

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.15
(629 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
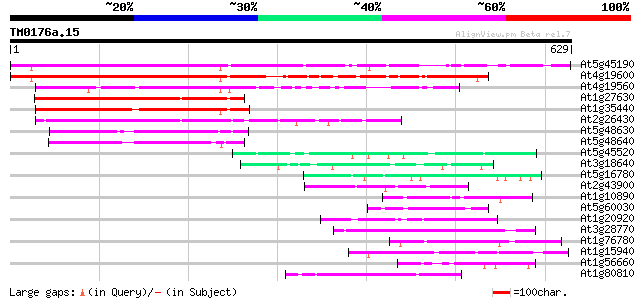
At5g45190 unknown protein 498 e-141
At4g19600 unknown protein 464 e-131
At4g19560 putative protein 241 7e-64
At1g27630 cyclin like protein 236 2e-62
At1g35440 hypothetical protein 224 1e-58
At2g26430 cyclin like protein 140 2e-33
At5g48630 cyclin C-like protein 83 5e-16
At5g48640 cyclin C-like protein 80 4e-15
At5g45520 putative protein 50 3e-06
At3g18640 hypothetical protein 49 8e-06
At5g16780 putative protein 48 1e-05
At2g43900 putative inositol polyphosphate 5'-phosphatase 45 9e-05
At1g10890 unknown protein 45 9e-05
At5g60030 KED - like protein 45 2e-04
At1g20920 putative RNA helicase 45 2e-04
At3g28770 hypothetical protein 44 3e-04
At1g76780 putative heat shock protein 44 3e-04
At1g15940 T24D18.4 44 3e-04
At1g56660 hypothetical protein 44 4e-04
At1g80810 unknown protein 43 5e-04
>At5g45190 unknown protein
Length = 579
Score = 498 bits (1281), Expect = e-141
Identities = 314/645 (48%), Positives = 390/645 (59%), Gaps = 85/645 (13%)
Query: 1 MAGLLLGDTSHDGTHQSGSQGCS---QENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLR 57
MAG+L G+ S+ + S S QE SRWY RKEIEENSPS+ DG+DLKKE YLR
Sbjct: 1 MAGVLAGECSYSESGVSSHSRNSHEKQEEVSRWYFGRKEIEENSPSRLDGIDLKKETYLR 60
Query: 58 KSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETP 117
KSYCT+LQDLGMRLKVPQVTIA+AIIFCHRFF RQSHAKNDRR IAT CMFLAGKVEETP
Sbjct: 61 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFFRQSHAKNDRRTIATVCMFLAGKVEETP 120
Query: 118 RPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYK 177
RPLKDVI VSYEII+KKDP A Q+IKQKEVYEQQKELIL GE++VL+TLGFD +V HPYK
Sbjct: 121 RPLKDVIFVSYEIINKKDPGASQKIKQKEVYEQQKELILNGEKIVLSTLGFDLNVYHPYK 180
Query: 178 PLVEAIKKLKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKL--- 234
PLVEAIKK KV+QNALAQVAWNFVNDGLRTSLCLQFKP HIAAGAIFLAAKFL VKL
Sbjct: 181 PLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD 240
Query: 235 -EKLWWQVFDVTLTAHQLEEVSNQMLELYEQNQIAPS--NDVEGTAGGGNRATP--KAST 289
EK+WWQ FDVT QLE+VSNQMLELYEQN++ S ++VE + GGG+ P + +
Sbjct: 241 GEKVWWQEFDVT--PRQLEDVSNQMLELYEQNRVPASQGSEVESSVGGGSAQRPGSRNAV 298
Query: 290 SNDEAATTNNNLYIRGPSPRLEASKLA-ISKNVVSSANHVGPVSNHGTNGSTEMIHLVEG 348
S DE + +R + + SK V++ N G N S + +E
Sbjct: 299 STDEHVGSRQTSSVRSTHEQSNSDNHGGSSKGVLNQNNENG--GGEAANVSVDNKEEIER 356
Query: 349 DAKGNQHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIGRSNM-KEDVELNDK---HS 404
+ K + + P HK+N++EA R E P G+ N +E EL D H
Sbjct: 357 ETKESSLHLESHPAHKDNVREAPHNSRP---LVEGP----GKDNSEREGGELQDDGAVHK 409
Query: 405 SKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVREL 464
S+N++ D + + K+ RDKVKA EK KK +G T+K +LMD++DL E
Sbjct: 410 SRNVDVGDALISQSPKDL--KLLRDKVKAKREKAKKLLGERTRKKDLMDEDDLIE----- 462
Query: 465 EDGIELAAQSEKNKQDDEDLIERVRELEDGIELAAQSEKNKQDGQKSLSKP-VDRSAFEN 523
RELED ++LA + EK K+ +K S+P + S
Sbjct: 463 ------------------------RELED-VQLAVEDEKTKE--RKVQSRPKAENSDLMG 495
Query: 524 MQQGKHQDHSEEHLHLVKPSHETDLSAVEEGEVSALDDIDLGRKSSNHKRKAESSPDKFA 583
+ G+ D E + EEGE+ + ++ + KRK S P+K +
Sbjct: 496 TEHGEILDVKGE------------VKNTEEGEMV---NNNVSPMMHSRKRKMGSPPEKQS 540
Query: 584 EGKKRHNYSFGSTHHNRIDYIEDQNNVNLLGHAERDSKRPVQENH 628
EGK+RHN G H + + H +R+ +R QEN+
Sbjct: 541 EGKRRHNSENGEESH--------KTSRGSSHHGDREHRRHSQENN 577
>At4g19600 unknown protein
Length = 541
Score = 464 bits (1194), Expect = e-131
Identities = 286/551 (51%), Positives = 352/551 (62%), Gaps = 51/551 (9%)
Query: 1 MAGLLLGDTSHDGTHQSGSQGCS---QENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLR 57
MAG+L GD S + S S Q+ +RWY RKEIEENSPS+ D +DLKKE YLR
Sbjct: 1 MAGVLAGDCSFGESGVSSYSRNSNEKQDEVARWYFGRKEIEENSPSRLDSIDLKKETYLR 60
Query: 58 KSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETP 117
KSYCT+LQDLGMRLKVPQVTIA+AIIFCHRFF+RQSHA+NDRR IAT CMFLAGKVEETP
Sbjct: 61 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFIRQSHARNDRRTIATVCMFLAGKVEETP 120
Query: 118 RPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYK 177
RPLKDVI+VSYEIIHKKDP Q+IKQKEVYEQQKELIL GE++VL+TLGFDF+V HPYK
Sbjct: 121 RPLKDVIVVSYEIIHKKDPTTAQKIKQKEVYEQQKELILNGEKIVLSTLGFDFNVYHPYK 180
Query: 178 PLVEAIKKLKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKL--- 234
PLVEAIKK KV+QNALAQVAWNFVNDGLRTSLCLQFKP HIAAGAIFLAAKFL VKL
Sbjct: 181 PLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD 240
Query: 235 -EKLWWQVFDVTLTAHQLEEVSNQMLELYEQNQIAPS--NDVEGTAGGGNRATPKASTSN 291
EK+WWQ FDV T QLE+VSNQMLELYEQN++ S ++VE + GGG+ +
Sbjct: 241 GEKVWWQEFDV--TPRQLEDVSNQMLELYEQNRVPASQVSEVESSVGGGSAHHVGSR--- 295
Query: 292 DEAATTNNNLYIRGPSPRLEASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAK 351
PS RL + + S N+ S SN NGS E ++ + K
Sbjct: 296 --------------PSARL-THEHSNSDNLGGSTKATQNRSN--DNGSGEAGSVIT-EQK 337
Query: 352 GNQHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHSSKNLNHR 411
G + E + H E+ +SRSG E I K + S+ +
Sbjct: 338 GERDTETKDSMH----TESHPAHKSRSGVEAPGEDKI----EKAGAHFPEDDKSRIVGTA 389
Query: 412 DGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELA 471
D T + + K+ RDKVKA LE KK G T+K +L+D++DL E RELED +ELA
Sbjct: 390 DVTVSQSPKDI--KMFRDKVKAKLE-AKKVQGEKTRKKDLVDEDDLIE--RELED-VELA 443
Query: 472 AQSEKNKQDDEDLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFEN-----MQQ 526
+ +K+ Q+ + E+ DG L +E+ + S + P + E+ + +
Sbjct: 444 VEDDKDIQNKSSMGTEHGEILDGNNLVVNTEEGEMIDDVSSTMPSRKRKMESPCEKQLGE 503
Query: 527 GKHQDHSEEHL 537
GK + + E++
Sbjct: 504 GKRRHDNSENV 514
>At4g19560 putative protein
Length = 474
Score = 241 bits (616), Expect = 7e-64
Identities = 184/496 (37%), Positives = 253/496 (50%), Gaps = 95/496 (19%)
Query: 30 WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHR-- 87
W+ SR+EIE NSPS++DG+DLK E LR SYCT+L+ LG RLK+ C R
Sbjct: 31 WFFSREEIERNSPSRRDGIDLKTETRLRDSYCTFLEILGERLKMSLPDFIHDKTVCDRDQ 90
Query: 88 ---FFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQ 144
FF S IAT CM LAGKVEETP L+DVI+ SYE IHKKD A QR
Sbjct: 91 ALCFFPFGSMCMT----IATVCMLLAGKVEETPVTLEDVIIASYERIHKKDLAGAQR--- 143
Query: 145 KEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKV--SQNALAQVAWNFVN 202
KEVY+QQKEL+L+GE +VL+TL FD + HPYKPLVEAIKK V ++ LAQ AWNFVN
Sbjct: 144 KEVYDQQKELVLIGEELVLSTLNFDLCISHPYKPLVEAIKKYMVEDAKTQLAQFAWNFVN 203
Query: 203 DGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKLE---KLWWQVFDVT-----------LTA 248
D LRT+LCLQ++P HIAAGAI LAA+ V L+ ++ Q FD+T +
Sbjct: 204 DCLRTTLCLQYQPHHIAAGAILLAAELPTVDLQSYREVLCQEFDITPCQLEDLVDVVNLS 263
Query: 249 HQLEEVSNQMLELYEQNQIAPSNDVEGTAGGGNRATPKASTSNDEAATTNNNLYIRGPSP 308
++ Q+LELYE+ + + VE + G A S D A+T + PS
Sbjct: 264 FAFSDIRGQILELYERIPTSQESKVESS---GGVAVVHQPISRDMASTE------KCPSS 314
Query: 309 RLEASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQ 368
+E + N+ S +H S H + S EG + N E + +NLQ
Sbjct: 315 DIEGGSSQV--NLSQSDDH----SVHDGSRS-------EGIGEVNSESEAQ-----KNLQ 356
Query: 369 EAQDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHSSKNLNHRDGTFAPPSLEAIKKIDR 428
+ S E ++G +K+D++L+ +
Sbjct: 357 D-----HSVGNIMVEKSDDVGVVQLKKDLQLH---------------------------Q 384
Query: 429 DKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIERV 488
++V++ EK KK+ K +LMD++DL E E+ED I K Q + +V
Sbjct: 385 EEVESKQEKDKKSFEKDITKIDLMDEKDLTE--SEVEDEI------NKTMQTGRQIFMKV 436
Query: 489 RELEDGIELAAQSEKN 504
+ +D + + +N
Sbjct: 437 EDPDDNMTVEHSEIRN 452
>At1g27630 cyclin like protein
Length = 317
Score = 236 bits (603), Expect = 2e-62
Identities = 123/238 (51%), Positives = 164/238 (68%), Gaps = 5/238 (2%)
Query: 28 SRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHR 87
S+WY SR+EIE SPS++DG+DL KE++LR SYCT+LQ LGM+L V QVTI+ A++ CHR
Sbjct: 32 SKWYFSREEIERFSPSRKDGIDLVKESFLRSSYCTFLQRLGMKLHVSQVTISCAMVMCHR 91
Query: 88 FFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEV 147
F++RQSHAKND + IATS +FLA K E+ P L V++ SYEII++ DP+A RI Q E
Sbjct: 92 FYMRQSHAKNDWQTIATSSLFLACKAEDEPCQLSSVVVASYEIIYEWDPSASIRIHQTEC 151
Query: 148 YEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRT 207
Y + KE+IL GE ++L+T F ++ PYKPL A+ +L + LA AWNFV+D +RT
Sbjct: 152 YHEFKEIILSGESLLLSTSAFHLDIELPYKPLAAALNRLNAWPD-LATAAWNFVHDWIRT 210
Query: 208 SLCLQFKPQHIAAGAIFLAAKFLNVKL--EKLWWQVFDVTLTAHQLEEVSNQMLELYE 263
+LCLQ+KP IA + LAA F N K+ + WW F V T L+EV +M L E
Sbjct: 211 TLCLQYKPHVIATATVHLAATFQNAKVGSRRDWWLEFGV--TTKLLKEVIQEMCTLIE 266
>At1g35440 hypothetical protein
Length = 247
Score = 224 bits (570), Expect = 1e-58
Identities = 114/246 (46%), Positives = 165/246 (66%), Gaps = 15/246 (6%)
Query: 30 WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFF 89
WY +R+ IE+ SPS+ DG++LK+E + R SY ++LQ+LG RL PQ TIA+AI+ C RFF
Sbjct: 7 WYNTREAIEKTSPSRLDGINLKEETFQRWSYTSFLQELGQRLNNPQKTIATAIVLCQRFF 66
Query: 90 LRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYE 149
RQS KND + +A CMF+AGKVE +PRP DV+ VSY ++ K+P ++V+E
Sbjct: 67 TRQSLTKNDPKTVAIICMFIAGKVEGSPRPAGDVVFVSYRVLFNKEPL-------RDVFE 119
Query: 150 QQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNA--LAQVAWNFVNDGLRT 207
+ K +L GE++VL+TL D ++HPYK +++ +K+ +++ L Q A+NFVND LRT
Sbjct: 120 RLKMTVLTGEKLVLSTLECDLEIEHPYKLVMDWVKRSVKTEDGRRLCQAAFNFVNDSLRT 179
Query: 208 SLCLQFKPQHIAAGAIFLAAKFLNVKL----EKLWWQVFDVTLTAHQLEEVSNQMLELYE 263
SLCLQF P IA+ AI++ + L +K WW+ FDV T QL E+ +QML+LY
Sbjct: 180 SLCLQFGPSQIASAAIYIGLSMCKMTLPCDGDKAWWREFDV--TKRQLWEICDQMLDLYV 237
Query: 264 QNQIAP 269
Q+ + P
Sbjct: 238 QDFVVP 243
>At2g26430 cyclin like protein
Length = 416
Score = 140 bits (354), Expect = 2e-33
Identities = 112/424 (26%), Positives = 204/424 (47%), Gaps = 32/424 (7%)
Query: 30 WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFF 89
+YLS ++++ SPS++DG+D E LR C +Q+ G+ LK+PQ +A+ + RF+
Sbjct: 9 FYLSDEQLKA-SPSRKDGIDETTEISLRIYGCDLIQEGGILLKLPQAVMATGQVLFQRFY 67
Query: 90 LRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQ-KEVY 148
++S AK D +I+A SC++LA K+EE P+ + VI+V + + +++ ++ + + +
Sbjct: 68 CKKSLAKFDVKIVAASCVWLASKLEENPKKARQVIIVFHRMECRRENLPLEHLDMYAKKF 127
Query: 149 EQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTS 208
+ K + ER +L +GF HV+HP+K + + L+ L Q AWN ND LRT+
Sbjct: 128 SELKVELSRTERHILKEMGFVCHVEHPHKFISNYLATLETPPE-LRQEAWNLANDSLRTT 186
Query: 209 LCLQFKPQHIAAGAIFLAAKFLNVKLEK--LWWQVFDVTLTAHQLEEVSNQMLELYEQNQ 266
LC++F+ + +A G ++ AA+ V L + WW+ FD ++ ++EV + LY
Sbjct: 187 LCVRFRSEVVACGVVYAAARRFQVPLPENPPWWKAFDADKSS--IDEVCRVLAHLYS--- 241
Query: 267 IAPSNDVEGTAGGGNRATPKASTSNDEAATTNNNLYIRGPSPRLEASKLAISKN------ 320
P G T + + N + + +L + G ++ A S N
Sbjct: 242 -LPKAQYISVCKDGKPFTFSSRSGNSQGQSATKDL-LPGAGEAVDTKCTAGSANNDLKDG 299
Query: 321 -VVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHP----EQEPPPHKENLQEAQDTVR 375
V + GT +++ I GD+ + E+E KE +E +D R
Sbjct: 300 MVTTPHEKATDSKKSGTESNSQPI---VGDSSYERSKVGDRERESDREKERGRE-RDRGR 355
Query: 376 SRSGYSEEPEINIGRSNMKEDVELNDKHSSKNLNHRDGTFAPPSLEAIKKIDRDKVKAAL 435
S G + + + R +K+ H S++ G + S + DRD ++
Sbjct: 356 SHRGRDSDRDSDRERDKLKD----RSHHRSRDRLKDSGGHSDKSRHHSSR-DRDYRDSSK 410
Query: 436 EKRK 439
++R+
Sbjct: 411 DRRR 414
>At5g48630 cyclin C-like protein
Length = 253
Score = 82.8 bits (203), Expect = 5e-16
Identities = 54/223 (24%), Positives = 109/223 (48%), Gaps = 15/223 (6%)
Query: 45 QDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIAT 104
Q G+ ++ ++ Y+ L +K+ Q +A+A+ + R + R+S + + R++A
Sbjct: 29 QRGISVEDFRLIKLHMSNYISKLAQHIKIRQRVVATAVTYMRRVYTRKSLTEYEPRLVAP 88
Query: 105 SCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGERVVLA 164
+C++LA K EE+ K +LV Y ++++ E + + + IL E VL
Sbjct: 89 TCLYLACKAEESVVHAK--LLVFY----------MKKLYADEKFRYEIKDILEMEMKVLE 136
Query: 165 TLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAIF 224
L F V HPY+ L E ++ ++ ++ + W VND R L L P I I+
Sbjct: 137 ALNFYLVVFHPYRSLPEFLQDSGINDTSMTHLTWGLVNDTYRMDLILIHPPFLITLACIY 196
Query: 225 LAAKFLNVKLEKLWWQVFDVTLTAHQLEEVSNQMLELYEQNQI 267
+A+ + K W++ V + ++ ++ ++L+ YE +++
Sbjct: 197 IASVHKEKDI-KTWFEELSVDMNI--VKNIAMEILDFYENHRL 236
>At5g48640 cyclin C-like protein
Length = 253
Score = 79.7 bits (195), Expect = 4e-15
Identities = 54/223 (24%), Positives = 109/223 (48%), Gaps = 21/223 (9%)
Query: 44 KQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIA 103
K+ G+ + ++ ++ L +KV Q +A+AI + R ++R+S + + R++A
Sbjct: 28 KERGISIDDFKLIKFHMSNHIMKLAQHIKVRQRVVATAITYMRRVYIRKSMVEFEPRLVA 87
Query: 104 TSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGERVVL 163
+C++LA K EE+ ++++ ++R+ E + + + IL E VL
Sbjct: 88 LTCLYLASKAEESIVQARNLVFY------------IKRLYPDEYNKYELKDILGMEMKVL 135
Query: 164 ATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAI 223
L + V HPY+ L E ++ ++ + Q+ W VND + L L P IA I
Sbjct: 136 EALDYYLVVFHPYRSLSEFLQDAALNDVNMNQITWGIVNDTYKMDLILVHPPYRIALACI 195
Query: 224 FLAAKFLNVKLEK---LWWQVFDVTLTAHQLEEVSNQMLELYE 263
++A +V EK W++ D+ + ++ ++ ++L+ YE
Sbjct: 196 YIA----SVHREKDITAWFE--DLHEDMNLVKNIAMEILDFYE 232
>At5g45520 putative protein
Length = 1167
Score = 50.4 bits (119), Expect = 3e-06
Identities = 91/381 (23%), Positives = 139/381 (35%), Gaps = 61/381 (16%)
Query: 251 LEEVSNQMLELYEQNQIAPSNDVEGTAGGGNRATPKASTSNDEAATTNNNLYIRGPSPRL 310
L E + ++ Y Q +VE T G + N+EA + + G R
Sbjct: 582 LMEREDDQVQNYGQTSKEEKGNVEET--GKQEDGDQGDGINEEANLEDGKKHDEGKEERS 639
Query: 311 EASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQE- 369
S VV P + +TE GD KG + E + K +L+E
Sbjct: 640 LKSD-----EVVEEEKKTSP-----SEEATEKFQNKPGDQKGKSNVEGDGDKGKADLEEE 689
Query: 370 -AQDTVRSRSGYSEE-----------PEINIGRSNMKEDVELND----------KHSSKN 407
QD V + S+E ++ KE+ +L++ K S
Sbjct: 690 KKQDEVEAEKSKSDEIVEGEKKPDDKSKVEKKGDGDKENADLDEGKKRDEVEAKKSESGK 749
Query: 408 LNHRDGTFAPP--SLEAI-----------KKIDRDKVKAALEKRKK---AVGNITKKTEL 451
+ DG +PP S++ I K+ DRDK K E+ KK G I K
Sbjct: 750 VVEGDGKESPPQESIDTIQNMTDDQTKVEKEGDRDKGKVDPEEGKKHDEVEGGIWKSDNG 809
Query: 452 MDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIERVRELEDG-IELAAQSEKNKQDGQK 510
++ D RE D IE NK DD ++ E DG E E K D K
Sbjct: 810 VEGVDKASPSRESTDAIE-------NKPDDHQRGDKQEEKGDGEKEKVNLEEWKKHDEIK 862
Query: 511 SLSKPVDRSAFENMQQGKHQDHSEEHLHLVKPSHETDLSAVEEGEVSALDDIDLGRKSSN 570
S D ++++ ++ S++ + + + + E+G+V DL
Sbjct: 863 EESSKQDNVTGGDVKKSPPKE-SKDTMESKRDDQKENSKVQEKGDVDKGKAADLDEGKKE 921
Query: 571 HKRKAESS-PDKFAEGKKRHN 590
+ KAESS DK EG + N
Sbjct: 922 NDVKAESSKSDKVIEGDEEKN 942
>At3g18640 hypothetical protein
Length = 676
Score = 48.9 bits (115), Expect = 8e-06
Identities = 74/304 (24%), Positives = 117/304 (38%), Gaps = 45/304 (14%)
Query: 259 LELYEQNQIAPSNDVEGTAGGGNRATPKASTSNDEAATTNN----NLYIRGPSPRLEASK 314
+E + QN A TAGG + A+T+ + +N +Y +E K
Sbjct: 382 VETFNQNHNALPYQSSLTAGGSQQVLAAAATNFSVGSNLSNLESGKVYQDNHHSTVE--K 439
Query: 315 LAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEP----PPHKENLQEA 370
+ +N VS + ++N + L + A G P+ E P H E++Q
Sbjct: 440 PVLVQNTVSR-EQIDQITNISAS-------LAQFLANGQPIPQLEQALQLPLHSESVQPN 491
Query: 371 QDTVRSRSGYSEEPEI-NIGRSNMKEDVELNDKHSSKNLNHRDGTFAPPSLEAIKKIDRD 429
Q T +S S ++ +G S E V N+ P KK D
Sbjct: 492 QATTQSNVVSSNPNQLWGLGMSTGAEGVPAVTASKISNVEEIQEVSLDPKENGDKKTD-- 549
Query: 430 KVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDED------ 483
+A+ E+ K G T E + DED EDG + + E K+ D
Sbjct: 550 --EASKEEEGKKTGEDTNDAENVVDED--------EDGDDDGSDEENKKEKDPKGMRAFK 599
Query: 484 --LIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQG---KHQDHSEEHLH 538
L+E V+EL ++ A + K +DG K++ K V MQ G + Q+ + +L
Sbjct: 600 FALVEVVKEL---LKPAWKEGKLNKDGYKNIVKKVAEKVTGTMQSGNVPQTQEKIDHYLS 656
Query: 539 LVKP 542
KP
Sbjct: 657 ASKP 660
>At5g16780 putative protein
Length = 820
Score = 48.1 bits (113), Expect = 1e-05
Identities = 75/316 (23%), Positives = 128/316 (39%), Gaps = 52/316 (16%)
Query: 330 PVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIG 389
PV H E H + K + H+ + ++ +D RSR +E+ EI+ G
Sbjct: 23 PVREHRDGRRKEKDHRSKDKEKDYDREKIRDKDHRRDKEKERDRKRSRDEDTEK-EISRG 81
Query: 390 RSNMKED------VELNDKHSSKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVG 443
R +E V+ DK +N H+D + E K DR +VK K+
Sbjct: 82 RDKEREKDKSRDRVKEKDKEKERN-RHKDRENERDN-EKEKDKDRARVKERASKKSHEDD 139
Query: 444 NITKKT----ELMDDEDLFE---RVRELEDGIELAAQSEKNK--QDDEDLIERVRELEDG 494
+ T K E D+ L E V G E +A +N+ + E+ ++ + D
Sbjct: 140 DETHKAAERYEHSDNRGLNEGGDNVDAASSGKEASALDLQNRILKMREERKKKAEDASDA 199
Query: 495 IELAAQSEK--NKQDGQKSLSKPVDR--SAFENMQQGKHQDHSE-EHLHLVKPSHE---- 545
+ A+S K K++ +K ++ + R +N+ QG+++D + EHL VK H
Sbjct: 200 LSWVARSRKIEEKRNAEKQRAQQLSRIFEEQDNLNQGENEDGEDGEHLSGVKVLHGLEKV 259
Query: 546 ----------TDLSAVEEG----EVSALDDIDLGRKSSNH------KRKAESSPDKF--- 582
D S + +G E+ L+++++G + + K+K DKF
Sbjct: 260 VEGGAVILTLKDQSVLTDGDVNNEIDMLENVEIGEQKRRNEAYEAAKKKKGIYDDKFNDD 319
Query: 583 --AEGKKRHNYSFGST 596
AE K Y +T
Sbjct: 320 PGAEKKMLPQYDEAAT 335
Score = 31.2 bits (69), Expect = 1.8
Identities = 40/166 (24%), Positives = 62/166 (37%), Gaps = 31/166 (18%)
Query: 385 EINIGRSNMKEDVELNDKHSSKNLNHRDGTFAPP---SLEAIKKIDRDKVKAALEKRKKA 441
E+ +S + E D S HRDG S + K DR+K++ +R K
Sbjct: 2 EVEKSKSRHEIREERADYEGSPVREHRDGRRKEKDHRSKDKEKDYDREKIRDKDHRRDKE 61
Query: 442 VGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIERVRELEDGIELAAQS 501
K++ D E R R+ E EK+K D RV+E
Sbjct: 62 KERDRKRSRDEDTEKEISRGRDKE--------REKDKSRD-----RVKE----------- 97
Query: 502 EKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEHLHLVKPSHETD 547
+D +K ++ DR + ++ K +D + K SHE D
Sbjct: 98 ----KDKEKERNRHKDRENERDNEKEKDKDRARVKERASKKSHEDD 139
>At2g43900 putative inositol polyphosphate 5'-phosphatase
Length = 1305
Score = 45.4 bits (106), Expect = 9e-05
Identities = 37/189 (19%), Positives = 85/189 (44%), Gaps = 12/189 (6%)
Query: 331 VSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIGR 390
+ ++ +N ++ + EGD+ + + + ++ +++ S+S + + N +
Sbjct: 1101 IDSNPSNSKSQSLKKNEGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSN-SK 1159
Query: 391 SNMKEDVELNDKHSSKNLNHRDGTFAPPS-----LEAIKKIDRDKVKAALEKRKKAVGNI 445
S+ K D + N K S K+ + + S ++ KK D D + +KK+ G+
Sbjct: 1160 SSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSCS---KSQKKSDGDT 1216
Query: 446 TKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIERVRELEDGIELAAQSEKNK 505
K++ D D + + DG + S+ +K++D D + + DG + +K++
Sbjct: 1217 NSKSQKKGDGDSSSKSHKKNDG---DSSSKSHKKNDGDSSSKSHKKSDGDSSSKSHKKSE 1273
Query: 506 QDGQKSLSK 514
D SL++
Sbjct: 1274 GDSSLSLTR 1282
>At1g10890 unknown protein
Length = 592
Score = 45.4 bits (106), Expect = 9e-05
Identities = 43/173 (24%), Positives = 89/173 (50%), Gaps = 13/173 (7%)
Query: 419 SLEAIKKIDRDKVKAALEKRKKAVGNITKKTEL-MDDEDLFERVRE-LEDGIELAAQSEK 476
+LE+ K + +K+K E+RK+ ++ EL + +E+ +RV E + +E + QSEK
Sbjct: 51 TLESAKNRNGEKLKREEEERKRR----QREAELKLIEEETVKRVEEAIRKKVEESLQSEK 106
Query: 477 NKQDDEDLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEH 536
K + L+E R+ + E+AAQ E+ K + SL + ++ E ++ + + +EE+
Sbjct: 107 IKMEILTLLEEGRKRLNE-EVAAQLEEEK---EASLIEAKEKEEREQQEKEERERIAEEN 162
Query: 537 LHLVKPSHETDL---SAVEEGEVSALDDIDLGRKSSNHKRKAESSPDKFAEGK 586
L V+ + + EE L+++ ++ + ++KAE ++ + K
Sbjct: 163 LKRVEEAQRKEAMERQRKEEERYRELEELQRQKEEAMRRKKAEEEEERLKQMK 215
>At5g60030 KED - like protein
Length = 292
Score = 44.7 bits (104), Expect = 2e-04
Identities = 36/137 (26%), Positives = 66/137 (47%), Gaps = 14/137 (10%)
Query: 402 KHSSKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERV 461
K K N++D E +K+ D+ K+A K +K KK++ +DED+ +
Sbjct: 151 KEKKKKKNNKDEDVVD---EKVKEKLEDEQKSADRKERKK-----KKSKKNNDEDVVDEK 202
Query: 462 RELEDGIELAAQSEKNKQDDEDLI-ERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSA 520
+LED + A EK K DED++ E+ +E + + + + +K K+ +KS D
Sbjct: 203 EKLEDEQKSAEIKEKKKNKDEDVVDEKEKEKLEDEQRSGERKKEKKKKRKS-----DEEI 257
Query: 521 FENMQQGKHQDHSEEHL 537
++ K + S+E +
Sbjct: 258 VSEERKSKKKRKSDEEM 274
>At1g20920 putative RNA helicase
Length = 1166
Score = 44.7 bits (104), Expect = 2e-04
Identities = 43/199 (21%), Positives = 82/199 (40%), Gaps = 17/199 (8%)
Query: 349 DAKGNQHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHSSKNL 408
D ++ E+ +E + +D V+ RS + S+ ++DVE D+ + +
Sbjct: 64 DRDDDEEREKRKEKERERRRRDKDRVKRRSERRKS-------SDSEDDVEEEDERDKRRV 116
Query: 409 NHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGI 468
N ++ + K RD+ + E+RK K+ E D + ER RE E
Sbjct: 117 NEKERGHREHERDRGKDRKRDRER---EERK------DKEREREKDRERRERERE-EREK 166
Query: 469 ELAAQSEKNKQDDEDLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGK 528
E + E+ +++D + R RE E G + E++++ G + V R ++G
Sbjct: 167 ERVKERERREREDGERDRREREKERGSRRNRERERSREVGNEESDDDVKRDLKRRRKEGG 226
Query: 529 HQDHSEEHLHLVKPSHETD 547
+ E + + S D
Sbjct: 227 ERKEKEREKSVGRSSRHED 245
Score = 33.9 bits (76), Expect = 0.28
Identities = 47/245 (19%), Positives = 92/245 (37%), Gaps = 27/245 (11%)
Query: 354 QHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHSSKNLNHRDG 413
+ +E + + +E + SR E +G +DV+ + K K R
Sbjct: 171 ERERREREDGERDRREREKERGSRRNRERERSREVGNEESDDDVKRDLKRRRKEGGERK- 229
Query: 414 TFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQ 473
E K + R KRK N KK + +E+L + ++L++ +E +
Sbjct: 230 -----EKEREKSVGRSSRHEDSPKRKSVEDNGEKKEKKTREEELEDEQKKLDEEVE---K 281
Query: 474 SEKNKQDDEDLIERVRELE-------DGIELAA------QSEKNKQDGQKSLSKPVDRSA 520
+ Q+ ++L + E E DG E A + E + ++G P ++S
Sbjct: 282 RRRRVQEWQELKRKKEEAESESKGDADGNEPKAGKAWTLEGESDDEEGH-----PEEKSE 336
Query: 521 FENMQQGKHQDHSEEHLHLVKPSHETDLSAVEEGEVSALDDIDLGRKSSNHKRKAESSPD 580
E + + ++ +V +ET + E G A+D+ ++ + +
Sbjct: 337 TEMDVDEETKPENDGDAKMVDLENETAATVSESGGDGAVDEEEIDPLDAFMNTMVLPEVE 396
Query: 581 KFAEG 585
KF G
Sbjct: 397 KFCNG 401
>At3g28770 hypothetical protein
Length = 2081
Score = 43.9 bits (102), Expect = 3e-04
Identities = 48/228 (21%), Positives = 99/228 (43%), Gaps = 16/228 (7%)
Query: 364 KENLQEAQDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHSSK-NLNHRDGTFAPPSLEA 422
+E++++ ++ V+ R+ S E+ +NM DV+ S K + + + +
Sbjct: 874 EESMKKKREEVQ-RNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDT 932
Query: 423 IKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDE 482
I + K K +K+K++ + KK E E + +++ ED + +SE +K +E
Sbjct: 933 INTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEE 992
Query: 483 DLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEHLHLVKP 542
+ + ++ E + EK + + +KS +K + + Q K ++ E K
Sbjct: 993 NKDNKEKK-ESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKE 1051
Query: 543 SHET-DLSAVEEGEVSALDDIDLGRKSSNHKRKAESSPDKFAEGKKRH 589
E+ DL A ++ E + K+++E+ K E KK H
Sbjct: 1052 KEESRDLKAKKKEE------------ETKEKKESENHKSKKKEDKKEH 1087
Score = 41.2 bits (95), Expect = 0.002
Identities = 60/327 (18%), Positives = 130/327 (39%), Gaps = 34/327 (10%)
Query: 317 ISKNVVSSANHVGPVSNHGT----NGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQEAQD 372
+ +N SS V +N+ GS E + + + K E + + + Q+ +D
Sbjct: 884 VQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKDTINTSSKQKGKD 943
Query: 373 TVRSRSGYSEEPEINIGRSNMKEDV-------ELNDKHSSKNLNHR---DGTFAPPSLEA 422
+ + S+ + + KE V E N K ++K+ N + + E+
Sbjct: 944 KKKKKKE-SKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKES 1002
Query: 423 IKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDE 482
+++ K E++K KK + + + RE +D E +S+K K++
Sbjct: 1003 EDSASKNREKKEYEEKKSKTKEEAKKEKKKSQD----KKREEKDSEE--RKSKKEKEESR 1056
Query: 483 DLIERVRELEDGIELAAQSEKNKQ-------DGQKSLSKPVDRSAFENMQQGKHQDHSEE 535
DL + +E E + +++ K+K+ + KS+ K D+ + ++ K + E+
Sbjct: 1057 DLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEED 1116
Query: 536 HLHLVK-PSHETDLSAVEEGEVSALDDIDLGRKSSNHKRKAESSPDKFAEGKKRHNYSFG 594
+ K ++ ++ E + L +K S+ K K E+ E +
Sbjct: 1117 KKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENE-----EKSETKEIESS 1171
Query: 595 STHHNRIDYIEDQNNVNLLGHAERDSK 621
+ N +D E +++ + E++ K
Sbjct: 1172 KSQKNEVDKKEKKSSKDQQKKKEKEMK 1198
Score = 36.6 bits (83), Expect = 0.043
Identities = 45/251 (17%), Positives = 96/251 (37%), Gaps = 18/251 (7%)
Query: 347 EGDAKGNQHPEQEPPPHKENLQEAQD--TVRSRSGYSEEPEINIGRSNMKED-----VEL 399
E + K + E KE+ +E +D +++ E+ + +S KE+ +L
Sbjct: 1064 EEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKL 1123
Query: 400 NDKHSSKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFE 459
D++S+K ++ ++ +KK K K E++ + TK+ E +
Sbjct: 1124 EDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSE-----TKEIESSKSQKNEV 1178
Query: 460 RVRELEDGIELAAQSEKNKQDDEDLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRS 519
+E + + + EK ++ E+ + E + + + + K +++ +K +KP D
Sbjct: 1179 DKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDK 1238
Query: 520 AFENMQQGKHQDHSEEHLHLVKPSHETDLSAVEEGEVSALDDIDLGRKSSNHKRKAESSP 579
Q G ++ E S E + + A D +A+S
Sbjct: 1239 KNTTKQSGGKKESMESE------SKEAENQQKSQATTQADSDESKNEILMQADSQADSHS 1292
Query: 580 DKFAEGKKRHN 590
D A+ + N
Sbjct: 1293 DSQADSDESKN 1303
Score = 31.2 bits (69), Expect = 1.8
Identities = 44/259 (16%), Positives = 105/259 (39%), Gaps = 19/259 (7%)
Query: 341 EMIHLVEGDAKGNQHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIGRSNMKEDVELN 400
EM E K N+ ++ +EN ++ ++T + ++ ++ + +S K++ +
Sbjct: 1196 EMKESEEKKLKKNEEDRKKQTSVEENKKQ-KETKKEKNKPKDDKKNTTKQSGGKKESMES 1254
Query: 401 DKHSSKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELM-------- 452
+ ++N T S E+ +I A + K E++
Sbjct: 1255 ESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQAT 1314
Query: 453 ----DDEDLFERVRELEDGIELAAQSEKNKQDDE--DLIERVRELEDGIELAAQSEKNKQ 506
++ED ++ E+ + + EKNK D+ + ++ ++ +E ++ +N+Q
Sbjct: 1315 TQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQ 1374
Query: 507 DGQKSLSKPVDRSAFENMQQGKHQ--DHSEEHLHLVKPSHETDLSAVEEGEVSALDDIDL 564
Q + D S E + Q Q HS+ + +E + A + ++ D
Sbjct: 1375 KSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDR 1434
Query: 565 GRKSS--NHKRKAESSPDK 581
+++S +K++ E+ +K
Sbjct: 1435 KKQTSVAENKKQKETKEEK 1453
Score = 31.2 bits (69), Expect = 1.8
Identities = 28/144 (19%), Positives = 60/144 (41%), Gaps = 4/144 (2%)
Query: 449 TELMDDEDLFERVRELEDGIELAAQSEKNK--QDDEDLIERVRELEDGIELAAQSEKNKQ 506
T+ ++ED ++ E+ + + EKNK D ++ E+ ++ +E ++ +N+Q
Sbjct: 1426 TQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTEQSGGKKESMESESKEAENQQ 1485
Query: 507 DGQKSLSKPVDRSAFENMQQGKHQDHSEEHLHLVKPSHETDLSAVEEGEVSALDDIDLGR 566
Q + D S E + Q Q ++ H + S E+ + + + A D
Sbjct: 1486 KSQATTQGESDESKNEILMQADSQ--ADTHANSQGDSDESKNEILMQADSQADSQTDSDE 1543
Query: 567 KSSNHKRKAESSPDKFAEGKKRHN 590
+ +A+S D + + N
Sbjct: 1544 SKNEILMQADSQADSQTDSDESKN 1567
>At1g76780 putative heat shock protein
Length = 1871
Score = 43.9 bits (102), Expect = 3e-04
Identities = 47/211 (22%), Positives = 85/211 (40%), Gaps = 27/211 (12%)
Query: 426 IDRDKVKAALE-----------KRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQS 474
+D DK+KA + KR K + + E ++E+ ERV E E ++
Sbjct: 28 VDLDKIKARFDDDDATLTITMPKRVKGISGFKIEEEEEEEEEEEERVDVSE--AEHKEET 85
Query: 475 EKNKQDDEDLIERVRELEDGIELA-AQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHS 533
EK + D D +E+ ++++ IE ++ NK+ SL KP D E +Q +H++
Sbjct: 86 EKGELKD-DYLEKSHQIDERIEEEKGLADSNKESVDSSLRKPPDIEGRECHEQTRHEEQE 144
Query: 534 EEHLHLVKPSHETD------LSAVEEGEVSALDDIDLGRKSSNHKRKAESSPDKFAEGKK 587
+ S ++D +EE E LD ++S + D +G +
Sbjct: 145 NNKQLVQAESDDSDDFGSRAFEEIEEQESDVLD------RTSTSGAMEKEMTDDVGDGLR 198
Query: 588 RHNYSFGSTHHNRIDYIEDQNNVNLLGHAER 618
+ HN I + + GH ++
Sbjct: 199 KVQGIEEPERHNEESKISEMVDGETSGHEKK 229
Score = 39.7 bits (91), Expect = 0.005
Identities = 51/215 (23%), Positives = 88/215 (40%), Gaps = 34/215 (15%)
Query: 331 VSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIGR 390
++ GT EMI + E + P E +++ V+S++ E+ E I
Sbjct: 532 INEDGTRKVQEMI----------RQQELDEPARSEKENRSRELVKSKTNDEEKKEKEIAG 581
Query: 391 SNMKEDVELNDKHSSKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNIT-KKT 449
+ KE K S + R+ E ++ DK K ++ K I K+
Sbjct: 582 TERKE------KESDRPKILREQ-------EVADEVAEDKTKFSIYGEVKEEEEIAGKEK 628
Query: 450 ELMDDEDLFERVRELE--DGIELAAQSEKNKQDDEDLIERVRELEDGI----ELAAQSEK 503
E D+D+ VR+ E D + Q EK+ + L E+V + GI E A++ K
Sbjct: 629 EFGSDDDIARIVRDTEQLDSNAMKGQEEKDMIQELVLEEKVCDGGKGIIAVAETKAENNK 688
Query: 504 NKQ----DGQKSLSKPVDRSAFENMQQGKHQDHSE 534
+K+ + QK + F+ + +G+ DH E
Sbjct: 689 SKRVQETEEQKLDKEDTCGKHFQKLIEGEISDHGE 723
Score = 38.5 bits (88), Expect = 0.011
Identities = 42/229 (18%), Positives = 88/229 (38%), Gaps = 11/229 (4%)
Query: 355 HPEQEPPPHKENLQEAQDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHSSKNLNHRDGT 414
H +++ EN+ + R + E + N + E ++ S ++ H
Sbjct: 989 HNDRKEEEQNENVTAEVELETERVSSKKVQEGKMEDDNSGKFHEFEERKSYEDWTHEKRE 1048
Query: 415 FAPPSLEAIKKIDRDKVKAALE----KRKKAVGNITKKTELMDDEDLFERVRELE--DGI 468
+E + +DK + K ++ N+ K EL +ED F++V E+E D
Sbjct: 1049 KRKVLVEEEETYPKDKHTGGEDHNDHKEEEQKENVIAKAELNTEEDSFKKVEEIEKQDHG 1108
Query: 469 ELAAQSEKNKQDDEDLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGK 528
EL + K+ + + ++ R +E + K +DG + + ++
Sbjct: 1109 ELKRSMVQAKRQETEEKDKTRAMEK--NETVERRKQTKDGSLGKLREGEDPELGGHERRG 1166
Query: 529 HQDHSEEHLHLVKPSHETDLSAVEEGEVSALDD---IDLGRKSSNHKRK 574
+D EE + H+ + +E + D +DLG + K++
Sbjct: 1167 EEDRIEELVETEISDHKEKVKKKDEDYILRSQDTGKVDLGERERRSKQR 1215
Score = 32.7 bits (73), Expect = 0.63
Identities = 34/159 (21%), Positives = 67/159 (41%), Gaps = 20/159 (12%)
Query: 348 GDAKGNQHPEQEPPPHKENLQEAQDTVRS-----RSGYSEEPEINIGRSNMKEDVELNDK 402
G AK N + E+ P K + + R G SE+ + + +KE VE +
Sbjct: 1703 GKAKENLNDEE---PTKTETKATDNESRKIHQIKEQGTSEQERLK-EQGRIKELVE-DRT 1757
Query: 403 HSSKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVR 462
H + +R+ + S + I++ID+++ ++ + ++T D+E+L
Sbjct: 1758 HFCREKENRETEYEDGSSKMIQEIDKEE----------SIEPVDRETSEDDEEELEIEFE 1807
Query: 463 ELEDGIELAAQSEKNKQDDEDLIERVRELEDGIELAAQS 501
+ E+ E E + D D I +++ + G L S
Sbjct: 1808 DEEEDWEAEVIQETDSDSDNDKIRQIKRIRLGFRLVGGS 1846
Score = 30.0 bits (66), Expect = 4.1
Identities = 42/235 (17%), Positives = 89/235 (37%), Gaps = 25/235 (10%)
Query: 325 ANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQ-EAQDTVRSRSGYSEE 383
A V + NGS+ + +E +++ +H EQ P N + +D R ++E
Sbjct: 1233 AEEAAAVVSRNENGSSRKVQTIEEESE--KHKEQNKIPETSNPEVNEEDEERVVEKETKE 1290
Query: 384 PEINIGRSNMK-EDVELNDKHSSKNLNHRDGTFAPPSLEA----------IKKIDRDKVK 432
E ++ K E+ + +D + + G A L ++KI K +
Sbjct: 1291 VEAHVQELEGKTENCKDDDGEGRREERGKQGMTAENMLRQRFKTKSDDGIVRKIQETKEE 1350
Query: 433 AALEKRKKAV-----------GNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDD 481
EK+ + G++ E + E ++ +L++ E + K +
Sbjct: 1351 EPDEKKSQESSSHVVKLVAEDGSLRNGLEFSEKESTVSKMLKLDESKEKEEHKKIRKPTE 1410
Query: 482 EDLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEH 536
E V E + + A + ++K D + + + + ++ G+H +E H
Sbjct: 1411 ERSNAPVIEKQGNKKNAEEEMQDKIDRRGKNQEIKGQEPYGVLRNGEHDKITEYH 1465
>At1g15940 T24D18.4
Length = 1012
Score = 43.9 bits (102), Expect = 3e-04
Identities = 51/258 (19%), Positives = 109/258 (41%), Gaps = 29/258 (11%)
Query: 381 SEEPEINIGRSNMKEDVELND--------KHSSKNLNHRDGTFAPPSLEAIKKI--DRDK 430
S PE+ MK+ + D K + K +++ + +L+++KK+ + DK
Sbjct: 687 SSSPEVRSSMQTMKKKDSVTDSIKQTKRTKGALKAVSNEPESTTGKNLKSLKKLNGEPDK 746
Query: 431 VKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIERVRE 490
+ K++K + +K E DE ++ ED ++L +S+ + D +E +E
Sbjct: 747 TRGRTGKKQKVTQAMHRKIEKDCDEQEDLETKDEEDSLKLGKESDA----EPDRMEDHQE 802
Query: 491 LEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEHLHLVKPSHETDLSA 550
L + + +++ +Q+ K + + E + + +EH L +P+ E S
Sbjct: 803 LPENHNVETKTDGEEQEAAKEPTAESKTNGEE--PNAEPETDGKEHKSLKEPNAEPK-SD 859
Query: 551 VEEGEVSALDDIDLGRKSSNHKRKAESSPDKFAEGKKRHNYSFGSTHHNRIDYIE--DQN 608
EE E + + +L N + E + ++ + ++ H D +E Q
Sbjct: 860 GEEQEAAKEPNAELKTDGENQEAAKELTAERKTDEEE----------HKVADEVEQKSQK 909
Query: 609 NVNLLGHAERDSKRPVQE 626
N+ AE + ++ V+E
Sbjct: 910 ETNVEPEAEGEEQKSVEE 927
Score = 33.1 bits (74), Expect = 0.48
Identities = 58/304 (19%), Positives = 106/304 (34%), Gaps = 63/304 (20%)
Query: 242 FDVTLTAHQLEEVSNQMLELYEQNQIAPSNDVEGTAGGGNRATPKASTSNDEAATTNNNL 301
F T + ++ N+ E + Q+ PS+ +E N + T + +A
Sbjct: 282 FATTQAHNDVKPKDNEADEKISEGQVVPSDSLEDKL---NLGLSRKGTRSKRSA------ 332
Query: 302 YIRGPSPRLEASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPP 361
RG + R + I+ +N G + ST+ A G+ P
Sbjct: 333 --RGGTRRANGDEKVIT-------------ANEGLSESTDA-----ETASGSTRKRGWKP 372
Query: 362 PHKENLQEAQDTVRSRSGYSEEPEIN-----------------IGRSNMKEDVELNDKHS 404
N +E S S +E E+ +G++N + L+
Sbjct: 373 KSLMNPEEGYSFKTSSSKKVQEKELGDSSLGKVAAKKVPLPSKVGQTNQSVVISLSSSGR 432
Query: 405 SKNLNHRDGTFAPPSLEAIKKIDRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVREL 464
++ + + S +++ D D A + KK KKT ++ V++
Sbjct: 433 ARTGSRKR------SRTKMEETDHDVSSVATQPAKKQT---VKKTNPAKEDLTKSNVKKH 483
Query: 465 EDGIELAAQSEKNKQDD--------EDLIERVRELEDGIELAAQSEKNKQDGQKSLSKPV 516
EDGI+ S+K K D+ + L E + G +L K K S+ P+
Sbjct: 484 EDGIKTGKSSKKEKADNGLAKTSAKKPLAETMMVKPSGKKLVHSDAKKKNSEGASMDTPI 543
Query: 517 DRSA 520
+S+
Sbjct: 544 PQSS 547
>At1g56660 hypothetical protein
Length = 522
Score = 43.5 bits (101), Expect = 4e-04
Identities = 49/171 (28%), Positives = 77/171 (44%), Gaps = 34/171 (19%)
Query: 436 EKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIERVRELEDGI 495
E K+ ++ KT+ +D ++ E V E +E+ A+S IE+V+ +D
Sbjct: 6 ENAKEEKLHVKIKTQELDPKEKGENV---EVEMEVKAKS----------IEKVKAKKD-- 50
Query: 496 ELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQD-------HSEEHLHL-VKPS---- 543
S K+K+D +K K VD E+ K +D H E H L VK S
Sbjct: 51 --EESSGKSKKDKEKKKGKNVDSEVKEDKDDDKKKDGKMVSKKHEEGHGDLEVKESDVKV 108
Query: 544 --HETDLSAVEEGEVSALDDIDLGRKSSNHKRKAESSPD---KFAEGKKRH 589
HE + +E + L++ G+K N K K ES P+ K A+ +K+H
Sbjct: 109 EEHEKEHKKGKEKKHEELEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKH 159
Score = 39.3 bits (90), Expect = 0.007
Identities = 66/352 (18%), Positives = 137/352 (38%), Gaps = 39/352 (11%)
Query: 252 EEVSNQMLELYEQN---QIAPSNDVEGTAGGGNRATPKASTSNDEAATTNNNLYIRGPSP 308
E+VS + EL E++ D GT + PK E + +N + ++G
Sbjct: 160 EDVSQEKEELEEEDGKKNKKKEKDESGTEE--KKKKPKKEKKQKEESKSNEDKKVKGKKE 217
Query: 309 RLEASKLAIS----KNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHK 364
+ E L K + + N E + K + P++E
Sbjct: 218 KGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKDESCAEEKK--KKPDKEKKEKD 275
Query: 365 ENLQEAQDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHSSKNLNHRDGTFAPPSLEAIK 424
E+ ++ ++ + G E+PE KE + + +H++G K
Sbjct: 276 ESTEKEDKKLKGKKGKGEKPEKEDEGKKTKEHDATEQEMDDEAADHKEGK---------K 326
Query: 425 KIDRDKVKAALEKRKKAVGNI-TKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDE- 482
K ++DK K K++ + + K+T+ DD++ + ++ + + + + EK+ ++D+
Sbjct: 327 KKNKDKAK----KKETVIDEVCEKETKDKDDDEGETKQKKNKKKEKKSEKGEKDVKEDKK 382
Query: 483 ---DLIERVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEEHLHL 539
L V + +E +K + D ++ V+ E +GK + ++ +
Sbjct: 383 KENPLETEVMSRDIKLEEPEAEKKEEDDTEEKKKSKVEGGESE---EGKKKKKKDKKKNK 439
Query: 540 VKPSHETDLSAVEEGEVSALDDIDL-GRKSSNHKRKAESSPDKFAEGKKRHN 590
K + E ++ EE + D+ + G K+ K+ DK + KK N
Sbjct: 440 KKDTKEPKMTEDEEEKKDDSKDVKIEGSKAKEEKK------DKDVKKKKGGN 485
>At1g80810 unknown protein
Length = 826
Score = 43.1 bits (100), Expect = 5e-04
Identities = 44/199 (22%), Positives = 87/199 (43%), Gaps = 9/199 (4%)
Query: 310 LEASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENLQE 369
++ K+ SKNV S V P S+ G S+ L++ D + + E +NL+
Sbjct: 634 MQRQKVKKSKNVAVS---VEPTSSSGVRSSSRT--LMKKDCGKRLNKQVEKTREGKNLRS 688
Query: 370 AQDTVRSRSGYSEEPEINIGRSN--MKEDVELNDKHSSKNLNHRDGTFAPPSLEAIKKID 427
++ +EE E+++ + E+ E D S K +D + E K+
Sbjct: 689 LKELNAETDRTAEEQEVSLEAESDDRSEEQEYEDDCSDKKEQSQDKGVEAETKEEEKQYP 748
Query: 428 RDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIER 487
+ ++ E + ++T+ M+D++ E E D +E A+ EK + DD++
Sbjct: 749 NSEGESEGEDSESEEEPKWRETDDMEDDE--EEEEEEIDHMEDEAEEEKEEVDDKEASAN 806
Query: 488 VRELEDGIELAAQSEKNKQ 506
+ E+E E + E+ ++
Sbjct: 807 MSEIEKEEEEEEEDEEKRK 825
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,658,531
Number of Sequences: 26719
Number of extensions: 657471
Number of successful extensions: 2433
Number of sequences better than 10.0: 189
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 167
Number of HSP's that attempted gapping in prelim test: 2250
Number of HSP's gapped (non-prelim): 313
length of query: 629
length of database: 11,318,596
effective HSP length: 105
effective length of query: 524
effective length of database: 8,513,101
effective search space: 4460864924
effective search space used: 4460864924
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0176a.15


