LP K D+
Sbjct: 536 LVDVSSFPGKSV--CSSVKNESEMWHKRLGHPSFAKIDTLSDVLM---LPKQKINKDSSH 590
Query: 177 CEACQKDKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTW 236
C C K +PFK+ N + ++ EL+HID +GP ++ RY + IVDD+SR TW
Sbjct: 591 CHVCHLSKQKHLPFKSVNHIR-EKAFELVHIDTWGPFSVPTVDSYRYFLTIVDDFSRATW 649
Query: 237 VKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSC 296
+ L +K + VF +F+ V+ + ++ VRSD+ + KF LF GI D C
Sbjct: 650 IYLLKQKSDVLTVFPSFLKMVETQYHTKVCSVRSDNAHEL---KFNELFAKEGIKADHPC 706
Query: 297 PRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPY 356
P TP+QN VVERK++ L +AR ++ ++G+ +W + V TA ++ NR+ I N+TPY
Sbjct: 707 PETPEQNFVVERKHQHLLNVARALMFQSGIPLEYWGDCVLTAVFLINRLLSPVINNETPY 766
Query: 357 ELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKT 416
E KP+ S FGC+CY + KFD ++ C+ LGY KG++ + + +
Sbjct: 767 ERLTKGKPDYSSLKAFGCLCYCSTSPKSRTKFDPRAKACIFLGYPMGYKGYKLLDIETYS 826
Query: 417 IEESIHVRF-DDKLDSDQSKLVEKFADLSINVSDKGKAPEE----VEPEEDEPE--EEAG 469
+ S HV F +D S + + D ++ +E V+ D P +E+
Sbjct: 827 VSISRHVIFYEDIFPFASSNITDAAKDFFPHIYLPAPNNDEHLPLVQSSSDAPHNHDESS 886
Query: 470 -----PSNSQTLKKSRITAAHPKELIMGNKDEPVRTRSAFRP----------SEETLLSL 514
PS ++ ++ ++ + H ++ N + P T+++ P SE +
Sbjct: 887 SMIFVPSEPKSTRQRKLPS-HLQDFHCYN-NTPTTTKTSPYPLTNYISYSYLSEPFGAFI 944
Query: 515 KGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKL 574
+ + P+ EA DK W AM +E++ F + WS+ P +G KW+ K
Sbjct: 945 NIITATKLPQKYSEARLDKVWNDAMGKEISAFVRTGTWSICDLPAGKVAVGCKWIITIKF 1004
Query: 575 NEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKS 634
G + R+KARLVA+GY+QQEGID+ TF+PVA++ +++L+S + LHQ+D+ +
Sbjct: 1005 LADGSIERHKARLVAKGYTQQEGIDFFNTFSPVAKMVTVKVLLSLAPKMKWYLHQLDISN 1064
Query: 635 AFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFER 694
A LNG + EE+Y+ PPG+ + + G + +P A +
Sbjct: 1065 ALLNGDLEEEIYMKLPPGYSE-------------IQGQEVSPNA---------------K 1096
Query: 695 GKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLG 754
D TLF K L+V +YVDDI+ S ++ E + + F++ +GE K+FLG
Sbjct: 1097 CHGDHTLFVKAQDGFFLVVLVYVDDILIASTTEAASAELTSQLSSFFQLRDLGEPKFFLG 1156
Query: 755 IQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGT 814
I++ + +G + Q KY +LL + + + PM P L K+ + K YR
Sbjct: 1157 IEIARNADGISLCQRKYVLDLLASSDFSDCKPSSIPMEPNQKLSKDTGTLLEDGKQYRRI 1216
Query: 815 IGSLLYLTASRPDILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYK 874
+G L YL +RPDI F+V A++ S P + HL A+ +ILRYLKGT GL Y + +
Sbjct: 1217 LGKLQYLCLTRPDINFAVSKLAQYSSAPTDIHLQALHKILRYLKGTIGQGLFYGADTNFD 1276
Query: 875 LSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQM 934
L G+ D+D+ R+ +G F+G++LVSW SK+Q +++S+AEAEY + ++ + ++
Sbjct: 1277 LRGFSDSDWQTCPDTRRCVTGFAIFVGNSLVSWRSKKQDVVSMSSAEAEYRAMSVATKEL 1336
Query: 935 LWMKH*LEDYQI-LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLL 993
+W+ + L ++I +YCDN AA+ ++ N + H R KHIE H +R+ ++ G+L
Sbjct: 1337 IWLGYILTAFKIPFTHPAYLYCDNEAALHIANNSVFHERTKHIENDCHKVRECIEAGILK 1396
Query: 994 LKFVDTDHQWADIFTKPL 1011
FV TD+Q AD TKPL
Sbjct: 1397 TIFVRTDNQLADTLTKPL 1414
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 526 bits (1355), Expect = e-149
Identities = 338/1071 (31%), Positives = 561/1071 (51%), Gaps = 85/1071 (7%)
Query: 4 APLKHQSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVD----S 59
A + +W LD+GCS HMT + + K G+V G + ++ G G + + S
Sbjct: 305 AEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKNEDGS 364
Query: 60 SPCIDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI 119
+ + +V + ++ NL+S+ L DKG ++K + + D +VL K+++ +Y +
Sbjct: 365 TILLTDVRYIPEMSKNLISLGTLEDKGC-WFESKKGILTIFKNDLTVL-TGKKESTLYFL 422
Query: 120 RLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQL-SKLNLVRGLPNLKFASDALCE 178
+ + L A + +E +WH RLGH + + L SK +L
Sbjct: 423 QGTTL-AGEANVIDKEKDETSLWHSRLGHIGAKGLQVLVSKGHL---------------- 465
Query: 179 ACQKDKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTE-SIGGKRYGMVIVDDYSRWTWV 237
DK + F A V T L+ +H DL+G SIG +Y + +DD++R TW+
Sbjct: 466 ----DKNIMISFGAAKHV-TKDKLDYVHSDLWGSTNVPFSIGKCQYFITFIDDFTRRTWI 520
Query: 238 KFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAHDFSCP 297
F+ KDE+ + F + Q++N++ ++ + +D+G +F N +F+S G+ +C
Sbjct: 521 YFIRTKDEAFSKFVEWKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKEGVIRHRTCA 580
Query: 298 RTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYE 357
TPQQNGV ER NRT+ R ML E+G+ K FWAEA +TA ++ N+ I P E
Sbjct: 581 YTPQQNGVAERMNRTIMNKVRCMLSESGLGKQFWAEAASTAVFLINKSPSSSIEFDIPEE 640
Query: 358 LWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKTI 417
W P+ FG V Y+ + + +L+ ++ K + LGY + K F+ + + +
Sbjct: 641 KWTGHPPDYKILKKFGSVAYIHSDQGKLNP---RAKKGIFLGYPDGVKRFKVWLLEDRKC 697
Query: 418 EESIHVRFDDKLDSDQSKLVEKFADLSIN-VSDKGKAPEEVE---------PEEDEPEEE 467
S + F + + + +L N +S++ K EVE +DE + E
Sbjct: 698 VVSRDIVFQEN---------QMYKELQKNDMSEEDKQLTEVERTLIELKNLSADDENQSE 748
Query: 468 AGPSNSQTLKKSRITAAHPKELIMGNKDEP---------------VRTRSAFRPSEETLL 512
G +++Q + +A+ K++ + D+ +R F +++L+
Sbjct: 749 GGDNSNQEQASTTRSASKDKQVEETDSDDDCLENYLLARDRIRRQIRAPQRFVEEDDSLV 808
Query: 513 SLKGLVS----LIEPKSIDEALQDKD---WILAMEEELNQFSKNDVWSLVKKPESVHVIG 565
++ + EP++ +EA++ + W A EE++ KND W ++ KPE VIG
Sbjct: 809 GFALTMTEDGEVYEPETYEEAMRSPECEKWKQATIEEMDSMKKNDTWDVIDKPEGKRVIG 868
Query: 566 TKWVFRNKLNEKG-DVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHN 624
KW+F+ K G + R KARLVA+G+SQ+EGIDY E F+PV + +IR L+S V +
Sbjct: 869 CKWIFKRKAGIPGVEPPRYKARLVAKGFSQREGIDYQEIFSPVVKHVSIRYLLSIVVQFD 928
Query: 625 IVLHQIDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLS 684
+ L Q+DVK+AFL+G + E + + QP G+EDE + V LKKSLYGLKQ+PR W +R
Sbjct: 929 MELEQLDVKTAFLHGNLDEYILMSQPEGYEDEDSTEKVCLLKKSLYGLKQSPRQWNQRFD 988
Query: 685 SFLLENEFERGKVDTTLFCKTYKDDILI-VQIYVDDIIFGSANQSLCKEFYEMMQAEFEM 743
SF++ + ++R K + ++ + D I + +YVDD++ S N+ ++ E + EFEM
Sbjct: 989 SFMINSGYQRSKYNPCVYTQQLNDGSYIYLLLYVDDMLIASQNKDQIQKLKESLNREFEM 1048
Query: 744 SMMGELKYFLGIQVDQTPEGTYIH--QSKYTKELLKKFNMLESTVAKTPMHPTCIL---- 797
+G + LG+++ + E + QS+Y +L+ F M +S V++TP+ L
Sbjct: 1049 KDLGPARKILGMEITRNREQGILDLSQSEYVAGVLRAFGMDQSKVSQTPLGAHFKLRAAN 1108
Query: 798 EKEDKSGKVCQKL--YRGTIGSLLY-LTASRPDILFSVHLCARF*SDPRETHLTAVKRIL 854
EK KL Y IGS++Y + SRPD+ + V + +RF S P + H AVK ++
Sbjct: 1109 EKTLARDAEYMKLVPYPNAIGSIMYSMIGSRPDLAYPVGVVSRFMSKPSKEHWQAVKWVM 1168
Query: 855 RYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQST 914
RY+KGT + L +KK ++++ GYCD+DYA D R+S +G G N +SW S Q
Sbjct: 1169 RYMKGTQDTCLRFKKDDKFEIRGYCDSDYATDLDRRRSITGFVFTAGGNTISWKSGLQRV 1228
Query: 915 IALSTAEAEYISAAICSTQMLWMKH*LEDYQILESNIPIYCDNTAAISLSKNPILHSRAK 974
+ALST EAEY++ A + +W++ + + + + CD+ +AI+LSKN + H R K
Sbjct: 1229 VALSTTEAEYMALAEAVKEAIWLRGLAAEMGFEQDAVEVMCDSQSAIALSKNSVHHERTK 1288
Query: 975 HIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1025
HI+V+YHFIR+ + G + + + T ADIFTK + + LK L +
Sbjct: 1289 HIDVRYHFIREKIADGEIQVVKISTTWNPADIFTKTVPVSKLQEALKLLRV 1339
>At4g03810 putative retrotransposon protein
Length = 964
Score = 488 bits (1256), Expect = e-138
Identities = 317/980 (32%), Positives = 502/980 (50%), Gaps = 62/980 (6%)
Query: 63 IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNN------- 115
+ N V + N++S+S L +G+ K C S + + S +N
Sbjct: 5 LKNCYYVPAINKNIISVSCLDMEGFHFSIKNKCC---SFDRDDMFYGSAPLDNGLHVLNQ 61
Query: 116 ---IYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFA 172
IY IR + ++ ++ ++WH RLGH + + I +L L L + +
Sbjct: 62 SMPIYNIRTKKFKSNDLN-------PTFLWHCRLGHINEKHIQKLHSDGL---LNSFDYE 111
Query: 173 SDALCEACQKDKFTKVPFKAKNVVSTSRSLELLHIDLFGPVKTESIGGKRYGMVIVDDYS 232
S CE+C K TK PF + S L L+H D+ GP+ T + G +Y + DD+S
Sbjct: 112 SYETCESCLLGKMTKAPFTGHSE-RASDLLGLIHTDVCGPMSTSARGNYQYFITFTDDFS 170
Query: 233 RWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSYGIAH 292
R+ +V + K +S F F +VQN+ I +RSD GG++ + F GI
Sbjct: 171 RYGYVYLMKHKSKSFENFKEFQNEVQNQFGKSIKALRSDRGGEYLSQVFSDHLRECGIVS 230
Query: 293 DFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILN 352
+ P TPQ NGV ER+NRTL +M R+M+ T + FW A+ T+ ++ NR + +
Sbjct: 231 QLTPPGTPQWNGVSERRNRTLLDMVRSMMSHTDLPSPFWGYALETSAFMLNRCPSKSV-E 289
Query: 353 KTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYN- 411
KTPYE+W PN+S+ +GC Y + K KS KC +GY + +KG+ FY+
Sbjct: 290 KTPYEIWTGKVPNLSFLKIWGCESYA--KRLITDKLGPKSDKCYFVGYPKETKGYYFYHP 347
Query: 412 TDAKTIEESIHVRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEEV-EPEEDEPEEEAGP 470
TD K + + + L +F LS S EEV EP+ D P +
Sbjct: 348 TDNKVF-----------VVRNGAFLEREF--LSKGTSGSKVLLEEVREPQGDVPTSQEEH 394
Query: 471 SNSQTLKKSRITAAHPKELIMGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEAL 530
I ++ EP R R ++ EP S +EAL
Sbjct: 395 QLDLRRVVEPILVEPEVRRSERSRHEPDRFRDWVMDDHALF-----MIESDEPTSYEEAL 449
Query: 531 QDKD---WILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARL 587
D W+ A + E+ S+N VW+LV P+ V I KW+F+ K++ G++ KA L
Sbjct: 450 MGPDSDKWLEAAKSEMESMSQNKVWTLVDLPDGVKPIECKWIFKKKIDMDGNIQIYKAGL 509
Query: 588 VAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQIDVKSAFLNGYISEEVYV 647
VA+GY Q GIDY ET++PVA L++IR+L++ + +++ + Q+DVK+AFLNG + E VY+
Sbjct: 510 VAKGYKQVHGIDYDETYSPVAMLKSIRILLATAAHYDYEIWQMDVKTAFLNGNLEEHVYM 569
Query: 648 HQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFERGKVDTTLFCKTYK 707
QP GF + V KL +S+YGLKQA R+W R + + E +F R + + ++ KT
Sbjct: 570 TQPEGFTVPEAARKVCKLHRSIYGLKQASRSWNLRFNEAIKEFDFIRNEEEPCVYKKTSG 629
Query: 708 DDILIVQIYVDDIIFGSANQSLCKEFYEMMQAEFEMSMMGELKYFLGIQV--DQTPEGTY 765
+ + +YVDDI+ + L + + + F M MGE Y LGI++ D+ +
Sbjct: 630 SAVAFLVLYVDDILLLGNDIPLLQSVKTWLGSCFSMKDMGEAAYILGIRIYRDRLNKIIG 689
Query: 766 IHQSKYTKELLKKFNMLESTVAKTPMHPTCILEK------EDKSGKVCQKLYRGTIGSLL 819
+ Q Y ++L +FNM +S PM L K D+ ++ + Y IGS++
Sbjct: 690 LSQDTYIDKVLHRFNMHDSKKGFIPMSHGITLSKTQCPSTHDERERMSKIPYASAIGSIM 749
Query: 820 Y-LTASRPDILFSVHLCARF*SDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGY 878
Y + +RPD+ ++ + +R+ SDP E+H V+ I +YL+ T + L+Y + E +SGY
Sbjct: 750 YAMLYTRPDVACALSMTSRYQSDPGESHWIVVRNIFKYLRRTKDKFLVYGGSEELVVSGY 809
Query: 879 CDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMK 938
DA + D+ + +S SG L VSW S +QST+A ST EAEYI+A+ + +++W++
Sbjct: 810 TDASFQTDKDDFRSQSGFFFCLNGGAVSWKSTKQSTVADSTTEAEYIAASEAAKEVVWIR 869
Query: 939 H*LEDYQILES---NIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLK 995
+ + ++ S I +YCDN AI+ +K P H ++KHI+ +YH IR+ + +G + +
Sbjct: 870 KFITELGVVPSISGPIDLYCDNNGAIAQAKEPKSHQKSKHIQRRYHLIREIIDRGDVKIS 929
Query: 996 FVDTDHQWADIFTKPLAEDR 1015
V TD AD FTKPL + +
Sbjct: 930 RVSTDANVADHFTKPLPQPK 949
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 474 bits (1220), Expect = e-133
Identities = 295/838 (35%), Positives = 449/838 (53%), Gaps = 63/838 (7%)
Query: 229 DDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGKFENDKFESLFDSY 288
+D+SR WV FL KDE+ A F+ + V+ + ++ +R+D+G +F N KF+ +
Sbjct: 319 NDWSRKVWVYFLKTKDEAFASFTEWKKMVETQSERKLKHLRTDNGLEFCNHKFDEVCKKE 378
Query: 289 GIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVR 348
GI +C TPQQNGV ER NRT+ R+ML E+G+ K FWA+A +TA Y+ NR
Sbjct: 379 GIVRHRTCTYTPQQNGVAERLNRTIMNKVRSMLSESGLDKKFWAKAASTAVYLINRSPSS 438
Query: 349 PILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFR 408
I NK P ELW + PN S FGCV YV + + +L D ++ K + +GY KGFR
Sbjct: 439 SIENKIPEELWTSAVPNFSGLKRFGCVVYVYSQEGKL---DPRAKKGVFVGYPNGVKGFR 495
Query: 409 FYNTDAKTIEESIHVRF------DDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEED 462
+ + + S +V F D L+ S + F + + A +ED
Sbjct: 496 VWMIEEERCSISRNVVFREDVMYKDILNQSTSGMSFDFPLATNRIPSFECAGNR---KED 552
Query: 463 EPEEEAGPSNSQTLKKSRITAAHP--------KELIMGNKDEPVRTRSA------FRPSE 508
E + G S+ T + S + + +D+P R + +E
Sbjct: 553 EISVQGGVSDDDTKQSSEESPISTGSSGQNSGQRTYQIARDKPKRQTKIPDKLRDYELNE 612
Query: 509 ETLLSLKGLVSLI-------EPKSIDEALQDKD---WILAMEEELNQFSKNDVWSLVKKP 558
E L + G +I EP +ALQD D W+ A++EE+ KN+ W LV +
Sbjct: 613 EVLDEIAGYAYMITEDGGNPEPNDYQKALQDSDYKMWLKAVDEEIESLLKNNTWVLVNRD 672
Query: 559 ESVHVIGTKWVFRNKLNEKG-DVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLI 617
+ IG KWVF+ K G + R KARLV +GYSQ+EGIDY E F+PV + +IRLL+
Sbjct: 673 QFQKPIGCKWVFKRKSGIVGVEKPRFKARLVVKGYSQKEGIDYQEIFSPVVKHVSIRLLL 732
Query: 618 SFSVNHNIVLHQIDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPR 677
S + ++ L Q+DVK+AFL+GY+ E +Y+ QP G+ ++ PD V LK+SLYGL+Q+PR
Sbjct: 733 SMVTHCDMELQQMDVKTAFLHGYLDETIYIEQPEGYVHKRYPDKVCLLKRSLYGLRQSPR 792
Query: 678 AWYERLSSFLLENEFERGKVDTTLFCKTYKD-DILIVQIYVDDIIFGSANQSLCKEFYEM 736
W R + F+ + +ER K D+ ++ K + + + + +YVDDI+ S ++ + +
Sbjct: 793 QWNNRFNEFMQKIGYERSKYDSCVYFKELQSGEYIYLLLYVDDILIASRDKRTVCDLKAL 852
Query: 737 MQAEFEMSMMGELKYFLGIQV--DQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPM--- 791
+ +EFEM +G+ K LG+++ D+ I Q Y ++L F M ++ TPM
Sbjct: 853 LNSEFEMKDLGDAKKILGMEIVRDRKAGTMSISQEGYLLKVLGNFGMDQAKPVFTPMGAH 912
Query: 792 ---HPTCILEKEDKSGKVCQKLYRGTIGSLLY-LTASRPDILFSVHLCARF*SDPRETHL 847
P E +S + Y+ +GSL+Y + +RPD+ SV L RF S P + H
Sbjct: 913 FKLKPATDEEVMRQSEVMRAVPYQSAVGSLMYSMIGTRPDLAHSVGLVCRFMSKPLKEHW 972
Query: 848 TAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSW 907
AVK ILRY++G+ + L YK E L GYCD+DYA D+ R+STSG
Sbjct: 973 QAVKWILRYIRGSIDRKLCYKNEGELILEGYCDSDYAADKEGRRSTSG------------ 1020
Query: 908 ASKRQSTIALSTAEAEYISAAICSTQMLWMKH*LEDYQILESNIPIYCDNTAAISLSKNP 967
+ALS+ EAEY++ + + +W+K + + ++ + I+CD+ +AI+L+KN
Sbjct: 1021 ----VKVVALSSTEAEYMALTDGAKEAIWLKGHVSELGFVQKTVNIHCDSQSAIALAKNA 1076
Query: 968 ILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1025
+ H R KHI+VKYHFIRD V G + + +DT+ ADIFTK L +F L+ L +
Sbjct: 1077 VYHERTKHIDVKYHFIRDLVNNGEVQVLKIDTEDNPADIFTKVLPVSKFQDALELLRV 1134
Score = 33.5 bits (75), Expect = 0.64
Identities = 13/47 (27%), Positives = 22/47 (46%)
Query: 9 QSWYLDSGCSRHMTGERRMFRELKLKPGGEVGFGGNEKGKIVGTGTI 55
+ W +D+GCS HMT + + G+V N ++ G G +
Sbjct: 264 EEWIMDTGCSFHMTPRKEYLMDFVEAKSGKVRMANNSFSEVKGIGKV 310
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,614,839
Number of Sequences: 26719
Number of extensions: 1047991
Number of successful extensions: 4001
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 120
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 3282
Number of HSP's gapped (non-prelim): 311
length of query: 1032
length of database: 11,318,596
effective HSP length: 109
effective length of query: 923
effective length of database: 8,406,225
effective search space: 7758945675
effective search space used: 7758945675
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0177.6


