

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0177.14
(805 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
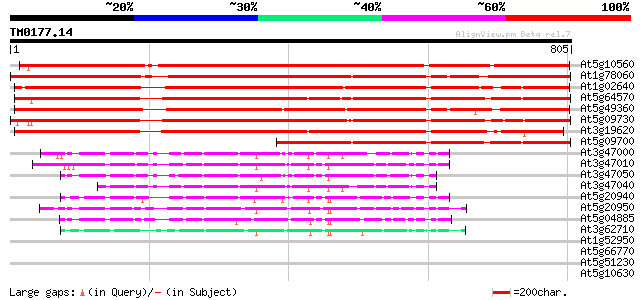
Score E
Sequences producing significant alignments: (bits) Value
At5g10560 beta-xylosidase - like protein 1037 0.0
At1g78060 putative protein 762 0.0
At1g02640 beta-xylosidase, putative 722 0.0
At5g64570 beta-xylosidase 708 0.0
At5g49360 xylosidase 704 0.0
At5g09730 beta-xylosidase - like protein 704 0.0
At3g19620 beta-xylosidase, putative 702 0.0
At5g09700 beta-glucosidase - like protein 340 2e-93
At3g47000 beta-D-glucan exohydrolase - like protein 134 3e-31
At3g47010 beta-D-glucan exohydrolase - like protein 126 4e-29
At3g47050 beta-D-glucan exohydrolase - like protein 120 3e-27
At3g47040 beta-D-glucan exohydrolase - like protein 114 2e-25
At5g20940 beta-glucosidase - like protein 107 2e-23
At5g20950 beta-D-glucan exohydrolase-like protein 103 5e-22
At5g04885 unknown protein 102 7e-22
At3g62710 beta-D-glucan exohydrolase-like protein 100 3e-21
At1g52950 replication protein A1, putative 31 2.4
At5g66770 SCARECROW gene regulator 30 5.4
At5g51230 putative protein 30 7.0
At5g10630 putative protein 30 7.0
>At5g10560 beta-xylosidase - like protein
Length = 792
Score = 1037 bits (2682), Expect = 0.0
Identities = 517/805 (64%), Positives = 622/805 (77%), Gaps = 36/805 (4%)
Query: 14 VVLIHFTTSIED----------FACKRPHHSRYKFCDTSLSIPTRAHSLVSLLTLPEKIQ 63
+ L+ FT++I + F CK PH S Y FC+ SLSI RA SLVSLL LPEKI
Sbjct: 8 ISLLFFTSAIAETFKNLDSHPQFPCKPPHFSSYPFCNVSLSIKQRAISLVSLLMLPEKIG 67
Query: 64 QLSNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTL 123
QLSN A+S+PRLGIP Y+WWSESLHG+A NGPGVSF+G++SAAT FPQVIVSAASFNRTL
Sbjct: 68 QLSNTAASVPRLGIPPYEWWSESLHGLADNGPGVSFNGSISAATSFPQVIVSAASFNRTL 127
Query: 124 WFLIASAVAVEARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYV 183
W+ I SAVAVE RAM+N GQAGLTFWAPN+NVFRDPRWGRGQETPGEDP V S Y V++V
Sbjct: 128 WYEIGSAVAVEGRAMYNGGQAGLTFWAPNINVFRDPRWGRGQETPGEDPKVVSEYGVEFV 187
Query: 184 RGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEK 243
RG Q K + KR SD D D D D LM+SACCKHFTAYDLEK
Sbjct: 188 RGFQEKKKRKVL---KRRFSD--------DVDDDRHDDDADGKLMLSACCKHFTAYDLEK 236
Query: 244 WGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLLGVA 303
WG F RY+FNAVV++QD+EDTYQPPF C++ GKASCLMCSYN VNGVPACA DLL A
Sbjct: 237 WGNFTRYDFNAVVTEQDMEDTYQPPFETCIRDGKASCLMCSYNAVNGVPACAQGDLLQKA 296
Query: 304 RNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQ 363
R WGF+GYITSDCDAVAT+F YQGY KS E+AVA+ +KAG DINCGTYMLRHT SA+EQ
Sbjct: 297 RVEWGFEGYITSDCDAVATIFAYQGYTKSPEEAVADAIKAGVDINCGTYMLRHTQSAIEQ 356
Query: 364 GKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGIV 423
GKV EE +DRALLNLF+VQLRLGLFDGDPR G+YGKLG +D+C+S+H+ LALEA RQGIV
Sbjct: 357 GKVSEELVDRALLNLFAVQLRLGLFDGDPRRGQYGKLGSNDICSSDHRKLALEATRQGIV 416
Query: 424 LLKNDKKFLPLNRNYGSSLAVIGPMAVT-NKLGGGYSGIPCSPKSLYEGLAEYAKKISYA 482
LLKND K LPLN+N+ SSLA++GPMA + +GG Y+G PC K+L+ L EY KK SYA
Sbjct: 417 LLKNDHKLLPLNKNHVSSLAIVGPMANNISNMGGTYTGKPCQRKTLFTELLEYVKKTSYA 476
Query: 483 SGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSI 542
SGCSD+SC+SD GF EA+ A+ ADFV++VAG+D + ETED DRVSL LPGKQ DLVS +
Sbjct: 477 SGCSDVSCDSDTGFGEAVAIAKGADFVIVVAGLDLSQETEDKDRVSLSLPGKQKDLVSHV 536
Query: 543 AAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFN 602
AA SK PVILVLTGGGP+DV+FA+ + I SI+W+GYPGE GG+ALAEIIFG+ FN
Sbjct: 537 AAVSKKPVILVLTGGGPVDVTFAKNDPRIGSIIWIGYPGETGGQALAEIIFGD-----FN 591
Query: 603 AAGRLPMTWYPESFTNVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYK 662
GRLP TWYPESFT+V M+DM MRA+ SRGYPGRTYRFYTG +VY FG GLSY+ F YK
Sbjct: 592 PGGRLPTTWYPESFTDVAMSDMHMRANSSRGYPGRTYRFYTGPQVYSFGTGLSYTKFEYK 651
Query: 663 FLSAPSKVSLSRI--TKGSLRKSLSDQAEKEVYGVDYVQVDELL--SCNSLSFPVHISVT 718
LSAP ++SLS + + S +K L E + Y+Q+D+++ SC SL F V + V+
Sbjct: 652 ILSAPIRLSLSELLPQQSSHKKQLQHGEE-----LRYLQLDDVIVNSCESLRFNVRVHVS 706
Query: 719 NLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFADE 778
N G++DGSHVVMLFSK P V+ G P+ QL+G+ RVH S++ +ET ++DPC+ LS A++
Sbjct: 707 NTGEIDGSHVVMLFSKMPPVLSGVPEKQLIGYDRVHVRSNEMMETVFVIDPCKQLSVAND 766
Query: 779 QGKRVFPLGNHVLSVGDVEHTVSIE 803
GKRV PLG+HVL +GD++H++S+E
Sbjct: 767 VGKRVIPLGSHVLFLGDLQHSLSVE 791
>At1g78060 putative protein
Length = 767
Score = 762 bits (1968), Expect = 0.0
Identities = 399/812 (49%), Positives = 523/812 (64%), Gaps = 53/812 (6%)
Query: 1 MSNQWRTLFFIFFVVLIHFTTSIEDFACKRPHHSRYKFCDTSLSIPTRAHSLVSLLTLPE 60
M+ Q L +F V + P Y+FC T L I RA LVS LT+ E
Sbjct: 1 MAKQLLLLLLLFIVHGVESAPPPHSCDPSNPTTKLYQFCRTDLPIGKRARDLVSRLTIDE 60
Query: 61 KIQQLSNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFN 120
KI QL N A IPRLG+PAY+WWSE+LHG+A GPG+ F+G V AAT FPQVI++AASF+
Sbjct: 61 KISQLVNTAPGIPRLGVPAYEWWSEALHGVAYAGPGIRFNGTVKAATSFPQVILTAASFD 120
Query: 121 RTLWFLIASAVAVEARAMFNVGQA-GLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYA 179
WF IA + EAR ++N GQA G+TFWAPN+N+FRDPRWGRGQETPGEDPM+ YA
Sbjct: 121 SYEWFRIAQVIGKEARGVYNAGQANGMTFWAPNINIFRDPRWGRGQETPGEDPMMTGTYA 180
Query: 180 VDYVRGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAY 239
V YVRGLQG + F ++ LS++ L SACCKHFTAY
Sbjct: 181 VAYVRGLQG-----DSFDGRKTLSNH---------------------LQASACCKHFTAY 214
Query: 240 DLEKWGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDL 299
DL++W RY FNA VS DL +TYQPPF+ C+++G+AS +MC+YN VNG+P+CA +L
Sbjct: 215 DLDRWKGITRYVFNAQVSLADLAETYQPPFKKCIEEGRASGIMCAYNRVNGIPSCADPNL 274
Query: 300 LG-VARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTA 358
L AR W F+GYITSDCDAV+ +++ QGY KS EDAVA+VLKAG D+NCG+Y+ +HT
Sbjct: 275 LTRTARGQWAFRGYITSDCDAVSIIYDAQGYAKSPEDAVADVLKAGMDVNCGSYLQKHTK 334
Query: 359 SAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAA 418
SA++Q KV E DIDRALLNLFSV++RLGLF+GDP YG + P++VC+ H+ LAL+AA
Sbjct: 335 SALQQKKVSETDIDRALLNLFSVRIRLGLFNGDPTKLPYGNISPNEVCSPAHQALALDAA 394
Query: 419 RQGIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAK 477
R GIVLLKN+ K LP ++ SSLAVIGP A V L G Y+G PC + + L Y K
Sbjct: 395 RNGIVLLKNNLKLLPFSKRSVSSLAVIGPNAHVVKTLLGNYAGPPCKTVTPLDALRSYVK 454
Query: 478 KISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMD 537
Y GC ++C S+ +A+ A+ AD VV++ G+D T E ED DRV L LPGKQ +
Sbjct: 455 NAVYHQGCDSVAC-SNAAIDQAVAIAKNADHVVLIMGLDQTQEKEDFDRVDLSLPGKQQE 513
Query: 538 LVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESN 597
L++S+A A+K PV+LVL GGP+D+SFA N I SI+W GYPGEAGG A++EIIFG+ N
Sbjct: 514 LITSVANAAKKPVVLVLICGGPVDISFAANNNKIGSIIWAGYPGEAGGIAISEIIFGDHN 573
Query: 598 PVGFNAAGRLPMTWYPESFTNVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYS 657
P GRLP+TWYP+SF N+ M DM MR+ + GYPGRTY+FY G +VY FGHGLSYS
Sbjct: 574 P-----GGRLPVTWYPQSFVNIQMTDMRMRS--ATGYPGRTYKFYKGPKVYEFGHGLSYS 626
Query: 658 GFSYKFLS-APSKVSLSRITKGSLRKSLSDQAEKEVYGVDYVQVDEL--LSCNSLSFPVH 714
+SY+F + A + + L++ +A+ V Y V E+ C+ V
Sbjct: 627 AYSYRFKTLAETNLYLNQ-----------SKAQTNSDSVRYTLVSEMGKEGCDVAKTKVT 675
Query: 715 ISVTNLGDLDGSHVVMLFSKWPKVVEGS--PQTQLVGFSRVHTVSSKSIETSILVDPCEH 772
+ V N G++ G H V++F++ + E + QLVGF + + + E + CEH
Sbjct: 676 VEVENQGEMAGKHPVLMFARHERGGEDGKRAEKQLVGFKSIVLSNGEKAEMEFEIGLCEH 735
Query: 773 LSFADEQGKRVFPLGNHVLSVGDVEHTVSIEI 804
LS A+E G V G + L+VGD E + + +
Sbjct: 736 LSRANEFGVMVLEEGKYFLTVGDSELPLIVNV 767
>At1g02640 beta-xylosidase, putative
Length = 768
Score = 722 bits (1864), Expect = 0.0
Identities = 377/803 (46%), Positives = 505/803 (61%), Gaps = 63/803 (7%)
Query: 8 LFFIFFV--VLIHFTTSIEDFAC--KRPHHSRYKFCDTSLSIPTRAHSLVSLLTLPEKIQ 63
LFF+ V +H S E FAC K + +FC S+ IP R L+ LTL EK+
Sbjct: 13 LFFLISSSSVCVH---SRETFACDTKDAATATLRFCQLSVPIPERVRDLIGRLTLAEKVS 69
Query: 64 QLSNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTL 123
L N A++IPRLGI Y+WWSE+LHG++ GPG F G AAT FPQVI + ASFN +L
Sbjct: 70 LLGNTAAAIPRLGIKGYEWWSEALHGVSNVGPGTKFGGVYPAATSFPQVITTVASFNASL 129
Query: 124 WFLIASAVAVEARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYV 183
W I V+ EARAM+N G GLT+W+PNVN+ RDPRWGRGQETPGEDP+VA YA YV
Sbjct: 130 WESIGRVVSNEARAMYNGGVGGLTYWSPNVNILRDPRWGRGQETPGEDPVVAGKYAASYV 189
Query: 184 RGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEK 243
RGLQG +D L V+ACCKHFTAYDL+
Sbjct: 190 RGLQG---------------------------------NDRSRLKVAACCKHFTAYDLDN 216
Query: 244 WGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLLG-V 302
W R++FNA VS+QD+EDT+ PFR CV++G + +MCSYN+VNGVP CA +LL
Sbjct: 217 WNGVDRFHFNAKVSKQDIEDTFDVPFRMCVKEGNVASIMCSYNQVNGVPTCADPNLLKKT 276
Query: 303 ARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVE 362
RN WG GYI SDCD+V +++ Q Y + E+A A+ +KAG D++CG ++ HT AV+
Sbjct: 277 IRNQWGLNGYIVSDCDSVGVLYDTQHYTGTPEEAAADSIKAGLDLDCGPFLGAHTIDAVK 336
Query: 363 QGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGI 422
+ ++E D+D AL+N +VQ+RLG+FDGD YG LGP VCT HK LALEAA+QGI
Sbjct: 337 KNLLRESDVDNALINTLTVQMRLGMFDGDIAAQPYGHLGPAHVCTPVHKGLALEAAQQGI 396
Query: 423 VLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAKKISY 481
VLLKN LPL+ ++AVIGP + T + G Y+G+ C S +G+ YA+ I +
Sbjct: 397 VLLKNHGSSLPLSSQRHRTVAVIGPNSDATVTMIGNYAGVACGYTSPVQGITGYARTI-H 455
Query: 482 ASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSS 541
GC D+ C D F A+E AR AD V+V G+D ++E E DR SLLLPGKQ +LVS
Sbjct: 456 QKGCVDVHCMDDRLFDAAVEAARGADATVLVMGLDQSIEAEFKDRNSLLLPGKQQELVSR 515
Query: 542 IAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGF 601
+A A+K PVILVL GGP+D+SFAE+++ IP+I+W GYPG+ GG A+A+I+FG +NP
Sbjct: 516 VAKAAKGPVILVLMSGGPIDISFAEKDRKIPAIVWAGYPGQEGGTAIADILFGSANP--- 572
Query: 602 NAAGRLPMTWYPESF-TNVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFS 660
G+LPMTWYP+ + TN+PM +MSMR S+ PGRTYRFY G VY FGHGLSY+ F+
Sbjct: 573 --GGKLPMTWYPQDYLTNLPMTEMSMRPVHSKRIPGRTYRFYDGPVVYPFGHGLSYTRFT 630
Query: 661 YKFLSAPSKVSLSRITKGSLRKSLSDQAEKEVYGVDYVQVDELLSCNSLSFPVHISVTNL 720
+ AP + ++ +G ++S ++ + + C+ LS VH+ VTN+
Sbjct: 631 HNIADAPKVIPIA--VRGR-NGTVSGKSIRVTHA----------RCDRLSLGVHVEVTNV 677
Query: 721 GDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFADEQG 780
G DG+H +++FS P E +P+ QLV F RVH + + + C++LS D G
Sbjct: 678 GSRDGTHTMLVFSA-PPGGEWAPKKQLVAFERVHVAVGEKKRVQVNIHVCKYLSVVDRAG 736
Query: 781 KRVFPLGNHVLSVGDVEHTVSIE 803
R P+G+H + +GD HTVS++
Sbjct: 737 NRRIPIGDHGIHIGDESHTVSLQ 759
>At5g64570 beta-xylosidase
Length = 784
Score = 708 bits (1828), Expect = 0.0
Identities = 384/807 (47%), Positives = 500/807 (61%), Gaps = 56/807 (6%)
Query: 8 LFFIFFVVLIHFTTSIED--FACK---RPHHSRYKFCDTSLSIPTRAHSLVSLLTLPEKI 62
+F FF+ ++F+ + FAC P + Y FC+T L I R LV+ LTL EKI
Sbjct: 24 IFLCFFLYFLNFSNAQSSPVFACDVAANPSLAAYGFCNTVLKIEYRVADLVARLTLQEKI 83
Query: 63 QQLSNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRT 122
L + A+ + RLGIP Y+WWSE+LHG++ GPG F V AT FPQVI++AASFN +
Sbjct: 84 GFLVSKANGVTRLGIPTYEWWSEALHGVSYIGPGTHFSSQVPGATSFPQVILTAASFNVS 143
Query: 123 LWFLIASAVAVEARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDY 182
L+ I V+ EARAM+NVG AGLT+W+PNVN+FRDPRWGRGQETPGEDP++AS YA Y
Sbjct: 144 LFQAIGKVVSTEARAMYNVGLAGLTYWSPNVNIFRDPRWGRGQETPGEDPLLASKYASGY 203
Query: 183 VRGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLE 242
V+GLQ DG D + L V+ACCKH+TAYD++
Sbjct: 204 VKGLQ------------------------------ETDGGDSNRLKVAACCKHYTAYDVD 233
Query: 243 KWGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLL-G 301
W RY+FNAVV+QQD++DTYQPPF+ CV G + +MCSYN+VNG P CA DLL G
Sbjct: 234 NWKGVERYSFNAVVTQQDMDDTYQPPFKSCVVDGNVASVMCSYNQVNGKPTCADPDLLSG 293
Query: 302 VARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAV 361
V R W GYI SDCD+V +++ Q Y K+ +A A + AG D+NCG+++ +HT AV
Sbjct: 294 VIRGEWKLNGYIVSDCDSVDVLYKNQHYTKTPAEAAAISILAGLDLNCGSFLGQHTEEAV 353
Query: 362 EQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQG 421
+ G V E ID+A+ N F +RLG FDG+P+ YG LGP DVCTS ++ LA +AARQG
Sbjct: 354 KSGLVNEAAIDKAISNNFLTLMRLGFFDGNPKNQIYGGLGPTDVCTSANQELAADAARQG 413
Query: 422 IVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAKKIS 480
IVLLKN LPL+ +LAVIGP A VT + G Y G PC + +GLA +
Sbjct: 414 IVLLKN-TGCLPLSPKSIKTLAVIGPNANVTKTMIGNYEGTPCKYTTPLQGLAG-TVSTT 471
Query: 481 YASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVS 540
Y GCS+++C + A A + A AD V+V G D ++E E DRV L LPG+Q +LV
Sbjct: 472 YLPGCSNVAC-AVADVAGATKLAATADVSVLVIGADQSIEAESRDRVDLHLPGQQQELVI 530
Query: 541 SIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVG 600
+A A+K PV+LV+ GG D++FA+ + I ILWVGYPGEAGG A+A+IIFG NP
Sbjct: 531 QVAKAAKGPVLLVIMSGGGFDITFAKNDPKIAGILWVGYPGEAGGIAIADIIFGRYNP-- 588
Query: 601 FNAAGRLPMTWYPESFT-NVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGF 659
+G+LPMTWYP+S+ VPM M+MR D + GYPGRTYRFYTG VY FG GLSY+ F
Sbjct: 589 ---SGKLPMTWYPQSYVEKVPMTIMNMRPDKASGYPGRTYRFYTGETVYAFGDGLSYTKF 645
Query: 660 SYKFLSAPSKVSLSRITKGSLRKSLSDQAEKEVYGVDYV--QVDELLSCNSLSFPVHISV 717
S+ + APS VSL R S E +D + + +S +F VHI V
Sbjct: 646 SHTLVKAPSLVSLGLEENHVCRSS-------ECQSLDAIGPHCENAVSGGGSAFEVHIKV 698
Query: 718 TNLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFAD 777
N GD +G H V LF+ P + GSP+ LVGF ++ + V+ C+ LS D
Sbjct: 699 RNGGDREGIHTVFLFTT-PPAIHGSPRKHLVGFEKIRLGKREEAVVRFKVEICKDLSVVD 757
Query: 778 EQGKRVFPLGNHVLSVGDVEHTVSIEI 804
E GKR LG H+L VGD++H++SI I
Sbjct: 758 EIGKRKIGLGKHLLHVGDLKHSLSIRI 784
>At5g49360 xylosidase
Length = 774
Score = 704 bits (1817), Expect = 0.0
Identities = 371/808 (45%), Positives = 501/808 (61%), Gaps = 64/808 (7%)
Query: 8 LFFIFFVVLIHFTTSIED-FACKRPHH--SRYKFCDTSLSIPTRAHSLVSLLTLPEKIQQ 64
+ +F + L+H + S+ FAC + +FC ++ I R L+ LTL EKI+
Sbjct: 16 VILVFLLCLVHSSESLRPLFACDPANGLTRTLRFCRANVPIHVRVQDLLGRLTLQEKIRN 75
Query: 65 LSNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTLW 124
L NNA+++PRLGI Y+WWSE+LHGI+ GPG F GA AT FPQVI +AASFN++LW
Sbjct: 76 LVNNAAAVPRLGIGGYEWWSEALHGISDVGPGAKFGGAFPGATSFPQVITTAASFNQSLW 135
Query: 125 FLIASAVAVEARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVR 184
I V+ EARAM+N G AGLT+W+PNVN+ RDPRWGRGQETPGEDP+VA+ YA YVR
Sbjct: 136 EEIGRVVSDEARAMYNGGVAGLTYWSPNVNILRDPRWGRGQETPGEDPIVAAKYAASYVR 195
Query: 185 GLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKW 244
GLQG G+ L V+ACCKH+TAYDL+ W
Sbjct: 196 GLQGTAA--------------------------------GNRLKVAACCKHYTAYDLDNW 223
Query: 245 GQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLL-GVA 303
R++FNA V+QQDLEDTY PF+ CV +GK + +MCSYN+VNG P CA E+LL
Sbjct: 224 NGVDRFHFNAKVTQQDLEDTYNVPFKSCVYEGKVASVMCSYNQVNGKPTCADENLLKNTI 283
Query: 304 RNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQ 363
R W GYI SDCD+V F Q Y + E+A A +KAG D++CG ++ T AV++
Sbjct: 284 RGQWRLNGYIVSDCDSVDVFFNQQHYTSTPEEAAARSIKAGLDLDCGPFLAIFTEGAVKK 343
Query: 364 GKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGIV 423
G + E DI+ AL N +VQ+RLG+FDG+ G Y LGP DVCT HK LALEAA QGIV
Sbjct: 344 GLLTENDINLALANTLTVQMRLGMFDGN--LGPYANLGPRDVCTPAHKHLALEAAHQGIV 401
Query: 424 LLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAKKISYA 482
LLKN + LPL+ ++AVIGP + VT + G Y+G C+ S +G++ YA+ + A
Sbjct: 402 LLKNSARSLPLSPRRHRTVAVIGPNSDVTETMIGNYAGKACAYTSPLQGISRYARTLHQA 461
Query: 483 SGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSI 542
GC+ ++C + GF A AR+AD V+V G+D ++E E DR LLLPG Q DLV+ +
Sbjct: 462 -GCAGVACKGNQGFGAAEAAAREADATVLVMGLDQSIEAETRDRTGLLLPGYQQDLVTRV 520
Query: 543 AAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFN 602
A AS+ PVILVL GGP+DV+FA+ + + +I+W GYPG+AGG A+A IIFG +NP
Sbjct: 521 AQASRGPVILVLMSGGPIDVTFAKNDPRVAAIIWAGYPGQAGGAAIANIIFGAANP---- 576
Query: 603 AAGRLPMTWYPESF-TNVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSY 661
G+LPMTWYP+ + VPM M+MRA S YPGRTYRFY G V+ FG GLSY+ F++
Sbjct: 577 -GGKLPMTWYPQDYVAKVPMTVMAMRA--SGNYPGRTYRFYKGPVVFPFGFGLSYTTFTH 633
Query: 662 KFLSAP---SKVSLSRITKGSLRKSLSDQAEKEVYGVDYVQVDELLSCNSL-SFPVHISV 717
+P VSLS + + + S + K + +CNS P+H+ V
Sbjct: 634 SLAKSPLAQLSVSLSNLNSANTILNSSSHSIKVSH----------TNCNSFPKMPLHVEV 683
Query: 718 TNLGDLDGSHVVMLFSKWP-KVVEG-SPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSF 775
+N G+ DG+H V +F++ P ++G QL+ F +VH ++ + VD C+HL
Sbjct: 684 SNTGEFDGTHTVFVFAEPPINGIKGLGVNKQLIAFEKVHVMAGAKQTVQVDVDACKHLGV 743
Query: 776 ADEQGKRVFPLGNHVLSVGDVEHTVSIE 803
DE GKR P+G H L +GD++HT+ ++
Sbjct: 744 VDEYGKRRIPMGEHKLHIGDLKHTILVQ 771
>At5g09730 beta-xylosidase - like protein
Length = 773
Score = 704 bits (1816), Expect = 0.0
Identities = 383/820 (46%), Positives = 510/820 (61%), Gaps = 63/820 (7%)
Query: 1 MSNQWRTLF-----FIFFVVLIHFTTSIED---FACK---RPHHSRYKFCDTSLSIPTRA 49
M+++ R LF F+ F+V I ++ + FAC P + +FC+ LSI R
Sbjct: 1 MASRNRALFSVSTLFLCFIVCISEQSNNQSSPVFACDVTGNPSLAGLRFCNAGLSIKARV 60
Query: 50 HSLVSLLTLPEKIQQLSNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDF 109
LV LTL EKI L++ A + RLGIP+Y+WWSE+LHG++ G G F G V AT F
Sbjct: 61 TDLVGRLTLEEKIGFLTSKAIGVSRLGIPSYKWWSEALHGVSNVGGGSRFTGQVPGATSF 120
Query: 110 PQVIVSAASFNRTLWFLIASAVAVEARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPG 169
PQVI++AASFN +L+ I V+ EARAM+NVG AGLTFW+PNVN+FRDPRWGRGQETPG
Sbjct: 121 PQVILTAASFNVSLFQAIGKVVSTEARAMYNVGSAGLTFWSPNVNIFRDPRWGRGQETPG 180
Query: 170 EDPMVASAYAVDYVRGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMV 229
EDP ++S YAV YV+GLQ DG D + L V
Sbjct: 181 EDPTLSSKYAVAYVKGLQ------------------------------ETDGGDPNRLKV 210
Query: 230 SACCKHFTAYDLEKWGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVN 289
+ACCKH+TAYD++ W R FNAVV+QQDL DT+QPPF+ CV G + +MCSYN+VN
Sbjct: 211 AACCKHYTAYDIDNWRNVNRLTFNAVVNQQDLADTFQPPFKSCVVDGHVASVMCSYNQVN 270
Query: 290 GVPACASEDLL-GVARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDIN 348
G P CA DLL GV R W GYI SDCD+V +F Q Y K+ E+AVA+ L AG D+N
Sbjct: 271 GKPTCADPDLLSGVIRGQWQLNGYIVSDCDSVDVLFRKQHYAKTPEEAVAKSLLAGLDLN 330
Query: 349 CGTYMLRHTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTS 408
C + +H AV+ G V E ID+A+ N F+ +RLG FDGDP+ YG LGP DVCT+
Sbjct: 331 CDHFNGQHAMGAVKAGLVNETAIDKAISNNFATLMRLGFFDGDPKKQLYGGLGPKDVCTA 390
Query: 409 EHKTLALEAARQGIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKS 467
+++ LA + ARQGIVLLKN LPL+ + +LAVIGP A T + G Y G+PC +
Sbjct: 391 DNQELARDGARQGIVLLKNSAGSLPLSPSAIKTLAVIGPNANATETMIGNYHGVPCKYTT 450
Query: 468 LYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRV 527
+GLAE +Y GC +++C D A++ A AD VV+V G D ++E E HDRV
Sbjct: 451 PLQGLAETVSS-TYQLGC-NVAC-VDADIGSAVDLAASADAVVLVVGADQSIEREGHDRV 507
Query: 528 SLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKA 587
L LPGKQ +LV+ +A A++ PV+LV+ GG D++FA+ ++ I SI+WVGYPGEAGG A
Sbjct: 508 DLYLPGKQQELVTRVAMAARGPVVLVIMSGGGFDITFAKNDKKITSIMWVGYPGEAGGLA 567
Query: 588 LAEIIFGESNPVGFNAAGRLPMTWYPESFT-NVPMNDMSMRADPSRGYPGRTYRFYTGSR 646
+A++IFG NP +G LPMTWYP+S+ VPM++M+MR D S+GYPGR+YRFYTG
Sbjct: 568 IADVIFGRHNP-----SGNLPMTWYPQSYVEKVPMSNMNMRPDKSKGYPGRSYRFYTGET 622
Query: 647 VYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSLRKSLSDQAEKEVYGVDYV--QVDELL 704
VY F L+Y+ F ++ + AP VSLS R S E +D + + +
Sbjct: 623 VYAFADALTYTKFDHQLIKAPRLVSLSLDENHPCRSS-------ECQSLDAIGPHCENAV 675
Query: 705 SCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHTVSSKSIETS 764
S F VH++V N GD GSH V LF+ P+ V GSP QL+GF ++ S+
Sbjct: 676 EGGS-DFEVHLNVKNTGDRAGSHTVFLFTTSPQ-VHGSPIKQLLGFEKIRLGKSEEAVVR 733
Query: 765 ILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIEI 804
V+ C+ LS DE GKR LG+H+L VG ++H+++I +
Sbjct: 734 FNVNVCKDLSVVDETGKRKIALGHHLLHVGSLKHSLNISV 773
>At3g19620 beta-xylosidase, putative
Length = 781
Score = 702 bits (1813), Expect = 0.0
Identities = 376/798 (47%), Positives = 498/798 (62%), Gaps = 59/798 (7%)
Query: 8 LFFIFFVVLIHFTTSIEDFAC--KRPHHSRYKFCDTSLSIPTRAHSLVSLLTLPEKIQQL 65
L + ++ S ++FAC P ++Y FC+ SLS RA LVS L+L EK+QQL
Sbjct: 9 LSLLIIALVSSLCESQKNFACDISAPATAKYGFCNVSLSYEARAKDLVSRLSLKEKVQQL 68
Query: 66 SNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTLWF 125
N A+ +PRLG+P Y+WWSE+LHG++ GPGV F+G V AT FP I++AASFN +LW
Sbjct: 69 VNKATGVPRLGVPPYEWWSEALHGVSDVGPGVHFNGTVPGATSFPATILTAASFNTSLWL 128
Query: 126 LIASAVAVEARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVRG 185
+ V+ EARAM NVG AGLT+W+PNVNVFRDPRWGRGQETPGEDP+V S YAV+YV+G
Sbjct: 129 KMGEVVSTEARAMHNVGLAGLTYWSPNVNVFRDPRWGRGQETPGEDPLVVSKYAVNYVKG 188
Query: 186 LQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWG 245
LQ D D L VS+CCKH+TAYDL+ W
Sbjct: 189 LQ-----------------------------DVHDAGKSRRLKVSSCCKHYTAYDLDNWK 219
Query: 246 QFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLL-GVAR 304
R++F+A V++QDLEDTYQ PF+ CV++G S +MCSYN VNG+P CA +LL GV R
Sbjct: 220 GIDRFHFDAKVTKQDLEDTYQTPFKSCVEEGDVSSVMCSYNRVNGIPTCADPNLLRGVIR 279
Query: 305 NNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQG 364
W GYI SDCD++ F Y K+ EDAVA LKAG ++NCG ++ ++T +AV+
Sbjct: 280 GQWRLDGYIVSDCDSIQVYFNDIHYTKTREDAVALALKAGLNMNCGDFLGKYTENAVKLK 339
Query: 365 KVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGIVL 424
K+ D+D AL+ + V +RLG FDGDP++ +G LGP DVC+ +H+ LALEAA+QGIVL
Sbjct: 340 KLNGSDVDEALIYNYIVLMRLGFFDGDPKSLPFGNLGPSDVCSKDHQMLALEAAKQGIVL 399
Query: 425 LKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYA-KKISYA 482
L+N + LPL + LAVIGP A T + Y+G+PC S +GL +Y +KI Y
Sbjct: 400 LEN-RGDLPLPKTTVKKLAVIGPNANATKVMISNYAGVPCKYTSPIQGLQKYVPEKIVYE 458
Query: 483 SGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSI 542
GC D+ C + A++ +AD V+V G+D T+E E DRV+L LPG Q LV +
Sbjct: 459 PGCKDVKCGDQTLISAAVKAVSEADVTVLVVGLDQTVEAEGLDRVNLTLPGYQEKLVRDV 518
Query: 543 AAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFN 602
A A+K V+LV+ GP+D+SFA+ I ++LWVGYPGEAGG A+A++IFG+ NP
Sbjct: 519 ANAAKKTVVLVIMSAGPIDISFAKNLSTIRAVLWVGYPGEAGGDAIAQVIFGDYNP---- 574
Query: 603 AAGRLPMTWYPESFTN-VPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSY 661
+GRLP TWYP+ F + V M DM+MR + + G+PGR+YRFYTG +Y FG+GLSYS FS
Sbjct: 575 -SGRLPETWYPQEFADKVAMTDMNMRPNSTSGFPGRSYRFYTGKPIYKFGYGLSYSSFST 633
Query: 662 KFLSAPSKVSLSRITKGSLRKSLSDQAEKEVYGVDYVQVDELLSCNSLSFPVHISVTNLG 721
LSAPS + + +L K+ S VD V +C+ L + I V N G
Sbjct: 634 FVLSAPSIIHIKTNPIMNLNKTTS---------VDISTV----NCHDLKIRIVIGVKNHG 680
Query: 722 DLDGSHVVMLFSKWPK-----VVEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFA 776
GSHVV++F K PK V G P TQLVGF RV S + + ++ D C+ LS
Sbjct: 681 LRSGSHVVLVFWKPPKCSKSLVGGGVPLTQLVGFERVEVGRSMTEKFTVDFDVCKALSLV 740
Query: 777 DEQGKRVFPLGNHVLSVG 794
D GKR G+H L +G
Sbjct: 741 DTHGKRKLVTGHHKLVIG 758
>At5g09700 beta-glucosidase - like protein
Length = 411
Score = 340 bits (871), Expect = 2e-93
Identities = 187/425 (44%), Positives = 255/425 (60%), Gaps = 17/425 (4%)
Query: 383 LRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGIVLLKNDKKFLPLNRNYGSSL 442
+RLG FDG+P+ YG LGP DVCT E++ LA+E ARQGIVLLKN LPL+ + +L
Sbjct: 1 MRLGFFDGNPKNQPYGGLGPKDVCTVENRELAVETARQGIVLLKNSAGSLPLSPSAIKTL 60
Query: 443 AVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAKKISYASGCSDISCNSDGGFAEAIE 501
AVIGP A VT + G Y G+ C + +GL Y GC +++C ++ A
Sbjct: 61 AVIGPNANVTKTMIGNYEGVACKYTTPLQGLERTVLTTKYHRGCFNVTC-TEADLDSAKT 119
Query: 502 TARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLD 561
A AD V+V G D T+E E DR+ L LPGKQ +LV+ +A A++ PV+LV+ GG D
Sbjct: 120 LAASADATVLVMGADQTIEKETLDRIDLNLPGKQQELVTQVAKAARGPVVLVIMSGGGFD 179
Query: 562 VSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFT-NVP 620
++FA+ ++ I SI+WVGYPGEAGG A+A++IFG NP +G+LPMTWYP+S+ VP
Sbjct: 180 ITFAKNDEKITSIMWVGYPGEAGGIAIADVIFGRHNP-----SGKLPMTWYPQSYVEKVP 234
Query: 621 MNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSL 680
M +M+MR D S GY GRTYRFY G VY FG GLSY+ FS++ + AP VSL+ S
Sbjct: 235 MTNMNMRPDKSNGYLGRTYRFYIGETVYAFGDGLSYTNFSHQLIKAPKFVSLNLDESQSC 294
Query: 681 RKSLSDQAEKEVYGVDYVQVD-ELLSCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKVV 739
R E +D + E F V + V N+GD +G+ V LF+ P V
Sbjct: 295 R-------SPECQSLDAIGPHCEKAVGERSDFEVQLKVRNVGDREGTETVFLFTT-PPEV 346
Query: 740 EGSPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHT 799
GSP+ QL+GF ++ + VD C+ L DE GKR LG+H+L VG ++H+
Sbjct: 347 HGSPRKQLLGFEKIRLGKKEETVVRFKVDVCKDLGVVDEIGKRKLALGHHLLHVGSLKHS 406
Query: 800 VSIEI 804
+I +
Sbjct: 407 FNISV 411
>At3g47000 beta-D-glucan exohydrolase - like protein
Length = 608
Score = 134 bits (336), Expect = 3e-31
Identities = 172/653 (26%), Positives = 278/653 (42%), Gaps = 141/653 (21%)
Query: 45 IPTRAHSLVSLLTLPEKIQQLS---------------------NNASSIP---------- 73
+ R L+S +TLPEKI Q++ N S+P
Sbjct: 17 VEARVKDLLSRMTLPEKIGQMTQIERRVASPSAFTDFFIGSVLNAGGSVPFEDAKSSDWA 76
Query: 74 -------------RLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFN 120
RLGIP + ++++HG + V AT FP I A+ +
Sbjct: 77 DMIDGFQRSALASRLGIPII-YGTDAVHG----------NNNVYGATVFPHNIGLGATRD 125
Query: 121 RTLWFLIASAVAVEARAMFNVGQAGLTF-WAPNVNVFRDPRWGRGQETPGEDPMVASAYA 179
L I +A A+E RA +G+ + ++P V V RDPRWGR E+ GEDP +
Sbjct: 126 ADLVRRIGAATALEVRA------SGVHWAFSPCVAVLRDPRWGRCYESYGEDPELVCEMT 179
Query: 180 VDYVRGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAY 239
V GLQG ++ G + G N V AC KHF
Sbjct: 180 -SLVSGLQGVPPEEHPNGYP-----FVAGRNN-----------------VVACVKHFVGD 216
Query: 240 DLEKWGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDL 299
G N + S ++LE + PP+ C+ QG S +M SY+ NG A L
Sbjct: 217 GGTDKGI---NEGNTIASYEELEKIHIPPYLKCLAQG-VSTVMASYSSWNGTRLHADRFL 272
Query: 300 LG-VARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTY----ML 354
L + + GFKG++ SD + + + E QG + + + AG D+ + +
Sbjct: 273 LTEILKEKLGFKGFLVSDWEGLDRLSEPQG--SNYRYCIKTAVNAGIDMVMVPFKYEQFI 330
Query: 355 RHTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLA 414
+ VE G++ I+ A+ + V+ GLF G P T + L P C EH+ LA
Sbjct: 331 QDMTDLVESGEIPMARINDAVERILRVKFVAGLF-GHPLTDR--SLLPTVGC-KEHRELA 386
Query: 415 LEAARQGIVLLKN----DKKFLPLNRNYGSSLAVIGPMA--VTNKLGG------GYSGIP 462
EA R+ +VLLK+ DK FLPL+RN + V G A + + GG G SG
Sbjct: 387 QEAVRKSLVLLKSGKNADKPFLPLDRN-AKRILVTGTHADDLGYQCGGWTKTWFGLSGRI 445
Query: 463 CSPKSLYEGLAEYA---KKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTL 519
+L + + E ++ Y S + S GF+ AI + + +
Sbjct: 446 TIGTTLLDAIKEAVGDETEVIYEKTPSKETLASSEGFSYAIVAVGEPPYAETMG------ 499
Query: 520 ETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGP--LDVSFAERNQLIPSILWV 577
D L +P D+V+++A P +++L G P L+ + E+ + + + W+
Sbjct: 500 -----DNSELRIPFNGTDIVTAVAEII--PTLVILISGRPVVLEPTVLEKTEALVA-AWL 551
Query: 578 GYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMNDMSMRADP 630
PG G+ +A+++FG+ ++ G+LP++W+ + ++P++ + DP
Sbjct: 552 --PG-TEGQGVADVVFGD-----YDFKGKLPVSWF-KHVEHLPLDAHANSYDP 595
>At3g47010 beta-D-glucan exohydrolase - like protein
Length = 609
Score = 126 bits (317), Expect = 4e-29
Identities = 161/645 (24%), Positives = 272/645 (41%), Gaps = 103/645 (15%)
Query: 34 SRYKFCDTSLSIPTRAHSLVSLLTLPEKIQQLSNNASSIPRLGI-----------PAYQW 82
S + + + + R L+S +TLPEKI Q++ S+ + A W
Sbjct: 7 SSWVYKNRDAPVEARVKDLLSRMTLPEKIGQMTQIERSVASPQVITNSFIGSVQSGAGSW 66
Query: 83 ---------WSESLHG-------------IAINGPGVSFDGAVSAATDFPQVIVSAASFN 120
W++ + G I V + V AT FP I A+ +
Sbjct: 67 PLEDAKSSDWADMIDGFQRSALASRLGIPIIYGTDAVHGNNNVYGATVFPHNIGLGATRD 126
Query: 121 RTLWFLIASAVAVEARAMFNVGQAGLTF-WAPNVNVFRDPRWGRGQETPGEDPMVASAYA 179
L I +A A+E RA +G+ + +AP V V DPRWGR E+ E + +
Sbjct: 127 ADLVKRIGAATALEIRA------SGVHWTFAPCVAVLGDPRWGRCYESYSEAAKIVCEMS 180
Query: 180 VDYVRGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAY 239
+ + GLQG ++ +G + G N V AC KHF
Sbjct: 181 L-LISGLQGEPPEEHPYGYP-----FLAGRNN-----------------VIACAKHFVGD 217
Query: 240 DLEKWGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDL 299
+ G N + S +DLE + P+ C+ QG S +M S++ NG + L
Sbjct: 218 GGTEKGLSEG---NTITSYEDLEKIHVAPYLNCIAQG-VSTVMASFSSWNGSRLHSDYFL 273
Query: 300 LG-VARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTY----ML 354
L V + GFKG++ SD D + T+ E +G + + V + AG D+ + +
Sbjct: 274 LTEVLKQKLGFKGFLVSDWDGLETISEPEG--SNYRNCVKLGINAGIDMVMVPFKYEQFI 331
Query: 355 RHTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLA 414
+ VE G++ ++ A+ + V+ GLF+ P + LG V EH+ +A
Sbjct: 332 QDMTDLVESGEIPMARVNDAVERILRVKFVAGLFE-HPLADR-SLLG--TVGCKEHREVA 387
Query: 415 LEAARQGIVLLKN----DKKFLPLNRNYGSSLAVIGPMAVTNKLG---GGYSGIPCSPKS 467
EA R+ +VLLKN D FLPL+RN + V+G A N LG GG++ I
Sbjct: 388 REAVRKSLVLLKNGKNADTPFLPLDRN-AKRILVVGMHA--NDLGNQCGGWTKIKSGQSG 444
Query: 468 LYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRV 527
I A G E + ++ + ++ G E + D
Sbjct: 445 RITIGTTLLDSIKAAVGDKTEVIFEKTPTKETLASSDGFSYAIVAVGEPPYAEMKG-DNS 503
Query: 528 SLLLPGKQMDLVSSIAAASKNPVILVLTGGGP--LDVSFAERNQLIPSILWVGYPGEAGG 585
L +P ++++ A A K P +++L G P L+ + E+ + +++ +PG G
Sbjct: 504 ELTIPFNGNNIIT--AVAEKIPTLVILFSGRPMVLEPTVLEKTE---ALVAAWFPG-TEG 557
Query: 586 KALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMNDMSMRADP 630
+ ++++IFG+ ++ G+LP++W+ + +P+N + DP
Sbjct: 558 QGMSDVIFGD-----YDFKGKLPVSWF-KRVDQLPLNAEANSYDP 596
>At3g47050 beta-D-glucan exohydrolase - like protein
Length = 612
Score = 120 bits (301), Expect = 3e-27
Identities = 148/551 (26%), Positives = 237/551 (42%), Gaps = 76/551 (13%)
Query: 74 RLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTLWFLIASAVAV 133
RLGIP + +++HG + V AT FP I A+ + L I +A A+
Sbjct: 90 RLGIPII-YGIDAVHG----------NNDVYGATIFPHNIGLGATRDADLVKRIGAATAL 138
Query: 134 EARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVRGLQGAGGVK 193
E RA +AP V V +DPRWGR E+ GE + S V GLQG
Sbjct: 139 EVRAC-----GAHWAFAPCVAVVKDPRWGRCYESYGEVAQIVSEMT-SLVSGLQGEPSKD 192
Query: 194 NVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYNFN 253
+ G + G N V AC KHF + A N
Sbjct: 193 HTNGYP-----FLAGRKN-----------------VVACAKHFVG---DGGTNKAINEGN 227
Query: 254 AVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLLG-VARNNWGFKGY 312
++ +DLE + P++ C+ QG S +M SY+ NG + LL + + GFKGY
Sbjct: 228 TILRYEDLERKHIAPYKKCISQG-VSTVMASYSSWNGDKLHSHYFLLTEILKQKLGFKGY 286
Query: 313 ITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTAS----AVEQGKVKE 368
+ SD + + + + G + + V + AG D+ + + VE G+V
Sbjct: 287 VVSDWEGLDRLSDPPG--SNYRNCVKIGINAGIDMVMVPFKYEQFRNDLIDLVESGEVSM 344
Query: 369 EDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGIVLLKND 428
++ A+ + V+ GLF+ P T + L P C EH+ LA EA R+ +VLLKN
Sbjct: 345 ARVNDAVERILRVKFVAGLFEF-PLTDR--SLLPTVGC-KEHRELAREAVRKSLVLLKNG 400
Query: 429 K--KFLPLNRNYGSSLAVIGPMAVTNKLG---GGYSGIPCSPKSLYEGLAEYAKKISYAS 483
+ +FLPLN N + V+G A + LG GG++ I A
Sbjct: 401 RYGEFLPLNCN-AERILVVGTHA--DDLGYQCGGWTKTMYGQSGRITDGTTLLDAIKAAV 457
Query: 484 GCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSIA 543
G E + + + + ++ G ET D L++P ++++++
Sbjct: 458 GDETEVIYEKSPSEETLASGYRFSYAIVAVGESPYAETMG-DNSELVIPFNGSEIITTV- 515
Query: 544 AASKNPVILVLTGGGP--LDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGF 601
A K P +++L G P L+ E+ + + + W+ PG G+ +A++IFG+ +
Sbjct: 516 -AEKIPTLVILFSGRPMFLEPQVLEKAEALVA-AWL--PG-TEGQGIADVIFGD-----Y 565
Query: 602 NAAGRLPMTWY 612
+ G+LP TW+
Sbjct: 566 DFRGKLPATWF 576
>At3g47040 beta-D-glucan exohydrolase - like protein
Length = 636
Score = 114 bits (285), Expect = 2e-25
Identities = 140/508 (27%), Positives = 223/508 (43%), Gaps = 83/508 (16%)
Query: 127 IASAVAVEARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVRGL 186
+ +A A+E RA +AP V RDPRWGR E+ EDP + + V GL
Sbjct: 157 VGAATALEVRAC-----GAHWAFAPCVAALRDPRWGRSYESYSEDPDIICELS-SLVSGL 210
Query: 187 QGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQ 246
QG ++ G + G N V AC KHF G
Sbjct: 211 QGEPPKEHPNGYP-----FLAGRNN-----------------VVACAKHFVGDGGTDKGI 248
Query: 247 FARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLLG-VARN 305
N +VS ++LE + P+ C+ QG S +M SY+ NG + LL + +
Sbjct: 249 ---NEGNTIVSYEELEKIHLAPYLNCLAQG-VSTVMASYSSWNGSKLHSDYFLLTELLKQ 304
Query: 306 NWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTY----MLRHTASAV 361
GFKG++ SD +A+ + E G + + V + AG D+ + ++ V
Sbjct: 305 KLGFKGFVISDWEALERLSEPFG--SNYRNCVKISVNAGVDMVMVPFKYEQFIKDLTDLV 362
Query: 362 EQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQG 421
E G+V ID A+ + V+ GLF+ P T + LG V EH+ LA E+ R+
Sbjct: 363 ESGEVTMSRIDDAVERILRVKFVAGLFE-HPLTDR-SLLGT--VGCKEHRELARESVRKS 418
Query: 422 IVLLKN----DKKFLPLNRNYGSSLAVIGPMA--VTNKLGG------GYSGIPCSPKSLY 469
+VLLKN +K FLPL+RN + V G A + + GG G SG +L
Sbjct: 419 LVLLKNGTNSEKPFLPLDRNV-KRILVTGTHADDLGYQCGGWTKAWFGLSGRITIGTTLL 477
Query: 470 EGLAEYA---KKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDR 526
+ + E ++ Y S+ E + + ++ + ++ G ET D
Sbjct: 478 DAIKEAVGDKTEVIYEKTPSE----------ETLASLQRFSYAIVAVGETPYAETLG-DN 526
Query: 527 VSLLLPGKQMDLVSSIAAASKNPVILVLTGGGP--LDVSFAERNQLIPSILWVGYPGEAG 584
L +P D+V+ A A K P ++VL G P L+ E+ + + + W+ PG
Sbjct: 527 SELTIPLNGNDIVT--ALAEKIPTLVVLFSGRPLVLEPLVLEKAEALVA-AWL--PG-TE 580
Query: 585 GKALAEIIFGESNPVGFNAAGRLPMTWY 612
G+ + ++IFG+ ++ G+LP++W+
Sbjct: 581 GQGMTDVIFGD-----YDFEGKLPVSWF 603
>At5g20940 beta-glucosidase - like protein
Length = 626
Score = 107 bits (268), Expect = 2e-23
Identities = 151/584 (25%), Positives = 250/584 (41%), Gaps = 109/584 (18%)
Query: 74 RLGIPAYQWWSESLHGI-AINGPGVSFDGAVSAATDFPQVIVSAASFNRTLWFLIASAVA 132
RLGIP ++GI A++G ++ AT FP + + + L I A A
Sbjct: 112 RLGIPI-------IYGIDAVHGHNTVYN-----ATIFPHNVGLGVTRDPGLVKRIGEATA 159
Query: 133 VEARAMFNVGQAGLTF-WAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVRGLQG--- 188
+E RA G+ + +AP + V RDPRWGR E+ ED + + + GLQG
Sbjct: 160 LEVRA------TGIQYVFAPCIAVCRDPRWGRCYESYSEDHKIVQQMT-EIIPGLQGDLP 212
Query: 189 --AGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQ 246
GV V G+ + V+AC KHF G
Sbjct: 213 TGQKGVPFVAGKTK----------------------------VAACAKHFVGDGGTLRGM 244
Query: 247 FARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLL-GVARN 305
A N V++ L + P + V +G A+ +M SY+ +NG+ A++ L+ G +N
Sbjct: 245 NAN---NTVINSNGLLGIHMPAYHDAVNKGVAT-VMVSYSSINGLKMHANKKLITGFLKN 300
Query: 306 NWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCG----TYMLRHTASAV 361
F+G + SD V + G + +V AG D+ G T ++ S V
Sbjct: 301 KLKFRGIVISDYLGVDQINTPLG--ANYSHSVYAATTAGLDMFMGSSNLTKLIDELTSQV 358
Query: 362 EQGKVKEEDIDRALLNLFSVQLRLGLFDG---DPRTGKYGKLGPHDVCTSEHKTLALEAA 418
++ + ID A+ + V+ +GLF+ D K KLG + EH+ LA EA
Sbjct: 359 KRKFIPMSRIDDAVKRILRVKFTMGLFENPIADHSLAK--KLG-----SKEHRELAREAV 411
Query: 419 RQGIVLLKN----DKKFLPLNRNYGSSLAVIGPMAVTNKLG---GGYS----GIPCSPKS 467
R+ +VLLKN DK LPL + + + V G A + LG GG++ G+ + +
Sbjct: 412 RKSLVLLKNGENADKPLLPLPKK-ANKILVAGTHA--DNLGYQCGGWTITWQGLNGNNLT 468
Query: 468 LYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLE-TEDHDR 526
+ + KK + N D F + A D+ ++ G E D
Sbjct: 469 IGTTILAAVKKTVDPKTQVIYNQNPDTNFVK----AGDFDYAIVAVGEKPYAEGFGDSTN 524
Query: 527 VSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGK 586
+++ PG + ++ A+ K ++V+ G P+ + + + L+ + L PG G+
Sbjct: 525 LTISEPGP--STIGNVCASVK--CVVVVVSGRPVVMQISNIDALVAAWL----PG-TEGQ 575
Query: 587 ALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMNDMSMRADP 630
+A+++FG+ + G+L TW+ ++ +PMN DP
Sbjct: 576 GVADVLFGD-----YGFTGKLARTWF-KTVDQLPMNVGDPHYDP 613
>At5g20950 beta-D-glucan exohydrolase-like protein
Length = 624
Score = 103 bits (256), Expect = 5e-22
Identities = 153/631 (24%), Positives = 264/631 (41%), Gaps = 111/631 (17%)
Query: 44 SIPTRAHSLVSLLTLPEKIQQLSNNASSIPRLGIPAYQWWSESLHGIAINGPGVSFDGAV 103
S+P+ + + + + +IQ+ AS RLGIP + +++HG V
Sbjct: 80 SVPSEKATPETWVNMVNEIQK----ASLSTRLGIPMI-YGIDAVHG----------HNNV 124
Query: 104 SAATDFPQVIVSAASFNRTLWFLIASAVAVEARAMFNVGQAGLTF-WAPNVNVFRDPRWG 162
AT FP + + + L I +A A+E RA G+ + +AP + V RDPRWG
Sbjct: 125 YGATIFPHNVGLGVTRDPNLVKRIGAATALEVRA------TGIPYAFAPCIAVCRDPRWG 178
Query: 163 RGQETPGEDPMVASAYAVDYVRGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDDGD 222
R E+ ED + + + GLQG ++ +++ + GG
Sbjct: 179 RCYESYSEDYRIVQ-QMTEIIPGLQG-----DLPTKRKGVPFVGG--------------- 217
Query: 223 DGDSLMVSACCKHFTAYDLEKWGQFARYNFNAVVSQQDLEDTYQPPFRGCVQQGKASCLM 282
V+AC KHF G N V+ + L + P + V +G A+ +M
Sbjct: 218 ---KTKVAACAKHFVGDGGTVRGIDEN---NTVIDSKGLFGIHMPGYYNAVNKGVAT-IM 270
Query: 283 CSYNEVNGVPACASEDLL-GVARNNWGFKGYITSDCDAV--ATVFEYQGYVKSAEDAVAE 339
SY+ NG+ A+++L+ G +N F+G++ SD + T + Y S V
Sbjct: 271 VSYSAWNGLRMHANKELVTGFLKNKLKFRGFVISDWQGIDRITTPPHLNYSYS----VYA 326
Query: 340 VLKAGTDINCGTY----MLRHTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTG 395
+ AG D+ Y + +S +++ + ID AL + V+ +GLF+
Sbjct: 327 GISAGIDMIMVPYNYTEFIDEISSQIQKKLIPISRIDDALKRILRVKFTMGLFEEPLADL 386
Query: 396 KYGKLGPHDVCTSEHKTLALEAARQGIVLLKND----KKFLPLNRNYGSSLAVIGPMAVT 451
+ + + + EH+ LA EA R+ +VLLKN K LPL + G L V G A
Sbjct: 387 SFA----NQLGSKEHRELAREAVRKSLVLLKNGKTGAKPLLPLPKKSGKIL-VAG--AHA 439
Query: 452 NKLG---GGYS----GIPCSPKSLYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETAR 504
+ LG GG++ G+ + ++ + K + S N D F + +
Sbjct: 440 DNLGYQCGGWTITWQGLNGNDHTVGTTILAAVKNTVAPTTQVVYSQNPDANFVK----SG 495
Query: 505 QADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSF 564
+ D+ ++V G E D +L + ++ ++ + K ++V+ G P V
Sbjct: 496 KFDYAIVVVGEPPYAEMFG-DTTNLTISDPGPSIIGNVCGSVK--CVVVVVSGRP--VVI 550
Query: 565 AERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMNDM 624
I +++ PG G+ +A+ +FG+ + G+L TW+ +S +PMN
Sbjct: 551 QPYVSTIDALVAAWLPG-TEGQGVADALFGD-----YGFTGKLARTWF-KSVKQLPMNVG 603
Query: 625 SMRADPSRGYPGRTYRFYTGSRVYGFGHGLS 655
DP +Y FG GL+
Sbjct: 604 DRHYDP----------------LYPFGFGLT 618
>At5g04885 unknown protein
Length = 665
Score = 102 bits (255), Expect = 7e-22
Identities = 151/581 (25%), Positives = 240/581 (40%), Gaps = 89/581 (15%)
Query: 72 IPRLGIPAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTLWFLIASAV 131
+ RLGIP + +++HG V AT FP + A+ + L I +A
Sbjct: 108 VSRLGIPMI-YGIDAVHG----------HNNVYNATIFPHNVGLGATRDPDLVKRIGAAT 156
Query: 132 AVEARAMFNVGQAGLTF-WAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVRGLQGAG 190
AVE RA G+ + +AP + V RDPRWGR E+ ED V D + GLQG
Sbjct: 157 AVEVRA------TGIPYTFAPCIAVCRDPRWGRCYESYSEDHKVVED-MTDVILGLQG-- 207
Query: 191 GVKNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARY 250
+ K + GG V+AC KH+ G
Sbjct: 208 --EPPSNYKHGVPFVGGRD------------------KVAACAKHYVGDGGTTRGVNEN- 246
Query: 251 NFNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLL-GVARNNWGF 309
N V L + P + V +G S +M SY+ NG A+ +L+ G + F
Sbjct: 247 --NTVTDLHGLLSVHMPAYADAVYKG-VSTVMVSYSSWNGEKMHANTELITGYLKGTLKF 303
Query: 310 KGYITSDCDAVATV-----FEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQG 364
KG++ SD V + Y V++A A +++ + T + + V+
Sbjct: 304 KGFVISDWQGVDKISTPPHTHYTASVRAAIQAGIDMVMVPFNF---TEFVNDLTTLVKNN 360
Query: 365 KVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGIVL 424
+ ID A+ + V+ +GLF+ + ++ + H+ LA EA R+ +VL
Sbjct: 361 SIPVTRIDDAVRRILLVKFTMGLFENPLADYSFSS----ELGSQAHRDLAREAVRKSLVL 416
Query: 425 LKNDKK---FLPLNRNYGSSLAVIGPMAVTNKLG---GGYS----GIPCSPKSLYEGLAE 474
LKN K LPL R S + V G A + LG GG++ G + + L
Sbjct: 417 LKNGNKTNPMLPLPRK-TSKILVAGTHA--DNLGYQCGGWTITWQGFSGNKNTRGTTLLS 473
Query: 475 YAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETE-DHDRVSLLLPG 533
K S N D F ++ A + +I G ET D D++++L PG
Sbjct: 474 AVKSAVDQSTEVVFRENPDAEFIKSNNFA----YAIIAVGEPPYAETAGDSDKLTMLDPG 529
Query: 534 KQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIF 593
++SS A K ++V+ G PL + + I +++ PG G+ + + +F
Sbjct: 530 PA--IISSTCQAVK--CVVVVISGRPLVMEPYVAS--IDALVAAWLPG-TEGQGITDALF 582
Query: 594 GESNPVGFNAAGRLPMTWYPESFTNVPMNDMSMRADPSRGY 634
G+ GF +G+LP+TW+ + +PM+ DP Y
Sbjct: 583 GDH---GF--SGKLPVTWF-RNTEQLPMSYGDTHYDPLFAY 617
>At3g62710 beta-D-glucan exohydrolase-like protein
Length = 650
Score = 100 bits (249), Expect = 3e-21
Identities = 157/613 (25%), Positives = 247/613 (39%), Gaps = 121/613 (19%)
Query: 74 RLGIPAYQWWSESLHGI-AINGPGVSFDGAVSAATDFPQVIVSAASFNRTLWFLIASAVA 132
RLGIP L+ + A++G D AT FP + A+ + L I + A
Sbjct: 125 RLGIPL-------LYAVDAVHGHNTFID-----ATIFPHNVGLGATRDPQLVKKIGAITA 172
Query: 133 VEARAMFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVRGLQGAGGV 192
E RA V QA +AP V V RDPRWGR E+ EDP V + + GLQ
Sbjct: 173 QEVRAT-GVAQA----FAPCVAVCRDPRWGRCYESYSEDPAVVNMMTESIIDGLQ----- 222
Query: 193 KNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYN- 251
GN + +D + V+ C KHF G N
Sbjct: 223 -----------------GNAPYLAD-------PKINVAGCAKHFVG----DGGTINGINE 254
Query: 252 FNAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLL-GVARNNWGFK 310
N V L + PPF V++G AS +M SY+ +NGV A+ ++ +N F+
Sbjct: 255 NNTVADNATLFGIHMPPFEIAVKKGIAS-IMASYSSLNGVKMHANRAMITDYLKNTLKFQ 313
Query: 311 GYITSDCDAV--ATVFEYQGYVKSAEDAVAEVLKAGTDINCGTY----MLRHTASAVEQG 364
G++ SD + T E Y S E ++ AG D+ + L + V G
Sbjct: 314 GFVISDWLGIDKITPIEKSNYTYSIEASI----NAGIDMVMVPWAYPEYLEKLTNLVNGG 369
Query: 365 KVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAARQGIVL 424
+ ID A+ + V+ +GLF+ KL + + H+ + EA R+ +VL
Sbjct: 370 YIPMSRIDDAVRRILRVKFSIGLFENSLAD---EKLPTTEFGSEAHREVGREAVRKSMVL 426
Query: 425 LKNDK----KFLPLNRNYGSSLAVIGPMAVTNKLG---GGYS-----------GIPCSPK 466
LKN K K +PL + + V G A N +G GG+S +P + K
Sbjct: 427 LKNGKTDADKIVPLPKKV-KKIVVAGRHA--NDMGWQCGGFSLTWQGFNGTGEDMPTNTK 483
Query: 467 -SLYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETAR---QADFVVIVAGIDTTLET- 521
L G + + D + +TA+ A + ++V G ET
Sbjct: 484 HGLPTGKIKGTTILEAIQKAVDPTTEVVYVEEPNQDTAKLHADAAYTIVVVGETPYAETF 543
Query: 522 EDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPG 581
D + + PG D +S + +++++TG + + + + ++ W+ PG
Sbjct: 544 GDSPTLGITKPGP--DTLSHTCGSGMKCLVILVTGRPLVIEPYIDMLDAL-AVAWL--PG 598
Query: 582 EAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMNDMSMRADPSRGYPGRTYRF 641
G+ +A+++FG+ +P G LP TW + T +PMN DP
Sbjct: 599 -TEGQGVADVLFGD-HPF----TGTLPRTWM-KHVTQLPMNVGDKNYDP----------- 640
Query: 642 YTGSRVYGFGHGL 654
+Y FG+G+
Sbjct: 641 -----LYPFGYGI 648
>At1g52950 replication protein A1, putative
Length = 566
Score = 31.2 bits (69), Expect = 2.4
Identities = 25/95 (26%), Positives = 43/95 (44%), Gaps = 12/95 (12%)
Query: 654 LSYSGFSYKFLSAPSKVSLSRITKGSLRKSLSDQAEKEVYGVDYVQVDELLSCNSLSFPV 713
L YS + Y + KV S+ K+LS A+K +Y D D ++ + +
Sbjct: 298 LDYSWYYYAHIKCNKKVFKSK-------KTLSSGAKKIIYRCDKCATD--VTSIEARYWL 348
Query: 714 HISVTNLGDLDGSHVVMLFSKWPKVVEGSPQTQLV 748
H+ + D G +MLF + + + G P T+L+
Sbjct: 349 HLDIM---DNTGESKLMLFDSFVEPIIGCPATELL 380
>At5g66770 SCARECROW gene regulator
Length = 584
Score = 30.0 bits (66), Expect = 5.4
Identities = 11/16 (68%), Positives = 14/16 (86%)
Query: 202 LSDYGGGSGNGSFDSD 217
LSD+GGG+G G F+SD
Sbjct: 99 LSDFGGGTGGGEFESD 114
>At5g51230 putative protein
Length = 388
Score = 29.6 bits (65), Expect = 7.0
Identities = 62/266 (23%), Positives = 100/266 (37%), Gaps = 57/266 (21%)
Query: 553 VLTGGGPLD-VSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVGF---------- 601
+LTGG +D VS A+ N L+P + + ++G A+ I F + F
Sbjct: 131 ILTGGLGVDGVSQAQANFLLPDMNRLALEAKSGSLAILFISFAGAQNSQFGIDSGKIHSG 190
Query: 602 NAAG-----RLPMTWYPESFTNVPMNDMSMRADPS----------RGYPGRTYRFYTGSR 646
N G ++P+ S+ P D+ R D + P + +
Sbjct: 191 NIGGHCLWSKIPLQSLYASWQKSPNMDLGQRVDTVSLVEMQPCFIKSSPQQVQVTISAEE 250
Query: 647 VYGFGHGLSYSGFSYKFLSAPSKVSLSRITKGSLRKSLSDQAEKEVYGVDYVQVDELLSC 706
V G YS FSY +S+ S + + R+ G++ V+ Y +
Sbjct: 251 V-GSTEKSPYSSFSYNDISSSSLLQIIRLRTGNV-----------VFNYRYYNNKLQKTE 298
Query: 707 NSLSFPVH--ISVTNLGDLDGSHVVMLFSKWPKVVEG----------SPQTQLVGFSRVH 754
LSFP + V + + +FS P V G SPQ +++ F V+
Sbjct: 299 GGLSFPFSFILLVKDTAVVTSLCSDAIFSFSPNHVVGPEISLAINPRSPQFRVLEFQAVN 358
Query: 755 ------TVSSKSIETSILVDPCEHLS 774
T+ SK + TS L D C +S
Sbjct: 359 VSLKTETMISK-VRTSCLFDLCSLVS 383
>At5g10630 putative protein
Length = 804
Score = 29.6 bits (65), Expect = 7.0
Identities = 25/96 (26%), Positives = 40/96 (41%), Gaps = 23/96 (23%)
Query: 210 GNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYNFNAVVSQQDLEDTYQPPF 269
G +FD D DDG D D A+D Y+++ + + + E +P
Sbjct: 115 GLSNFD-DYDDGFDDDD----------DAFD---------YDYDVDIDEHEEEAAAEPKE 154
Query: 270 RGCVQQGKASCLMCSYNEVNGVPACASEDLLGVARN 305
QG C +C+Y+ V + C D+ GV R+
Sbjct: 155 EIAKTQGLWRCAICTYDNVETMFVC---DICGVLRH 187
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,560,560
Number of Sequences: 26719
Number of extensions: 840810
Number of successful extensions: 2488
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 2384
Number of HSP's gapped (non-prelim): 44
length of query: 805
length of database: 11,318,596
effective HSP length: 107
effective length of query: 698
effective length of database: 8,459,663
effective search space: 5904844774
effective search space used: 5904844774
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0177.14


