

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
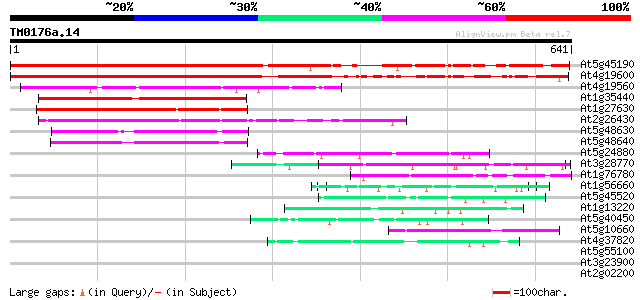
Query= TM0176a.14
(641 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g45190 unknown protein 566 e-161
At4g19600 unknown protein 519 e-147
At4g19560 putative protein 264 1e-70
At1g35440 hypothetical protein 252 5e-67
At1g27630 cyclin like protein 249 4e-66
At2g26430 cyclin like protein 147 1e-35
At5g48630 cyclin C-like protein 89 6e-18
At5g48640 cyclin C-like protein 84 2e-16
At5g24880 glutamic acid-rich protein 52 1e-06
At3g28770 hypothetical protein 50 3e-06
At1g76780 putative heat shock protein 49 7e-06
At1g56660 hypothetical protein 47 3e-05
At5g45520 putative protein 45 1e-04
At1g13220 putative nuclear matrix constituent protein 44 4e-04
At5g40450 unknown protein 43 5e-04
At5g10660 vacuolar calcium binding protein - like 43 5e-04
At4g37820 unknown protein 43 6e-04
At5g55100 unknown protein 41 0.002
At3g23900 unknown protein 40 0.003
At2g02200 putative protein on transposon FARE2.10 (cds2) 40 0.003
>At5g45190 unknown protein
Length = 579
Score = 566 bits (1459), Expect = e-161
Identities = 337/648 (52%), Positives = 416/648 (64%), Gaps = 81/648 (12%)
Query: 1 MAGLLLGDISHHGSYQSGSQGGSQDNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLR 60
MAG+L G+ S+ S S S + QE SRWYF RKEIEENSPS+ DG+DLKKE YLR
Sbjct: 1 MAGVLAGECSYSESGVSSHSRNSHEKQEEVSRWYFGRKEIEENSPSRLDGIDLKKETYLR 60
Query: 61 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETP 120
KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFF RQSHAKNDRR IATVCMFLAGKVEETP
Sbjct: 61 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFFRQSHAKNDRRTIATVCMFLAGKVEETP 120
Query: 121 RPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYK 180
RPLKDVI VSYEII+KKDP A Q+IKQK+VYEQQKELIL GE++VL+TLGFDLNV+HPYK
Sbjct: 121 RPLKDVIFVSYEIINKKDPGASQKIKQKEVYEQQKELILNGEKIVLSTLGFDLNVYHPYK 180
Query: 181 PLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD 240
PLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD
Sbjct: 181 PLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD 240
Query: 241 GEKVWWQEFDVTPRQLEEVSNQMLELYEQNRM--AQSNDVEGTTGGGNRATPKAPTSNDE 298
GEKVWWQEFDVTPRQLE+VSNQMLELYEQNR+ +Q ++VE + GGG+ P +
Sbjct: 241 GEKVWWQEFDVTPRQLEDVSNQMLELYEQNRVPASQGSEVESSVGGGSAQRP----GSRN 296
Query: 299 AASTNRNLHIGGPSSTLETSKPATSKNVFVSSANHVGRPVSNHG---VSGSTEVKHPVED 355
A ST+ ++ SS T + + S N SS + + N G + S + K +E
Sbjct: 297 AVSTDEHVGSRQTSSVRSTHEQSNSDNHGGSSKGVLNQNNENGGGEAANVSVDNKEEIER 356
Query: 356 DVK-GNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRS 414
+ K +LH + P A+++N++EA N P E
Sbjct: 357 ETKESSLHLESHP-AHKDNVREAPH-----------NSRPLVE----------------- 387
Query: 415 RSGYGKEQESNVGRSNIKEEDVELNDK---HSSKNLNHRDATFASPNLEGIKKIDRDKVK 471
G+ N + E EL D H S+N++ DA + + K+ RDKVK
Sbjct: 388 ----------GPGKDNSEREGGELQDDGAVHKSRNVDVGDALISQSPKD--LKLLRDKVK 435
Query: 472 AALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSLSKSLDRS 531
A EK KK G ++K +L+D++DLIE RELED ++LA + EK K+ +S K+ + S
Sbjct: 436 AKREKAKKLLGERTRKKDLMDEDDLIE--RELED-VQLAVEDEKTKERKVQSRPKA-ENS 491
Query: 532 AFENMQHGKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKRKVESSP 591
+HG+ D + ++ EEGE + + ++ P ++KRK+ S P
Sbjct: 492 DLMGTEHGEILD------------VKGEVKNTEEGE---MVNNNVSPMMHSRKRKMGSPP 536
Query: 592 DNFVEGKKRHNYGSGSTHHNRIDYIEDRNKVNRLGHAERDSKRHIQEN 639
+ EGK+RHN +G H + H +R+ +RH QEN
Sbjct: 537 EKQSEGKRRHNSENGEESH--------KTSRGSSHHGDREHRRHSQEN 576
>At4g19600 unknown protein
Length = 541
Score = 519 bits (1336), Expect = e-147
Identities = 322/647 (49%), Positives = 405/647 (61%), Gaps = 116/647 (17%)
Query: 1 MAGLLLGDISHHGSYQSGSQGGSQDNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLR 60
MAG+L GD S S S S + Q+ +RWYF RKEIEENSPS+ D +DLKKE YLR
Sbjct: 1 MAGVLAGDCSFGESGVSSYSRNSNEKQDEVARWYFGRKEIEENSPSRLDSIDLKKETYLR 60
Query: 61 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETP 120
KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFF+RQSHA+NDRR IATVCMFLAGKVEETP
Sbjct: 61 KSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFIRQSHARNDRRTIATVCMFLAGKVEETP 120
Query: 121 RPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYK 180
RPLKDVI+VSYEIIHKKDP Q+IKQK+VYEQQKELIL GE++VL+TLGFD NV+HPYK
Sbjct: 121 RPLKDVIVVSYEIIHKKDPTTAQKIKQKEVYEQQKELILNGEKIVLSTLGFDFNVYHPYK 180
Query: 181 PLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD 240
PLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD
Sbjct: 181 PLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD 240
Query: 241 GEKVWWQEFDVTPRQLEEVSNQMLELYEQNRM--AQSNDVEGTTGGGNRATPKAPTSNDE 298
GEKVWWQEFDVTPRQLE+VSNQMLELYEQNR+ +Q ++VE + GGG+
Sbjct: 241 GEKVWWQEFDVTPRQLEDVSNQMLELYEQNRVPASQVSEVESSVGGGSAH---------- 290
Query: 299 AASTNRNLHIGGPSSTLETSKPATSKNVFVSSANHVGRPVSN-HGVSGS--TEVKHPVED 355
H+G S T + + S N+ S+ R N G +GS TE K +
Sbjct: 291 --------HVGSRPSARLTHEHSNSDNLGGSTKATQNRSNDNGSGEAGSVITEQKGERDT 342
Query: 356 DVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSR 415
+ K ++H + P+ +SRSG + P E+K+++A
Sbjct: 343 ETKDSMHTESHPAH------------KSRSGV--------EAPG--EDKIEKA------G 374
Query: 416 SGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALE 475
+ + ++ +S + + DV ++ S K++ K+ RDKVKA LE
Sbjct: 375 AHFPEDDKSRI----VGTADVTVS--QSPKDI----------------KMFRDKVKAKLE 412
Query: 476 KRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSLSKSLDRSAFEN 535
KK G ++K +LVD++DLIE RELED +ELA + +K+ Q+ +
Sbjct: 413 -AKKVQGEKTRKKDLVDEDDLIE--RELED-VELAVEDDKDIQN-----------KSSMG 457
Query: 536 MQHGKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKRKVESSPDNFV 595
+HG+ D + ++ EEGE+ +DD+ ++KRK+ES +
Sbjct: 458 TEHGEILDGNNLVVN------------TEEGEM--IDDV--SSTMPSRKRKMESPCEK-- 499
Query: 596 EGKKRHNYGSGSTHHNRIDYIEDRNKVNRLG----HAERDSKRHIQE 638
G G H+ + +E+ K N G + +R+ +RH QE
Sbjct: 500 ------QLGEGKRRHDNSENVEEGQKTNPGGSSHSYGDREPRRHSQE 540
>At4g19560 putative protein
Length = 474
Score = 264 bits (674), Expect = 1e-70
Identities = 177/397 (44%), Positives = 230/397 (57%), Gaps = 45/397 (11%)
Query: 13 GSYQSGSQGGSQDNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGM 72
GS S S + + W+FSR+EIE NSPS++DG+DLK E LR SYCTFL+ LG
Sbjct: 11 GSESDASSVASNLHDDEIIPWFFSREEIERNSPSRRDGIDLKTETRLRDSYCTFLEILGE 70
Query: 73 RLKVPQVTIATAIIFCHR-----FFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVI 127
RLK+ C R FF S IATVCM LAGKVEETP L+DVI
Sbjct: 71 RLKMSLPDFIHDKTVCDRDQALCFFPFGSMCMT----IATVCMLLAGKVEETPVTLEDVI 126
Query: 128 LVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIK 187
+ SYE IHKKD A QR K+VY+QQKEL+L+GE +VL+TL FDL + HPYKPLVEAIK
Sbjct: 127 IASYERIHKKDLAGAQR---KEVYDQQKELVLIGEELVLSTLNFDLCISHPYKPLVEAIK 183
Query: 188 KFKV--AQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVW 245
K+ V A+ LAQ AWNFVND LRT+LCLQ++PHHIAAGAI LAA+ V L S E V
Sbjct: 184 KYMVEDAKTQLAQFAWNFVNDCLRTTLCLQYQPHHIAAGAILLAAELPTVDLQSYRE-VL 242
Query: 246 WQEFDVTPRQLE-------------EVSNQMLELYEQNRMAQSNDVEGTTG--------- 283
QEFD+TP QLE ++ Q+LELYE+ +Q + VE + G
Sbjct: 243 CQEFDITPCQLEDLVDVVNLSFAFSDIRGQILELYERIPTSQESKVESSGGVAVVHQPIS 302
Query: 284 GGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSKPATSKNV-FVSSANHVGRPVSNHG 342
+T K P+S+ E S+ NL S + S+ S+ + V+S + + + +H
Sbjct: 303 RDMASTEKCPSSDIEGGSSQVNLSQSDDHSVHDGSR---SEGIGEVNSESEAQKNLQDHS 359
Query: 343 VSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQD 379
V K DDV G + K++ ++E ++ Q+
Sbjct: 360 VGNIMVEK---SDDV-GVVQLKKDLQLHQEEVESKQE 392
Score = 31.6 bits (70), Expect = 1.4
Identities = 39/190 (20%), Positives = 81/190 (42%), Gaps = 13/190 (6%)
Query: 356 DVKGNLHPKQE--PSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVR 413
D++G + E P++ E ++ + + ++ ++ PS+ E ++ +
Sbjct: 268 DIRGQILELYERIPTSQESKVESSGGVAVVHQPISRDMASTEKCPSSDIEGGSSQVNLSQ 327
Query: 414 S--RSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLN-HRDATFASPNLEGIKKIDRDKV 470
S S + + +G N + E + HS N+ + L+ ++ +++V
Sbjct: 328 SDDHSVHDGSRSEGIGEVNSESEAQKNLQDHSVGNIMVEKSDDVGVVQLKKDLQLHQEEV 387
Query: 471 KAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSLSKSLDR 530
++ EK KK+ K +L+D++DL E E+ED I K Q GR+ K D
Sbjct: 388 ESKQEKDKKSFEKDITKIDLMDEKDLTE--SEVEDEI------NKTMQTGRQIFMKVEDP 439
Query: 531 SAFENMQHGK 540
++H +
Sbjct: 440 DDNMTVEHSE 449
>At1g35440 hypothetical protein
Length = 247
Score = 252 bits (643), Expect = 5e-67
Identities = 123/240 (51%), Positives = 169/240 (70%), Gaps = 9/240 (3%)
Query: 33 WYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFF 92
WY +R+ IE+ SPS+ DG++LK+E + R SY +FLQ+LG RL PQ TIATAI+ C RFF
Sbjct: 7 WYNTREAIEKTSPSRLDGINLKEETFQRWSYTSFLQELGQRLNNPQKTIATAIVLCQRFF 66
Query: 93 LRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYE 152
RQS KND + +A +CMF+AGKVE +PRP DV+ VSY ++ K+P +DV+E
Sbjct: 67 TRQSLTKNDPKTVAIICMFIAGKVEGSPRPAGDVVFVSYRVLFNKEPL-------RDVFE 119
Query: 153 QQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNA--LAQVAWNFVNDGLRT 210
+ K +L GE++VL+TL DL + HPYK +++ +K+ ++ L Q A+NFVND LRT
Sbjct: 120 RLKMTVLTGEKLVLSTLECDLEIEHPYKLVMDWVKRSVKTEDGRRLCQAAFNFVNDSLRT 179
Query: 211 SLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYEQN 270
SLCLQF P IA+ AI++ K+ LP DG+K WW+EFDVT RQL E+ +QML+LY Q+
Sbjct: 180 SLCLQFGPSQIASAAIYIGLSMCKMTLPCDGDKAWWREFDVTKRQLWEICDQMLDLYVQD 239
>At1g27630 cyclin like protein
Length = 317
Score = 249 bits (635), Expect = 4e-66
Identities = 127/241 (52%), Positives = 169/241 (69%), Gaps = 3/241 (1%)
Query: 31 SRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHR 90
S+WYFSR+EIE SPS++DG+DL KE++LR SYCTFLQ LGM+L V QVTI+ A++ CHR
Sbjct: 32 SKWYFSREEIERFSPSRKDGIDLVKESFLRSSYCTFLQRLGMKLHVSQVTISCAMVMCHR 91
Query: 91 FFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDV 150
F++RQSHAKND + IAT +FLA K E+ P L V++ SYEII++ DP+A RI Q +
Sbjct: 92 FYMRQSHAKNDWQTIATSSLFLACKAEDEPCQLSSVVVASYEIIYEWDPSASIRIHQTEC 151
Query: 151 YEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRT 210
Y + KE+IL GE ++L+T F L++ PYKPL A+ + A LA AWNFV+D +RT
Sbjct: 152 YHEFKEIILSGESLLLSTSAFHLDIELPYKPLAAALNRLN-AWPDLATAAWNFVHDWIRT 210
Query: 211 SLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYEQN 270
+LCLQ+KPH IA + LAA F K+ S + WW EF VT + L+EV +M L E +
Sbjct: 211 TLCLQYKPHVIATATVHLAATFQNAKVGS--RRDWWLEFGVTTKLLKEVIQEMCTLIEVD 268
Query: 271 R 271
R
Sbjct: 269 R 269
>At2g26430 cyclin like protein
Length = 416
Score = 147 bits (372), Expect = 1e-35
Identities = 114/436 (26%), Positives = 201/436 (45%), Gaps = 50/436 (11%)
Query: 33 WYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFF 92
+Y S ++++ SPS++DG+D E LR C +Q+ G+ LK+PQ +AT + RF+
Sbjct: 9 FYLSDEQLKA-SPSRKDGIDETTEISLRIYGCDLIQEGGILLKLPQAVMATGQVLFQRFY 67
Query: 93 LRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQ-KDVY 151
++S AK D +I+A C++LA K+EE P+ + VI+V + + +++ ++ + +
Sbjct: 68 CKKSLAKFDVKIVAASCVWLASKLEENPKKARQVIIVFHRMECRRENLPLEHLDMYAKKF 127
Query: 152 EQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTS 211
+ K + ER +L +GF +V HP+K + + + L Q AWN ND LRT+
Sbjct: 128 SELKVELSRTERHILKEMGFVCHVEHPHKFISNYLATLETPPE-LRQEAWNLANDSLRTT 186
Query: 212 LCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYEQNR 271
LC++F+ +A G ++ AA+ +V LP + WW+ FD ++EV + LY +
Sbjct: 187 LCVRFRSEVVACGVVYAAARRFQVPLPEN--PPWWKAFDADKSSIDEVCRVLAHLYSLPK 244
Query: 272 MAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSKPATSKNVFVSSA 331
AQ V G T + + N + S ++L + G ++T A S N
Sbjct: 245 -AQYISV---CKDGKPFTFSSRSGNSQGQSATKDL-LPGAGEAVDTKCTAGSAN------ 293
Query: 332 NHVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGN 391
N G H D K S E N Q + S Y +
Sbjct: 294 --------NDLKDGMVTTPHEKATDSK--------KSGTESNSQP----IVGDSSYERSK 333
Query: 392 QHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEED--------------VE 437
++ S E++ +D RS G +++S+ R +K+ +
Sbjct: 334 VGDRERESDREKERGRERDRGRSHRGRDSDRDSDRERDKLKDRSHHRSRDRLKDSGGHSD 393
Query: 438 LNDKHSSKNLNHRDAT 453
+ HSS++ ++RD++
Sbjct: 394 KSRHHSSRDRDYRDSS 409
>At5g48630 cyclin C-like protein
Length = 253
Score = 89.4 bits (220), Expect = 6e-18
Identities = 56/225 (24%), Positives = 109/225 (47%), Gaps = 17/225 (7%)
Query: 48 QDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIAT 107
Q G+ ++ ++ ++ L +K+ Q +ATA+ + R + R+S + + R++A
Sbjct: 29 QRGISVEDFRLIKLHMSNYISKLAQHIKIRQRVVATAVTYMRRVYTRKSLTEYEPRLVAP 88
Query: 108 VCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVVLA 167
C++LA K EE+ K +LV Y ++++ + + + + IL E VL
Sbjct: 89 TCLYLACKAEESVVHAK--LLVFY----------MKKLYADEKFRYEIKDILEMEMKVLE 136
Query: 168 TLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIF 227
L F L V HPY+ L E ++ + ++ + W VND R L L P I I+
Sbjct: 137 ALNFYLVVFHPYRSLPEFLQDSGINDTSMTHLTWGLVNDTYRMDLILIHPPFLITLACIY 196
Query: 228 LAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYEQNRM 272
+A+ + + K W++E V ++ ++ ++L+ YE +R+
Sbjct: 197 IASVHKEKDI-----KTWFEELSVDMNIVKNIAMEILDFYENHRL 236
>At5g48640 cyclin C-like protein
Length = 253
Score = 84.0 bits (206), Expect = 2e-16
Identities = 52/225 (23%), Positives = 107/225 (47%), Gaps = 17/225 (7%)
Query: 47 KQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIA 106
K+ G+ + ++ + L +KV Q +ATAI + R ++R+S + + R++A
Sbjct: 28 KERGISIDDFKLIKFHMSNHIMKLAQHIKVRQRVVATAITYMRRVYIRKSMVEFEPRLVA 87
Query: 107 TVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVVL 166
C++LA K EE+ ++++ ++R+ + + + + IL E VL
Sbjct: 88 LTCLYLASKAEESIVQARNLVFY------------IKRLYPDEYNKYELKDILGMEMKVL 135
Query: 167 ATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAI 226
L + L V HPY+ L E ++ + + Q+ W VND + L L P+ IA I
Sbjct: 136 EALDYYLVVFHPYRSLSEFLQDAALNDVNMNQITWGIVNDTYKMDLILVHPPYRIALACI 195
Query: 227 FLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYEQNR 271
++A+ + + + W+++ ++ ++ ++L+ YE R
Sbjct: 196 YIASVHREKDITA-----WFEDLHEDMNLVKNIAMEILDFYENYR 235
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 51.6 bits (122), Expect = 1e-06
Identities = 64/293 (21%), Positives = 124/293 (41%), Gaps = 42/293 (14%)
Query: 284 GGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSKPATSKNVFVSSANHVGRPVSNHGV 343
GGN T K+P + + N SST SK S+NV + A+ + + +
Sbjct: 144 GGN--TAKSPPVAPKKSGLNS-------SSTSSKSKKEGSENVRIKKASDKEIALDSASM 194
Query: 344 SGSTEVKHPVE------DDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQE 397
S + E H E D ++ + H +EP +E + + +V++ + +
Sbjct: 195 SSAQE-DHQEEILKVESDHLQVSDHDIEEPKYEKEEKEVQEKVVQANESVEEKAESSGPT 253
Query: 398 P-------------STYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSS 444
P + EEK+ + +D + ++ KEQ++N + +EEDV+ K
Sbjct: 254 PVASPVGKDCNAVVAELEEKLIKNEDDIEEKTEEMKEQDNNQANKSEEEEDVK---KKID 310
Query: 445 KNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELE 504
+N S +E +++ ++K + E+ K+ K+ E V ++D E+V E E
Sbjct: 311 ENETPEKVDTESKEVESVEETTQEKEEEVKEEGKERVEEEEKEKEKVKEDDQKEKVEE-E 369
Query: 505 DGIELAAQSEKNK------QDGRKS---LSKSLDRSAFENMQHGKHQDHSEEH 548
+ ++ EK K +G+K K SA+ ++ K Q++ ++
Sbjct: 370 EKEKVKGDEEKEKVKEEESAEGKKKEVVKGKKESPSAYNDVIASKMQENPRKN 422
>At3g28770 hypothetical protein
Length = 2081
Score = 50.4 bits (119), Expect = 3e-06
Identities = 64/311 (20%), Positives = 135/311 (42%), Gaps = 31/311 (9%)
Query: 354 EDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVR 413
+DD K++ S + + ++ VR++ +GN+ ++ E+K E++D
Sbjct: 741 KDDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKK--ESKDAKS 798
Query: 414 SRSGYGKEQESNVGRSNIKEEDVELN--DKHSSKNLNHRDATFASPN-----LEGIKKID 466
+ K+ S R KE E N DK SK+ +A + N G K+
Sbjct: 799 VETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAKEKNENGGVDTNVGNKEDS 858
Query: 467 RD-KVKAALEKRKKATGNISKKTELV--DDEDLIERVRELEDGIEL-----AAQSEKNKQ 518
+D K ++E + ++ KK E V +D+ + VR+ + +++ + +S K K+
Sbjct: 859 KDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKK 918
Query: 519 DGRKSLSKSLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGP 578
D +K +K ++ K +D ++ K S +++ EE + +++ +L
Sbjct: 919 DEKKEGNKEENKDTINTSSKQKGKDKKKKK----KESKNSNMKKKEEDKKEYVNN-ELKK 973
Query: 579 KSSNQKRKVES-----SPDNFVEGKKRHNYGSGSTHHNRIDYIEDRNKVNRLGHAER--- 630
+ N+K +S +N +K+ + S S + + +Y E ++K E+
Sbjct: 974 QEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKS 1033
Query: 631 -DSKRHIQENQ 640
D KR ++++
Sbjct: 1034 QDKKREEKDSE 1044
Score = 44.7 bits (104), Expect = 2e-04
Identities = 66/410 (16%), Positives = 149/410 (36%), Gaps = 38/410 (9%)
Query: 254 RQLEEVS-NQMLELYEQNRMAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPS 312
++ EEV N E A + D++ G G K + N++
Sbjct: 879 KKREEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGNKEENKD------- 931
Query: 313 STLETS----------KPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVEDDVKGNLH 362
T+ TS K SKN + + N+ + + K L
Sbjct: 932 -TINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLK 990
Query: 363 PKQEPSAYEENMQEAQDMVRSRSGY----AKGNQHPKQEPSTYEEKMQEAQDMVRSRSGY 418
+ + + ++ +++ R + Y +K + K+E ++K +E +D +S
Sbjct: 991 EENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKK 1050
Query: 419 GKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRK 478
KE+ ++ +EE E + + K+ D N K+ D+ + K E +
Sbjct: 1051 EKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKS 1110
Query: 479 KATGNISKKTELVDDEDLIERVRELEDG--------IELAAQSEKNKQDGRKSLSKSLDR 530
+ K E ++D++ ++ + + ++ + ++ K++ KS +K ++
Sbjct: 1111 RKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIES 1170
Query: 531 SAFENMQHGKHQ-----DHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKR 585
S + + K + D ++ +K S E L EE + K + +
Sbjct: 1171 SKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEE--NKKQKETK 1228
Query: 586 KVESSPDNFVEGKKRHNYGSGSTHHNRIDYIEDRNKVNRLGHAERDSKRH 635
K ++ P + + + + G + + E++ K A+ D ++
Sbjct: 1229 KEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKN 1278
Score = 33.5 bits (75), Expect = 0.37
Identities = 54/311 (17%), Positives = 127/311 (40%), Gaps = 41/311 (13%)
Query: 364 KQEPSAYEEN--MQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQD----------- 410
K++ +E+N M++ +D + + + + K+E EK+++
Sbjct: 1081 KEDKKEHEDNKSMKKEEDK-KEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKK 1139
Query: 411 ------MVRSRSGYGKEQESNVGRSNIKE--------EDVELNDKHSSKN-LNHRDATFA 455
+V+ S KE++ N +S KE +V+ +K SSK+ ++
Sbjct: 1140 KSQHVKLVKKESD-KKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMK 1198
Query: 456 SPNLEGIKKIDRDKVK-AALEKRKKATGNISKKTELVDD-EDLIERVRELEDGIELAAQS 513
+ +KK + D+ K ++E+ KK +K + DD ++ ++ ++ +E ++
Sbjct: 1199 ESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKE 1258
Query: 514 EKNKQDGRKSLSKSLDRSAFENMQHGKHQ--DHSEEHLHLVKPSHEADLSAVEEGEVSAL 571
+N+Q + + D S E + Q HS+ + +E + A +
Sbjct: 1259 AENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRN 1318
Query: 572 DDIDLGPKSS---NQKRK----VESSPDNFVEGKKRHNYGSGSTHHNRIDYIEDRNKVNR 624
++ D ++S N+K+K ++ P + + + + G + + E++ K
Sbjct: 1319 NEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQA 1378
Query: 625 LGHAERDSKRH 635
A+ D ++
Sbjct: 1379 TTQADSDESKN 1389
>At1g76780 putative heat shock protein
Length = 1871
Score = 49.3 bits (116), Expect = 7e-06
Identities = 50/257 (19%), Positives = 113/257 (43%), Gaps = 22/257 (8%)
Query: 390 GNQHPKQEPSTYE----EKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSK 445
G H +E +YE EK ++ + +V Y K++ + N +E+ + + +
Sbjct: 1028 GKFHEFEERKSYEDWTHEKREKRKVLVEEEETYPKDKHTGGEDHNDHKEEEQKENVIAKA 1087
Query: 446 NLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELED 505
LN + +F +E I+K D ++K ++ + K+ KT ++ + +ER ++ +D
Sbjct: 1088 ELNTEEDSFKK--VEEIEKQDHGELKRSMVQAKRQETEEKDKTRAMEKNETVERRKQTKD 1145
Query: 506 GIELAAQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADLSAVEE 565
G + ++ + G DR E + + DH E+ + K + L + +
Sbjct: 1146 GSLGKLREGEDPELGGHERRGEEDR--IEELVETEISDHKEK---VKKKDEDYILRSQDT 1200
Query: 566 GEVSALDDIDLGPKSSNQK-RKVESSPDNFVEGKKRHNYGSGSTHHNRIDYIEDRNKVNR 624
G+V DLG + K RK+ S ++ + ++ + + +R + +
Sbjct: 1201 GKV------DLGERERRSKQRKIHKSVEDEIGDQEDEDAEEAAAVVSR----NENGSSRK 1250
Query: 625 LGHAERDSKRHIQENQV 641
+ E +S++H ++N++
Sbjct: 1251 VQTIEEESEKHKEQNKI 1267
Score = 38.9 bits (89), Expect = 0.009
Identities = 66/319 (20%), Positives = 124/319 (38%), Gaps = 60/319 (18%)
Query: 355 DDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRS 414
D + + K + + N+ + Q+ S KG+ + EEK EA+ +R+
Sbjct: 289 DKINEGGNTKVRRHSEDRNLIKLQEKEEQHSKEQKGHSKEENMKELVEEKTPEAETTIRN 348
Query: 415 -------------------RSGYGKEQESNVGRSNIK------EEDVELNDKHSSKNLNH 449
S GKE++ N+ + IK E D ++ + +S N
Sbjct: 349 DILGPGQEIEVPEVDTLGKTSDEGKEKQ-NIVKKEIKNGDATEEIDAKMGEVFAS---NI 404
Query: 450 RDATFASPNLEGIKKIDRDKVKAALEKRKKATGN-----------ISKKTELVDDE---- 494
D S + E K D+V +EK+ + N ++K E+ + +
Sbjct: 405 ADTGMNSEDFESDKLESADEVDKMVEKKDRQEENDKVGAQSEDISLTKLQEIGEQQFQGQ 464
Query: 495 ---DLIERVRELEDGIELAAQSEKN------KQDGRKSLSKSLDRSAFENMQHGKHQDHS 545
D E ++EL +G A+++EKN K ++S K + F+ + + + ++
Sbjct: 465 KRHDKQENIKELREG--QASEAEKNIKNDILKPVQKRSEGKHKIQKTFQEETNKQPEGYN 522
Query: 546 EEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKRKVESSPDNFVEGKKRHNYGS 605
E+ + K +E V+E D + N+ R++ S N E K++ G+
Sbjct: 523 EKIMETGKKINEDGTRKVQEMIRQQELDEPARSEKENRSRELVKSKTNDEEKKEKEIAGT 582
Query: 606 GSTHHNRIDYIEDRNKVNR 624
R + DR K+ R
Sbjct: 583 -----ERKEKESDRPKILR 596
Score = 38.5 bits (88), Expect = 0.012
Identities = 51/214 (23%), Positives = 92/214 (42%), Gaps = 18/214 (8%)
Query: 402 EEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHS--SKNLNHRDATFASPNL 459
EE+ + D + KE +VG K + +E ++H+ SK D +
Sbjct: 169 EEQESDVLDRTSTSGAMEKEMTDDVGDGLRKVQGIEEPERHNEESKISEMVDGETSGHEK 228
Query: 460 EGIKKIDRDKVKAALEKRKKATG-----NISKKTELVDDEDLIERVRELE----DGIELA 510
+ + K+D+ K + E+ A G NI +T++V D+++ E + E D +E
Sbjct: 229 KKVVKMDK-KNRDVKEEVDGAMGEGFRPNID-RTQVVGDDEIAETEKNDEEFESDKLEAD 286
Query: 511 AQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADL--SAVEEGEV 568
+ N+ K S DR+ + + K + HS+E K + +L E E
Sbjct: 287 EVDKINEGGNTKVRRHSEDRNLIKLQE--KEEQHSKEQKGHSKEENMKELVEEKTPEAET 344
Query: 569 SALDDIDLGPKSSNQKRKVESSPDNFVEGKKRHN 602
+ +DI LGP + +V++ EGK++ N
Sbjct: 345 TIRNDI-LGPGQEIEVPEVDTLGKTSDEGKEKQN 377
Score = 32.0 bits (71), Expect = 1.1
Identities = 63/353 (17%), Positives = 128/353 (35%), Gaps = 35/353 (9%)
Query: 316 ETSKPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQ 375
ETS P ++ + V H + ++ +DD +G + + ENM
Sbjct: 1269 ETSNPEVNEEDEERVVEKETKEVEAHVQELEGKTENCKDDDGEGRREERGKQGMTAENML 1328
Query: 376 EAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYG--------KEQESNVG 427
+ +S G + Q K+E ++ + + +V+ + G E+ES V
Sbjct: 1329 RQRFKTKSDDGIVRKIQETKEEEPDEKKSQESSSHVVKLVAEDGSLRNGLEFSEKESTVS 1388
Query: 428 RSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISKK 487
+ +E E + + + +G KK ++++ +++R K ++
Sbjct: 1389 KMLKLDESKEKEEHKKIRKPTEERSNAPVIEKQGNKKNAEEEMQDKIDRRGKNQEIKGQE 1448
Query: 488 TELVDDEDLIERVRELEDGIELAA----------QSEKNKQDGRKSLSKSLDRSAFENMQ 537
V +++ E G E Q++ + RK S + + E M
Sbjct: 1449 PYGVLRNGEHDKITEYHRGEEKGTAENVSSTKIQQTKDELEKPRKPSEISENHNIHEFMD 1508
Query: 538 HGKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKRKVESSPDNFVEG 597
+ QD E K S +A+ A + ++D D + +R ++
Sbjct: 1509 SSQSQDIEE------KGSDQAEKYAKQNKIQEVMNDEDKKEEYHISERVRNEMAKRILQV 1562
Query: 598 KKRHNYGSGSTHHNRIDYIED---RNKVNRLGHAE------RDSKRHIQENQV 641
+ + N GS+ N + E R + R H E D K+ ++E++V
Sbjct: 1563 ESKAN--DGSSKKNETEGQESTGLRGRKKRENHQELVELETSDQKKGVKEDEV 1613
Score = 30.4 bits (67), Expect = 3.2
Identities = 69/318 (21%), Positives = 120/318 (37%), Gaps = 59/318 (18%)
Query: 362 HPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPS---TYEEKMQEAQDMVRSRSGY 418
H EP ++E + SG KG + K S T + + Q+ + R G
Sbjct: 911 HELHEPKIHKERDNNRVTGAKEPSGQEKGEKEEKIVESMTITENDNSIDVQETKKERPG- 969
Query: 419 GKEQESNVGRSNIKEEDVEL--ND-KHSSKNLNHRDATFASPNLEGIKKIDRDKVK---- 471
ES+ R I+E +E ND K +N N KK+ K++
Sbjct: 970 --RLESHDKRYKIQELLMEAGHNDRKEEEQNENVTAEVELETERVSSKKVQEGKMEDDNS 1027
Query: 472 ---AALEKRKKATGNISKKTE----LVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSL 524
E+RK +K E LV++E+ + + + E+ K++
Sbjct: 1028 GKFHEFEERKSYEDWTHEKREKRKVLVEEEETYPKDKHTGGEDHNDHKEEEQKENVIAKA 1087
Query: 525 SKSLDRSAFENMQHGKHQDHSEEHLHLV----KPSHEADLSAVEE--------------- 565
+ + +F+ ++ + QDH E +V + + E D + E
Sbjct: 1088 ELNTEEDSFKKVEEIEKQDHGELKRSMVQAKRQETEEKDKTRAMEKNETVERRKQTKDGS 1147
Query: 566 -GEVSALDDIDLGPKSSNQKRKVESSPDNFVEGKKRHNYGSGSTHHNRI-----DYI--- 616
G++ +D +LG +++R E + VE + S H ++ DYI
Sbjct: 1148 LGKLREGEDPELG---GHERRGEEDRIEELVETEI-------SDHKEKVKKKDEDYILRS 1197
Query: 617 EDRNKVNRLGHAERDSKR 634
+D KV+ LG ER SK+
Sbjct: 1198 QDTGKVD-LGERERRSKQ 1214
Score = 29.3 bits (64), Expect = 7.1
Identities = 50/244 (20%), Positives = 95/244 (38%), Gaps = 36/244 (14%)
Query: 420 KEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKK 479
KE E +V + + D DK ++ + DAT + +K I K++ E+ ++
Sbjct: 11 KEVEFSVFKKVFRIPDTVDLDKIKAR-FDDDDATLTITMPKRVKGISGFKIEEEEEEEEE 69
Query: 480 ATGNI----------SKKTELVDDEDLIERVRELEDGIELA-AQSEKNKQDGRKSLSKSL 528
+ ++K EL DD +E+ ++++ IE ++ NK+ SL K
Sbjct: 70 EEERVDVSEAEHKEETEKGELKDD--YLEKSHQIDERIEEEKGLADSNKESVDSSLRKPP 127
Query: 529 DRSAFENMQHGKHQDHSEEHLHLVKPSHEAD------LSAVEEGEVSALD--------DI 574
D E + +H++ + S ++D +EE E LD +
Sbjct: 128 DIEGRECHEQTRHEEQENNKQLVQAESDDSDDFGSRAFEEIEEQESDVLDRTSTSGAMEK 187
Query: 575 DLGPKSSNQKRKVE--SSPDNFVEGKKRHNYGSGSTHHNRIDYIEDRNKVNRLGHAERDS 632
++ + RKV+ P+ E K G T + ++ KV ++ RD
Sbjct: 188 EMTDDVGDGLRKVQGIEEPERHNEESKISEMVDGETSGH------EKKKVVKMDKKNRDV 241
Query: 633 KRHI 636
K +
Sbjct: 242 KEEV 245
>At1g56660 hypothetical protein
Length = 522
Score = 47.0 bits (110), Expect = 3e-05
Identities = 63/268 (23%), Positives = 106/268 (39%), Gaps = 45/268 (16%)
Query: 352 PVEDDVKGNLHPKQEPSAYEENMQEAQDMVRSR------SGYAKGNQHPKQEPSTYEEKM 405
P E + K + K E + E+ E +D +++ SG + + PK+E EE
Sbjct: 146 PEEKNKKADKEKKHEDVSQEKEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKKQKEESK 205
Query: 406 QEAQDMVRSRSGYGK----EQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEG 461
V+ + G+ E+E + E D E+ +K S KN
Sbjct: 206 SNEDKKVKGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKN--------------- 250
Query: 462 IKKIDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGR 521
KK ++D+ A EK+KK +K E + ED ++L+ G + + + + +G+
Sbjct: 251 -KKKEKDE-SCAEEKKKKPDKEKKEKDESTEKED-----KKLK-GKKGKGEKPEKEDEGK 302
Query: 522 KSLSKSLDRSAFENMQHGKHQDHSEEHLHLVK---PSHEADLSAVEEGEVSALDDIDLG- 577
K+ + A E + DH E K E + V E E DD D G
Sbjct: 303 ----KTKEHDATEQEMDDEAADHKEGKKKKNKDKAKKKETVIDEVCEKETKDKDD-DEGE 357
Query: 578 ---PKSSNQKRKVESSPDNFVEGKKRHN 602
K+ +++K E + E KK+ N
Sbjct: 358 TKQKKNKKKEKKSEKGEKDVKEDKKKEN 385
Score = 45.4 bits (106), Expect = 1e-04
Identities = 64/275 (23%), Positives = 110/275 (39%), Gaps = 31/275 (11%)
Query: 345 GSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEK 404
G EVK E DVK H K+ E+ +E ++ + G K N+ K E S EEK
Sbjct: 97 GDLEVK---ESDVKVEEHEKEHKKGKEKKHEELEE---EKEGKKKKNKKEKDE-SGPEEK 149
Query: 405 MQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKK 464
++A D + +E+E ++EED + N K +D + + KK
Sbjct: 150 NKKA-DKEKKHEDVSQEKEE------LEEEDGKKNKK------KEKDESGTEEKKKKPKK 196
Query: 465 IDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSL 524
+ K ++ + KK G +K E D E E ++ D E ++D +K+
Sbjct: 197 EKKQKEESKSNEDKKVKGK-KEKGEKGDLEKEDEEKKKEHD----ETDQEMKEKDSKKNK 251
Query: 525 SKSLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQ- 583
K D S E + ++ E+ K E ++G+ + D G K+
Sbjct: 252 KKEKDESCAEEKKKKPDKEKKEKDESTEK---EDKKLKGKKGKGEKPEKEDEGKKTKEHD 308
Query: 584 --KRKVESSPDNFVEGKKRHNYGSGSTHHNRIDYI 616
+++++ + EGKK+ N ID +
Sbjct: 309 ATEQEMDDEAADHKEGKKKKNKDKAKKKETVIDEV 343
Score = 44.3 bits (103), Expect = 2e-04
Identities = 52/248 (20%), Positives = 97/248 (38%), Gaps = 20/248 (8%)
Query: 363 PKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVR--SRSGYGK 420
PK+E EE+ V+ + + K++ +E + Q+M S+ K
Sbjct: 194 PKKEKKQKEESKSNEDKKVKGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKK 253
Query: 421 EQESNVGRSNIKEEDVELNDKHSS-----KNLNHRDATFASPNL--EGIKKIDRDKVKAA 473
E++ + K+ D E +K S K L + P EG K + D +
Sbjct: 254 EKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEKEDEGKKTKEHDATEQE 313
Query: 474 L--------EKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSLS 525
+ E +KK + +KK E V DE + E+ + +D E + +KNK+ +KS
Sbjct: 314 MDDEAADHKEGKKKKNKDKAKKKETVIDE-VCEKETKDKDDDEGETKQKKNKKKEKKSEK 372
Query: 526 KSLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKR 585
D + ++ + + L +P EA+ ++ E ++ G +K+
Sbjct: 373 GEKDVKEDKKKENPLETEVMSRDIKLEEP--EAEKKEEDDTEEKKKSKVEGGESEEGKKK 430
Query: 586 KVESSPDN 593
K + N
Sbjct: 431 KKKDKKKN 438
Score = 41.2 bits (95), Expect = 0.002
Identities = 53/286 (18%), Positives = 113/286 (38%), Gaps = 23/286 (8%)
Query: 361 LHPKQEPSAYEENMQ---EAQDMVRSR-----SGYAKGNQHPKQEPSTYEEKMQEAQDMV 412
L PK++ E M+ ++ + V+++ SG +K ++ K++ + +++E +D
Sbjct: 22 LDPKEKGENVEVEMEVKAKSIEKVKAKKDEESSGKSKKDKE-KKKGKNVDSEVKEDKDDD 80
Query: 413 RSRSG--YGKEQESNVGRSNIKEED--VELNDKHSSKNLNHRDATFASPNLEGIKKIDRD 468
+ + G K+ E G +KE D VE ++K K + KK ++
Sbjct: 81 KKKDGKMVSKKHEEGHGDLEVKESDVKVEEHEKEHKKGKEKKHEELEEEKEGKKKKNKKE 140
Query: 469 KVKAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKSLSKSL 528
K ++ E++ K K ++ +++ +E EDG + K K+ +
Sbjct: 141 KDESGPEEKNKKADKEKKHEDVSQEKEELEE----EDG-----KKNKKKEKDESGTEEKK 191
Query: 529 DRSAFENMQHGKHQDHSEEHLHLVKPSHE-ADLSAVEEGEVSALDDIDLGPKSSNQKRKV 587
+ E Q + + + ++ + K E DL +E + D+ D K + K+
Sbjct: 192 KKPKKEKKQKEESKSNEDKKVKGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNK 251
Query: 588 ESSPDNFVEGKKRHNYGSGSTHHNRIDYIEDRNKVNRLGHAERDSK 633
+ D +K+ + ED+ + G E+ K
Sbjct: 252 KKEKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEK 297
>At5g45520 putative protein
Length = 1167
Score = 45.1 bits (105), Expect = 1e-04
Identities = 68/280 (24%), Positives = 108/280 (38%), Gaps = 31/280 (11%)
Query: 353 VEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQ--EPSTYEEKMQEAQD 410
VE D G P QE +NM + Q V KG P++ + E + ++ +
Sbjct: 751 VEGD--GKESPPQESIDTIQNMTDDQTKVEKEGDRDKGKVDPEEGKKHDEVEGGIWKSDN 808
Query: 411 MVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKV 470
V +ES N K +D + DK K ++ NLE KK D++
Sbjct: 809 GVEGVDKASPSRESTDAIEN-KPDDHQRGDKQEEKGDGEKEKV----NLEEWKK--HDEI 861
Query: 471 KAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQD---------GR 521
K K+ TG KK+ + +D +E R +D E + EK D G+
Sbjct: 862 KEESSKQDNVTGGDVKKSPPKESKDTMESKR--DDQKENSKVQEKGDVDKGKAADLDEGK 919
Query: 522 KSLSKSLDRSAFENMQHG---KHQDHSEEHLHLVKPSHEADLSAVEE---GEVSA--LDD 573
K + S + + G K+ + + KP ++S V+E GE + L
Sbjct: 920 KENDVKAESSKSDKVIEGDEEKNPPQKSKDIIQSKPDDHREISKVQEKVDGEKNGDDLKK 979
Query: 574 IDLGPKSSNQK-RKVESSPDNFVEGKKRHNYGSGSTHHNR 612
+D G ++ QK RK N + KR N + H +
Sbjct: 980 LDGGGEAKTQKSRKTTKFGKNVSDHPKRDNTMAEDIHETQ 1019
Score = 37.0 bits (84), Expect = 0.034
Identities = 58/266 (21%), Positives = 100/266 (36%), Gaps = 36/266 (13%)
Query: 374 MQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKE 433
M+ D V++ +K + +E E+ Q + GK+ + ++K
Sbjct: 583 MEREDDQVQNYGQTSKEEKGNVEETGKQEDGDQGDGINEEANLEDGKKHDEGKEERSLKS 642
Query: 434 EDVELNDKHSSKNLNHRDATFASPNLEGIKKI------DRDKVKAALEKRKK---ATGNI 484
++V +K +S + +AT N G +K D DK KA LE+ KK
Sbjct: 643 DEVVEEEKKTSPS---EEATEKFQNKPGDQKGKSNVEGDGDKGKADLEEEKKQDEVEAEK 699
Query: 485 SKKTELVDDE---DLIERVRELEDGIELAAQSEKNKQ-----------------DGRKSL 524
SK E+V+ E D +V + DG + A ++ K+ DG++S
Sbjct: 700 SKSDEIVEGEKKPDDKSKVEKKGDGDKENADLDEGKKRDEVEAKKSESGKVVEGDGKESP 759
Query: 525 -SKSLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADLSAVEEG---EVSALDDIDLGPKS 580
+S+D K + + V P VE G + ++ +D S
Sbjct: 760 PQESIDTIQNMTDDQTKVEKEGDRDKGKVDPEEGKKHDEVEGGIWKSDNGVEGVDKASPS 819
Query: 581 SNQKRKVESSPDNFVEGKKRHNYGSG 606
+E+ PD+ G K+ G G
Sbjct: 820 RESTDAIENKPDDHQRGDKQEEKGDG 845
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 43.5 bits (101), Expect = 4e-04
Identities = 62/289 (21%), Positives = 112/289 (38%), Gaps = 27/289 (9%)
Query: 315 LETSKPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENM 374
L + N V+S N V N S +++ + L K+ +
Sbjct: 182 LSSEAKLVEANALVASVNGRSSDVENKIYSAESKLAEATRKSSELKLRLKEVETRESVLQ 241
Query: 375 QEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEE 434
QE + R Y Q ++ + +E+K+Q ++ + EQ+ N+ + K
Sbjct: 242 QERLSFTKERESYEGTFQKQREYLNEWEKKLQGKEESIT-------EQKRNLNQREEKVN 294
Query: 435 DVELNDKHSSKNL---NHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNIS---KKT 488
++E K K L N + S + E + I + + ++++ T I+ K+
Sbjct: 295 EIEKKLKLKEKELEEWNRKVDLSMSKSKETEEDITKRLEELTTKEKEAHTLQITLLAKEN 354
Query: 489 EL-VDDEDLIERV-----RELEDGIELAAQS----EKNKQDGRKSLSKSLDRSAFENMQH 538
EL +E LI R + ++D E+ E ++ RKSL K L R E +
Sbjct: 355 ELRAFEEKLIAREGTEIQKLIDDQKEVLGSKMLEFELECEEIRKSLDKELQRKIEELERQ 414
Query: 539 GKHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSNQKRKV 587
DHSEE L + V E E+ D++ K+ ++ K+
Sbjct: 415 KVEIDHSEEKLEKRNQAMNKKFDRVNEKEM----DLEAKLKTIKEREKI 459
Score = 28.9 bits (63), Expect = 9.2
Identities = 31/164 (18%), Positives = 68/164 (40%), Gaps = 4/164 (2%)
Query: 384 RSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHS 443
R K Q +E + ++ +++ + R+ + R N KE D+E K
Sbjct: 397 RKSLDKELQRKIEELERQKVEIDHSEEKLEKRN---QAMNKKFDRVNEKEMDLEAKLKTI 453
Query: 444 SKNLNHRDATFASPNLEGIKKI-DRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRE 502
+ A +LE + + D++ ++ ++ +K ++KK E++++E +++
Sbjct: 454 KEREKIIQAEEKRLSLEKQQLLSDKESLEDLQQEIEKIRAEMTKKEEMIEEECKSLEIKK 513
Query: 503 LEDGIELAAQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHSE 546
E L QSE Q + + + EN++ K + E
Sbjct: 514 EEREEYLRLQSELKSQIEKSRVHEEFLSKEVENLKQEKERFEKE 557
>At5g40450 unknown protein
Length = 2910
Score = 43.1 bits (100), Expect = 5e-04
Identities = 64/293 (21%), Positives = 119/293 (39%), Gaps = 32/293 (10%)
Query: 276 NDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSKPATSKNVFVSSANHVG 335
N VE T+ N+A +A TN + P E + A K + S H
Sbjct: 2566 NQVEETSFEFNKAQEDKKEETVDALITNVQVQ-DQPKEDFEAA--AIEKEI--SEQEHKL 2620
Query: 336 RPVSNHGVSGSTEVKHPVEDD-VKGNLHP-----KQEPSAYEENMQEAQDMVRSRSGYAK 389
+++ T VK V DD +KG+ H K+E S+ EE +E + + K
Sbjct: 2621 NDLTDVQEDIGTYVKVQVPDDEIKGDGHDSVAAQKEETSSIEEK-REVEHVKAEMEDAIK 2679
Query: 390 GNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNH 449
+++ +T E EA + G +++N+ + I+EE+ E+N + S N+
Sbjct: 2680 HEVSVEEKNNTSENIDHEAAKEIEQEEG----KQTNIVKEEIREEEKEIN-QESFNNVKE 2734
Query: 450 RDATF--ASPNLEGIKKID-----RDKVKAALE----KRKKATGNISKKTELVDDEDLIE 498
D P + I+ + +DK + E ++++ T + +E+L +
Sbjct: 2735 TDDAIDKTQPEIRDIESLSSVSKTQDKPEPEYEVPNQQKREITNEVPSLENSKIEEELQK 2794
Query: 499 RVRELEDGIELAAQSEKN----KQDGRKSLSKSLDRSAFENMQHGKHQDHSEE 547
+ E E+ +L + ++ K+ RKSLS + + +DH +
Sbjct: 2795 KDEESENTKDLFSVVKETEPTLKEPARKSLSDHIQKEPKTEEDENDDEDHEHK 2847
Score = 35.0 bits (79), Expect = 0.13
Identities = 49/238 (20%), Positives = 90/238 (37%), Gaps = 22/238 (9%)
Query: 333 HVGRPVSNHGVSGSTEVKHPVE-DDVKGNLHPKQEPSAYEENMQEAQDMV------RSRS 385
H P H V ++E PVE D L + EE+ +A +++ R S
Sbjct: 1861 HEEEPKEAHDVEATSERNLPVETSDADNTLSSQLVSETKEEHKLQAGEILPTEIIPRESS 1920
Query: 386 GYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQ------ESNVGRSNIKEEDVELN 439
A + +E + D VR + +E+ E +VG + KE + E+
Sbjct: 1921 DEALVSMLASREDDKVALQEDNCADDVRETNDIQEERSISVETEESVGETKPKEHEDEIR 1980
Query: 440 DKHSSKN-----LNHRDA-TFASPNLEGIKKIDRDKVKAALEKRKKATGNISKKTELVDD 493
D H L D+ T + +G ++I+ + AL+ ++ + + K E D
Sbjct: 1981 DAHVETPTAPIILEENDSETLIAEAKKGNEEINETERTVALDHEEEFVNHEAPKLEETKD 2040
Query: 494 E---DLIERVRELEDGIELAAQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHSEEH 548
E ++ E + E I+ ++ D SL D + ++ ++ E H
Sbjct: 2041 EKSQEIPETAKATETTIDQTLPIGTSQADQTPSLVSDKDDQTPKQVEEILEEETKETH 2098
>At5g10660 vacuolar calcium binding protein - like
Length = 407
Score = 43.1 bits (100), Expect = 5e-04
Identities = 40/196 (20%), Positives = 81/196 (40%), Gaps = 14/196 (7%)
Query: 433 EEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISKKTELVD 492
E DV ++D +D FA P+ G +K + V A+ E++K + K TE ++
Sbjct: 213 ETDVHISDHGEEPKEEDKDQ-FAQPDESGEEK-ETSPVAASTEEQKGELIDEDKSTEQIE 270
Query: 493 DEDLIERVRELEDGIELAAQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHSEEHLHLV 552
+ E + E E + + + ++ ++++ + D + N++ K ++ E +
Sbjct: 271 EPKEPENIEENNSEEEEEVKKKSDDEENSETVATTTDMNEAVNVEESKEEEKEEAEVK-- 328
Query: 553 KPSHEADLSAVEEGEVSALDDIDLGPKSSNQKRKVESSPDNFVEGKKRHNYGSGSTHHNR 612
EEGE SA + + ++ E + + V+GKK ++
Sbjct: 329 ----------EEEGESSAAKEETTETMAQVEELPEEGTKNEVVQGKKESPTAYNDVIASK 378
Query: 613 IDYIEDRNKVNRLGHA 628
+ +NKV L A
Sbjct: 379 MQENSKKNKVLALAGA 394
Score = 32.7 bits (73), Expect = 0.64
Identities = 62/280 (22%), Positives = 113/280 (40%), Gaps = 29/280 (10%)
Query: 274 QSNDVEGTTGGGNR--ATPKAPTSNDEAASTNRNLHIGGPSSTLETSKPATSKNVFVSSA 331
+S G++ GG R +T K+P +S + + G SS+ SK S NV S+
Sbjct: 131 RSTSFHGSSRGGLRGSSTVKSPPVASRGSSGVKKSGLSGNSSS--KSKKEGSGNVPKKSS 188
Query: 332 NHVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGN 391
P S+ S + + V+ + ++ E E+ Q AQ SG K
Sbjct: 189 GKEISPDSSPLASAHEDEEEIVKVETDVHISDHGEEPKEEDKDQFAQP---DESGEEK-- 243
Query: 392 QHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRD 451
++ EE+ E D +S + +E N EE+ E+ K + +
Sbjct: 244 -ETSPVAASTEEQKGELIDEDKSTEQIEEPKEPENIEENNSEEEEEVKKKSDDEENSETV 302
Query: 452 ATFASPNLEGIKKIDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAA 511
AT N + V+ + E+ K ++ E+ ++E E E+ E A
Sbjct: 303 ATTTDMN-------EAVNVEESKEEEK-------EEAEVKEEEG--ESSAAKEETTETMA 346
Query: 512 QSEKNKQDGRKS---LSKSLDRSAFENMQHGKHQDHSEEH 548
Q E+ ++G K+ K +A+ ++ K Q++S+++
Sbjct: 347 QVEELPEEGTKNEVVQGKKESPTAYNDVIASKMQENSKKN 386
>At4g37820 unknown protein
Length = 532
Score = 42.7 bits (99), Expect = 6e-04
Identities = 56/296 (18%), Positives = 119/296 (39%), Gaps = 36/296 (12%)
Query: 295 SNDEAASTNRNLHIGGPSSTLETSKPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVE 354
SN E TN L + T E+S S SS G + + +V+ E
Sbjct: 255 SNGELPETN--LSTSNATETTESSGSDESG----SSGKSTGYQQTKNEEDEKEKVQSSEE 308
Query: 355 DDV--KGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMV 412
+ + + K S+ +E+ +E + + ++G + ++EP E++ +Q+
Sbjct: 309 ESKVKESGKNEKDASSSQDESKEEKPERKKKEESSSQG-EGKEEEPEKREKEDSSSQEES 367
Query: 413 RSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKA 472
+ KE+E++ + + ++ E+ +K S + + +++ K
Sbjct: 368 KEEEPENKEKEASSSQEENEIKETEIKEKEESSSQEGNE--------------NKETEKK 413
Query: 473 ALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQSEKNKQDGRKS---LSKSLD 529
+ E ++K N KK E V+ D + E + + + N +++ SK+
Sbjct: 414 SSESQRKENTNSEKKIEQVESTDSSNTQKGDEQKTDESKRESGNDTSNKETEDDSSKTES 473
Query: 530 RSAFENMQHG---KHQDHSEEHLHLVKPSHEADLSAVEEGEVSALDDIDLGPKSSN 582
EN ++G + Q+ E+ ++ SH D+ A D++ P++SN
Sbjct: 474 EKKEENNRNGETEETQNEQEQTKSALEISHTQDVK-------DARTDLETLPETSN 522
Score = 37.0 bits (84), Expect = 0.034
Identities = 68/342 (19%), Positives = 121/342 (34%), Gaps = 56/342 (16%)
Query: 333 HVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQ 392
++GR + + +VK VED+ + S +EN E + + N+
Sbjct: 63 NLGRKDLRPRIEETKDVKDEVEDEEGSKNEGGGDVSTDKENGDEIVEREEEEKAVEENNE 122
Query: 393 HPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDA 452
K+ T E+ E + S K + + G + I E+ ++ +N++
Sbjct: 123 --KEAEGTGNEEGNEDSNNGESE----KVVDESEGGNEISNEE--------AREINYKGD 168
Query: 453 TFASPNLEGIKKIDRDKVKAALEKRKKATGNIS-------KKTELVD-----DEDLIERV 500
+S + G ++ +KV+ E + +T N+S K E+++ + L
Sbjct: 169 DASSEVMHGTEEKSNEKVEVEGESKSNSTENVSVHEDESGPKNEVLEGSVIKEVSLNTTE 228
Query: 501 RELEDGIELAAQSEKNKQDGRKSLSKS----------------------LDRSAFENMQH 538
+DG + +SE + + G K S S D S
Sbjct: 229 NGSDDGEQQETKSELDSKTGEKGFSDSNGELPETNLSTSNATETTESSGSDESGSSGKST 288
Query: 539 GKHQDHSEEHLHLVKPSHEADLSAVEEG--EVSALDDIDLGPKSSNQKRKVESSPDNFVE 596
G Q +EE S E + E G E A D + +++K E S E
Sbjct: 289 GYQQTKNEEDEKEKVQSSEEESKVKESGKNEKDASSSQDESKEEKPERKKKEESSSQ-GE 347
Query: 597 GK-----KRHNYGSGSTHHNRIDYIEDRNKVNRLGHAERDSK 633
GK KR S S ++ + E++ K E + K
Sbjct: 348 GKEEEPEKREKEDSSSQEESKEEEPENKEKEASSSQEENEIK 389
>At5g55100 unknown protein
Length = 844
Score = 41.2 bits (95), Expect = 0.002
Identities = 78/340 (22%), Positives = 129/340 (37%), Gaps = 50/340 (14%)
Query: 285 GNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSKPATSKN------------VFVSSAN 332
GN + + S D A ST ++L +S ++S+ SK +FV+
Sbjct: 498 GNDVSYPSSKSPDLAKSTGQSLSGSTAASEADSSEAGLSKEQKLKAERLKRAKMFVAKLK 557
Query: 333 HVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQ 392
+PV S S V+ P++ + G L K +A E + + S+ A
Sbjct: 558 PDAQPVQQAEPSRSISVE-PLDSGISG-LGAK---AAKERDSSSIPYVAESK--LADDGN 610
Query: 393 HPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDA 452
++ Y + Q +D + G+E+ES++ E+ + + KHS H+
Sbjct: 611 SERRSKRNYRSRSQRDED---GKMEQGEEEESSMDEVT---EETKTDKKHSCSRKRHKHK 664
Query: 453 TFASPNLEGIKKIDRDKVKA---------ALEKRKKATGNISKKTELVD--DEDLIERVR 501
T S + RDK K + + + S + EL D D + R R
Sbjct: 665 TRYSSK----DRHSRDKHKHESSSDDEYHSRSRHRHRHSKSSDRHELYDSSDNEGEHRHR 720
Query: 502 ELEDGIELAAQSEKNKQDGR-KSLSKSLDRSAFENMQHG------KHQDHSE-EHLHLVK 553
+ ++ +K R + K D S E+ H KH+D S+ EH H K
Sbjct: 721 SSKHSKDVDYSKDKRSHHHRSRKHEKHRDSSDDEHHHHRHRSSRRKHEDSSDVEHGHRHK 780
Query: 554 PSH--EADLSAVEEGEVSALDDIDLGPKSSNQKRKVESSP 591
S + D VEE VS D DL + +++ P
Sbjct: 781 SSKRIKKDEKTVEEETVSKSDQSDLKASPGDNIPYLQNEP 820
Score = 31.2 bits (69), Expect = 1.9
Identities = 80/405 (19%), Positives = 159/405 (38%), Gaps = 65/405 (16%)
Query: 271 RMAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSKPATSKNVFVSS 330
++ SN EG+ G A+ + ND A+ RN + + +E + T K V +
Sbjct: 261 KVTDSNGPEGSKGAAKIASKHSLPLNDHASFIKRNPSVSA-VNVVEKKQINTEKLV---T 316
Query: 331 ANHVGRPVSNHGVSGSTEVKHPVE-------------------DDVKGNLHPKQEPSA-- 369
++ + V +TE+K ++ DVK + P PS+
Sbjct: 317 SDKSQPKLELQIVEPTTEMKRVIDKIVDFIQKNGKELEATLVAQDVKYGMFPFLRPSSLY 376
Query: 370 ---YEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNV 426
Y + +QEA+++ G + KQ EKM A + S+ G+G S +
Sbjct: 377 HAYYRKVLQEAEELKSGDKGVIIRKEDVKQ------EKMGNA--VKDSKHGFG----SVL 424
Query: 427 GRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISK 486
+ K+E +++ L++ P + +D + A L+ ++ N
Sbjct: 425 PDDSAKKEKLKMVSDKPKVELHNEPFKPVQPQMR--VNVDANTAAAILQAARRGIRN--P 480
Query: 487 KTELVDDEDLIERVRELEDGIEL-AAQSEKNKQDGRKSLSKSLDRSAFENMQHG--KHQD 543
+ ++ + + E + L + + +++S + +SLS S S ++ + G K Q
Sbjct: 481 QLGILTGKPMDETSQTLGNDVSYPSSKSPDLAKSTGQSLSGSTAASEADSSEAGLSKEQK 540
Query: 544 HSEEHLHLVK---PSHEADLSAVEEGEVS---ALDDIDLGPKSSNQK--RKVESSPDNFV 595
E L K + D V++ E S +++ +D G K ++ +SS +V
Sbjct: 541 LKAERLKRAKMFVAKLKPDAQPVQQAEPSRSISVEPLDSGISGLGAKAAKERDSSSIPYV 600
Query: 596 EGKKRHNYGSGSTHHNRIDYIEDRNKVNRLGHAERDSKRHIQENQ 640
K + G+ E R+K N ++RD +++ +
Sbjct: 601 AESKLADDGNS----------ERRSKRNYRSRSQRDEDGKMEQGE 635
>At3g23900 unknown protein
Length = 989
Score = 40.4 bits (93), Expect = 0.003
Identities = 83/364 (22%), Positives = 133/364 (35%), Gaps = 58/364 (15%)
Query: 239 SDGEKVWWQEFDVTPRQLEEVSNQMLELYEQNRMAQSNDVEGTTGGGNRATPKAPTSNDE 298
S GEK + +PR+ +EV + R + N V+ T +R+ + D+
Sbjct: 619 SRGEKKSSRAGSRSPRRRKEVKSTP-------RDDEENKVKRRTRSRSRSVEDSADIKDK 671
Query: 299 AASTNRNLHIGGPSSTLETSKPAT---SKNVFVSSANHVGRPVSN--HGVSGSTEVKHPV 353
+ H S + T S+N + H R S +GS E
Sbjct: 672 SRDEELKHHKKRSRSRSREDRSKTRDTSRNSDEAKQKHRQRSRSRSLENDNGSHENVDVA 731
Query: 354 EDDVKGNLHPKQEPSAYEEN--MQEAQDMVRSRSGYAKGNQHPK----QEPSTYEEKMQE 407
+D+ + H K+ + +E+ M+E + RSRS K K ++ +T + +
Sbjct: 732 QDNDLNSRHSKRRSKSLDEDYDMKERRGRSRSRSLETKNRSSRKNKLDEDRNTGSRRRRS 791
Query: 408 AQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDR 467
V + Y KE S +S K + SS+ RD +SP KK
Sbjct: 792 RSKSVEGKRSYNKETRSRDKKS--KRRSGRRSRSPSSEGKQGRDIR-SSPGYSDEKK--- 845
Query: 468 DKVKAALEKRKKATGNISKKTELVDDEDLI-ERVRELEDGIELAAQSEKNKQDGRKSLSK 526
+ KR + +I KK D ER+R G ++K+ G +SLS
Sbjct: 846 -----SRHKRHSRSRSIEKKNSSRDKRSKRHERLRSSSPG--------RDKRRGDRSLSP 892
Query: 527 SLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADLSAVEE--GEVSALDDIDLGPKSSNQK 584
SE+H +K H S E+ + +DD D SS Q+
Sbjct: 893 V----------------SSEDHK--IKKRHSGSKSVKEKPHSDYEKVDDGDANSDSSQQE 934
Query: 585 RKVE 588
R +E
Sbjct: 935 RNLE 938
Score = 30.8 bits (68), Expect = 2.4
Identities = 35/145 (24%), Positives = 63/145 (43%), Gaps = 17/145 (11%)
Query: 509 LAAQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDH---SEEHLHLVKPSHEADLSAVEE 565
L Q + R+S S D S E+ ++G+ +H S V P S E+
Sbjct: 552 LRYQRRSTYEGRRRSYRDSRDIS--ESRRYGRSDEHHSSSSRRSRSVSPKKRK--SGQED 607
Query: 566 GEVSAL--DDIDLGPKSSNQ--------KRKVESSPDNFVEGKKRHNYGSGSTHHNRIDY 615
E+S L D G K S++ +++V+S+P + E K + S S
Sbjct: 608 SELSRLRRDSSSRGEKKSSRAGSRSPRRRKEVKSTPRDDEENKVKRRTRSRSRSVEDSAD 667
Query: 616 IEDRNKVNRLGHAERDSKRHIQENQ 640
I+D+++ L H ++ S+ +E++
Sbjct: 668 IKDKSRDEELKHHKKRSRSRSREDR 692
Score = 30.4 bits (67), Expect = 3.2
Identities = 43/196 (21%), Positives = 76/196 (37%), Gaps = 15/196 (7%)
Query: 350 KHPVEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQEPSTYEEKMQEAQ 409
K+ +++D ++ S E + RSR +K + + E K + +
Sbjct: 775 KNKLDEDRNTGSRRRRSRSKSVEGKRSYNKETRSRDKKSKRRSGRRSRSPSSEGK--QGR 832
Query: 410 DMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDK 469
D +RS GY E++S R + + N ++ H +SP + ++ DR
Sbjct: 833 D-IRSSPGYSDEKKSRHKRHSRSRSIEKKNSSRDKRSKRHERLRSSSPGRDK-RRGDRSL 890
Query: 470 VKAALEK---RKKATGNISKKT------ELVDDEDLIERVRELEDGIE--LAAQSEKNKQ 518
+ E +K+ +G+ S K E VDD D + E +E L + + Q
Sbjct: 891 SPVSSEDHKIKKRHSGSKSVKEKPHSDYEKVDDGDANSDSSQQERNLEGHLLSLDSMSSQ 950
Query: 519 DGRKSLSKSLDRSAFE 534
D KS S+ E
Sbjct: 951 DVEKSKENPPSSSSVE 966
>At2g02200 putative protein on transposon FARE2.10 (cds2)
Length = 586
Score = 40.4 bits (93), Expect = 0.003
Identities = 53/246 (21%), Positives = 101/246 (40%), Gaps = 36/246 (14%)
Query: 347 TEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQDMVRSRSGYAKGNQHPKQ--------EP 398
T + ED+ K KQE EE +++ + R G K + PKQ E
Sbjct: 202 TREEQKEEDEKKEQEEEKQEEEGKEEKLEKIE--YRGDEGTEK-QEIPKQGDEEMEGEEE 258
Query: 399 STYEEKMQEAQDMVRSRSGYG-----------------KEQESNVGRSNIKEEDVELNDK 441
EE +E ++ V R G +E++ G+ +EE VE D
Sbjct: 259 KQEEEGKEEEEEKVEYRGDEGTEKQEIPKQGDEEMEGEEEKQKEEGKEE-EEEKVEYRDH 317
Query: 442 HSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVR 501
HS+ N+ + +P +G ++++R++ K E++ N+ + E + ED V
Sbjct: 318 HSTCNVEETEKQ-ENPK-QGDEEMEREEGK---EEKVLKEENVEEHDEHDETEDQEAYVI 372
Query: 502 ELEDGIELAAQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHSEEHLHLVKPSHEADLS 561
+D A +EK + ++ ++ E +H +H + ++ +++ E + +
Sbjct: 373 LSDDEDNGTAPTEKESESQKEETTEVAKEENVE--EHDEHDETEDQEAYVILSDDEDNGT 430
Query: 562 AVEEGE 567
A E E
Sbjct: 431 APTEKE 436
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,988,982
Number of Sequences: 26719
Number of extensions: 685587
Number of successful extensions: 2632
Number of sequences better than 10.0: 199
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 172
Number of HSP's that attempted gapping in prelim test: 2304
Number of HSP's gapped (non-prelim): 346
length of query: 641
length of database: 11,318,596
effective HSP length: 106
effective length of query: 535
effective length of database: 8,486,382
effective search space: 4540214370
effective search space used: 4540214370
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0176a.14


