

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.1
(440 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
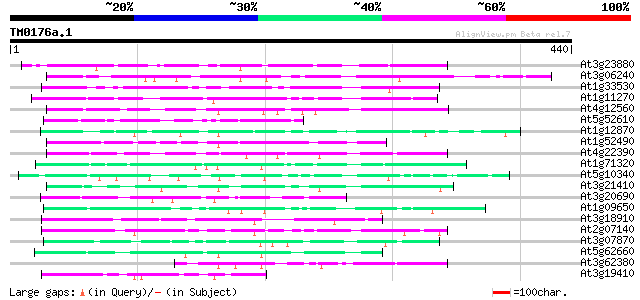
Score E
Sequences producing significant alignments: (bits) Value
At3g23880 unknown protein 104 8e-23
At3g06240 unknown protein 91 1e-18
At1g33530 hypothetical protein 79 5e-15
At1g11270 unknown protein (At1g11270) 77 2e-14
At4g12560 putative protein 73 4e-13
At5g52610 putative protein 69 4e-12
At1g12870 hypothetical protein 65 6e-11
At1g52490 F6D8.29 65 1e-10
At4g22390 unknown protein 62 6e-10
At1g71320 hypothetical protein 61 1e-09
At5g10340 putative protein 59 7e-09
At3g21410 hypothetical protein 58 1e-08
At3g20690 hypothetical protein 57 2e-08
At1g09650 hypothetical protein 57 2e-08
At3g18910 unknown protein 56 5e-08
At2g07140 pseudogene 56 5e-08
At3g07870 unknown protein 55 6e-08
At5g62660 putative protein 55 1e-07
At3g62380 putative protein 55 1e-07
At3g19410 hypothetical protein 53 3e-07
>At3g23880 unknown protein
Length = 364
Score = 104 bits (260), Expect = 8e-23
Identities = 97/341 (28%), Positives = 150/341 (43%), Gaps = 50/341 (14%)
Query: 10 TVLSMNTPHPDNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIK--- 66
T M +PH NLP +++ E+L LPVKSLTR KCVC SW SLI +K
Sbjct: 4 TTREMFSPH--NLPL-----EMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHAL 56
Query: 67 -LHLHRSALHGK-PHLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKK 124
L ++ K P+ ++ S + + C+ L +S V H+ L
Sbjct: 57 ILETSKATTSTKSPYGVI--TTSRYHLKSCCIHSLYNASTVYVSE------HDG--ELLG 106
Query: 125 GDRYHVVGCCNGLICLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFD--VKL 182
D Y VVG C+GL+C H S++ WNP +L ++L ++T + + V
Sbjct: 107 RDYYQVVGTCHGLVCFHVDYDKSLY------LWNPTIKL-QQRLSSSDLETSDDECVVTY 159
Query: 183 SFGYDNSTDTYKVVFFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVN 242
FGYD S D YKVV +++ V + T WR +FP D
Sbjct: 160 GFGYDESEDDYKVVAL----LQQRHQVKIETKIYSTRQKLWRSNTSFPSGVVVADK--SR 213
Query: 243 DGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVV 302
G++++GT+NW + S+ I+S D+ + + L P
Sbjct: 214 SGIYINGTLNWAATSSSSS------------WTIISYDMSRDEFKELPGPVCCGR-GCFT 260
Query: 303 PSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKV 343
++G L CL +G + +W MK+FG+ SW++LL +
Sbjct: 261 MTLGDLRGCLSMVCYCKGANADVWVMKEFGEVYSWSKLLSI 301
>At3g06240 unknown protein
Length = 427
Score = 91.3 bits (225), Expect = 1e-18
Identities = 110/441 (24%), Positives = 181/441 (40%), Gaps = 104/441 (23%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
++I E+L LP KS+ R +CV K + +L SD F K+HL L+L+ E+
Sbjct: 39 EIITEILLRLPAKSIGRFRCVSKLFCTLS-SDPGFAKIHLD---------LILRNESVRS 88
Query: 90 DHREFCVTHLSGSSLS-------------------LVENPWI-----------SLAHNPR 119
HR+ V+ + SL L ++P I L + R
Sbjct: 89 LHRKLIVSSHNLYSLDFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGDHLYDDRR 148
Query: 120 YRLKKGDRYH------VVGCCNGLICLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCV 173
LK + + +VG NGL+C+ +G + +NP T F
Sbjct: 149 VMLKLNAKSYRRNWVEIVGSSNGLVCISP-------GEGAVFLYNPTTGDSKRLPENFRP 201
Query: 174 KTKNFD----VKLSFGYDNSTDTYKVVFFEI--EKIRRASLVSVFTLDGCNGWNGWRDIQ 227
K+ ++ FG+D TD YK+V E I AS+ S+ + WR I
Sbjct: 202 KSVEYERDNFQTYGFGFDGLTDDYKLVKLVATSEDILDASVYSLKA-------DSWRRIC 254
Query: 228 NFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYT 287
N L + ++D GVH +G I+W+ + + Q +++ D++TE +
Sbjct: 255 N---LNYEHNDGSYTSGVHFNGAIHWVFTESRH-----------NQRVVVAFDIQTEEFR 300
Query: 288 LLSPPKGLDEVPSVVPS--VGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQL-LKVR 344
+ P ++ + VG L LC + H IW M ++G+AKSW+++ + +
Sbjct: 301 EMPVPDEAEDCSHRFSNFVVGSLNGRLCVVNSCYDVHDDIWVMSEYGEAKSWSRIRINLL 360
Query: 345 YQDLQREPLHSMFYCHYQLFPLCLHDGDTLILASYSDRYDGNEAILYDKRANTAERTALE 404
Y+ ++ PLC D +L DG + +LY+ N + +
Sbjct: 361 YRSMK---------------PLCSTKNDEEVLL----ELDG-DLVLYNFETNASSNLGI- 399
Query: 405 TESKYILGFSSFYYVESLVSP 425
K GF + YVESL+SP
Sbjct: 400 CGVKLSDGFEANTYVESLISP 420
>At1g33530 hypothetical protein
Length = 441
Score = 79.0 bits (193), Expect = 5e-15
Identities = 85/316 (26%), Positives = 136/316 (42%), Gaps = 51/316 (16%)
Query: 26 QLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRE 85
+L + L+ E+L LPVK L RLK + K W SLI SD HL L LL+++
Sbjct: 96 ELPDVLVEEILQRLPVKYLVRLKSISKGWKSLIESD------HLAEKHLR-----LLEKK 144
Query: 86 NSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSV 145
+ +E +T +S S+ + + D V G CNGL+C+ Y +
Sbjct: 145 ---YGLKEIKITVERSTSKSICIKFFSRRSGMNAINSDSDDLLRVPGSCNGLVCV--YEL 199
Query: 146 DSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKV-VFFEIEKIR 204
DS++ + NP T + + + + FG D T TYKV V + +++
Sbjct: 200 DSVY----IYLLNPMTGVTRT-----LTPPRGTKLSVGFGIDVVTGTYKVMVLYGFDRVG 250
Query: 205 RASLVSVFTLDGCNGWNGWRD-IQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPY 263
VF LD N WR + +P S + V ++G++ W+ + D S
Sbjct: 251 TV----VFDLD----TNKWRQRYKTAGPMPLSCIPTPERNPVFVNGSLFWLLASDFSE-- 300
Query: 264 YNPVTITVEQLGILSLDLRTETYTLLSPPKGLD--EVPSVVPSVGVLMNCLCFSHDLEGT 321
IL +DL TE + LS P +D +V S + L + LC S+ +G
Sbjct: 301 ------------ILVMDLHTEKFRTLSQPNDMDDVDVSSGYIYMWSLEDRLCVSNVRQGL 348
Query: 322 HFVIWQMKKFGDAKSW 337
H +W + + ++ W
Sbjct: 349 HSYVWVLVQDELSEKW 364
>At1g11270 unknown protein (At1g11270)
Length = 312
Score = 76.6 bits (187), Expect = 2e-14
Identities = 88/324 (27%), Positives = 137/324 (42%), Gaps = 44/324 (13%)
Query: 18 HPDNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGK 77
H ++ + L +D++ +L LPV+SL R KCV W S I S Q F + L R
Sbjct: 26 HCSSVVKLLLPHDVVGLILERLPVESLLRFKCVSNQWKSTIES-QCFQERQLIRRMESRG 84
Query: 78 PHLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGL 137
P +L+ + D ++ + GSS+ +R G C GL
Sbjct: 85 PDVLV--VSFADDEDKYGRKAVFGSSIV------------STFRFPTLHTLICYGSCEGL 130
Query: 138 ICLH-GYSVDSIFQQGWLSFWN---PATRLMSEKLGCFCVKTKNFDV-KLSFGYDNSTDT 192
IC++ YS + + + W+ P + L F K +F KL+FG D T
Sbjct: 131 ICIYCVYSPNIVVNPA--TKWHRSCPLSNLQQFLDDKFEKKEYDFPTPKLAFGKDKLNGT 188
Query: 193 YKVVF-FEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTI 251
YK V+ + + R + + D N N WR + P P+ +D+ D V+ G++
Sbjct: 189 YKQVWLYNSSEFRLDDVTTCEVFDFSN--NAWRYVH--PASPYRINDY--QDPVYSDGSV 242
Query: 252 NWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNC 311
+W+T +S ILS L TET+ +L L E V S+ +L N
Sbjct: 243 HWLTEGKESK--------------ILSFHLHTETFQVLCEAPFLRERDPVGDSMCILDNR 288
Query: 312 LCFSHDLEGTHFVIWQMKKFGDAK 335
LC S ++ G +IW + G K
Sbjct: 289 LCVS-EINGPAQLIWSLDSSGGNK 311
>At4g12560 putative protein
Length = 408
Score = 72.8 bits (177), Expect = 4e-13
Identities = 87/334 (26%), Positives = 142/334 (42%), Gaps = 55/334 (16%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
D++ ++ LP K+L R + + K LI +D FI+ HLHR G ++L R
Sbjct: 7 DIVNDIFLRLPAKTLVRCRALSKPCYHLI-NDPDFIESHLHRVLQTGDHLMILLRGAL-- 63
Query: 90 DHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSIF 149
R + V S S+S VE+P +K+G V G NGLI L D
Sbjct: 64 --RLYSVDLDSLDSVSDVEHP-----------MKRGGPTEVFGSSNGLIGLSNSPTD--- 107
Query: 150 QQGWLSFWNPATR----LMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVF---FEIEK 202
L+ +NP+TR L + + V GYD+ +D YKVV F+I+
Sbjct: 108 ----LAVFNPSTRQIHRLPPSSIDLPDGSSTRGYVFYGLGYDSVSDDYKVVRMVQFKIDS 163
Query: 203 IRRASL-----VSVFTLDGCNGWNGWRDIQN----FPLLPFSYDD--WGVNDGVHLSGTI 251
V VF+L N W+ I++ LL + Y + GV ++
Sbjct: 164 EDELGCSFPYEVKVFSLKK----NSWKRIESVASSIQLLFYFYYHLLYRRGYGVLAGNSL 219
Query: 252 NWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGL-DEVPSVVPSVGVLMN 310
+W+ R +N I+ DL E + ++ P+ + + + +GVL
Sbjct: 220 HWVLPRRPGLIAFNL---------IVRFDLALEEFEIVRFPEAVANGNVDIQMDIGVLDG 270
Query: 311 CLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVR 344
CLC + + ++ +W MK++ SWT++ V+
Sbjct: 271 CLCLMCNYDQSYVDVWMMKEYNVRDSWTKVFTVQ 304
>At5g52610 putative protein
Length = 351
Score = 69.3 bits (168), Expect = 4e-12
Identities = 67/205 (32%), Positives = 92/205 (44%), Gaps = 29/205 (14%)
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQREN 86
+ DL+ E+L LPVK L R CVCK W + II + FI L+ RS+ +P+++
Sbjct: 2 ISEDLLVEILLRLPVKPLARCLCVCKLW-ATIIRSRYFINLYQSRSSTR-QPYVMF---- 55
Query: 87 SHHDHREFCVTH-LSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSV 145
+ D C H S S SLV S ++ H C NGLIC+ S
Sbjct: 56 ALRDIFTSCRWHFFSSSQPSLVTKATCSANNSS----------HTPDCVNGLICVEYMS- 104
Query: 146 DSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKIRR 205
Q W+S NPATR L + F K GYD YKV+FF + +
Sbjct: 105 -----QLWIS--NPATR--KGVLVPQSAPHQKFR-KWYMGYDPINYQYKVLFFSKQYLLS 154
Query: 206 ASLVSVFTLDGCNGWNGWRDIQNFP 230
+ VFTL+G W +++N P
Sbjct: 155 PYKLEVFTLEGQGSWK-MIEVENIP 178
>At1g12870 hypothetical protein
Length = 416
Score = 65.5 bits (158), Expect = 6e-11
Identities = 92/405 (22%), Positives = 153/405 (37%), Gaps = 88/405 (21%)
Query: 25 TQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQR 84
+ L +D++ E+ LPVK+L R K + K W S + S + HL +
Sbjct: 31 SSLPDDVVEEIFLKLPVKALMRFKSLSKQWRSTL------------ESCYFSQRHLKI-A 77
Query: 85 ENSHHDHREFCV--------THLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVG--CC 134
E SH DH + + +S ++SL +S + + +G + + C
Sbjct: 78 ERSHVDHPKVMIITEKWNPDIEISFRTISLES---VSFLSSALFNFPRGFHHPIYASESC 134
Query: 135 NGLICLHGYSVDSIFQQGWLSFWNPATR-----------LMSEKLGCFCVKTKNFDVKLS 183
+G+ C+H I+ NPATR + KL ++
Sbjct: 135 DGIFCIHSPKTQDIY------VVNPATRWFRQLPPARFQIFMHKLNPTLDTLRDMIPVNH 188
Query: 184 FGYDNSTDTYKVVFFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVND 243
+ +TD YK+V+ R + VF N WR + P SY +
Sbjct: 189 LAFVKATD-YKLVWLYNSDASRVTKCEVFDFKA----NAWRYLTCIP----SYRIYHDQK 239
Query: 244 GVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVP 303
+GT+ W T YN ++ +++LD+ TE + LL P +
Sbjct: 240 PASANGTLYWFTET------YN------AEIKVIALDIHTEIFRLLPKPSLIASSEPSHI 287
Query: 304 SVGVLMNCLCFSHDLEGTHFV---IWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCH 360
+ ++ N LC ++ EG + IW++K DA W ++ + L S YCH
Sbjct: 288 DMCIIDNSLCM-YETEGDKKIIQEIWRLKSSEDA--WEKIYTIN--------LLSSSYCH 336
Query: 361 YQLFPLCLHDGDTLILASYSDRYDGNE-----AILYDKRANTAER 400
+ + DG L Y + D E AI DK+ + R
Sbjct: 337 FHVL-----DGYNLTRLCYWTQKDLVESSTPVAIYKDKKIILSHR 376
>At1g52490 F6D8.29
Length = 423
Score = 64.7 bits (156), Expect = 1e-10
Identities = 74/269 (27%), Positives = 109/269 (40%), Gaps = 32/269 (11%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
DLI E+L LP KSL R +CV K W+ +I + F++ + RS L PH L
Sbjct: 58 DLIVEILKKLPTKSLMRFRCVSKPWSFIISKRRDFVESIMARS-LRQPPHKLPVFIFHQC 116
Query: 90 DHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSIF 149
D F S+ S P +S+ P RY V G IC D +
Sbjct: 117 DPGTFFTV---SSTFSQSTKPKVSIM--PGRNHYNAFRYQYV---RGFICCSSSVYDLV- 167
Query: 150 QQGWLSFWNPATR--LMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKIRRAS 207
+ +NP TR L K+ + K FGYD+ + YKV+ +
Sbjct: 168 -----TIYNPTTRQCLPLPKIESMVLSPKRHK-HCYFGYDHVMNEYKVLAMVNDSQELTQ 221
Query: 208 LVSVFTLD-GCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPYYNP 266
VFTL C W R ++ L+ S GV + GTI ++ R K Y
Sbjct: 222 TFHVFTLGRDCPQWRKIRGNIDYELISVS------RAGVCIDGTIYYVAVRRKDNENYG- 274
Query: 267 VTITVEQLGILSLDLRTETYTLLSPPKGL 295
+L ++S D+++E + + P+ L
Sbjct: 275 ------ELFMMSFDVKSERFYHVRTPETL 297
>At4g22390 unknown protein
Length = 401
Score = 62.0 bits (149), Expect = 6e-10
Identities = 86/330 (26%), Positives = 141/330 (42%), Gaps = 53/330 (16%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
DLI E+ L +L + + + K SLI S + F+ HL R G+ ++L R
Sbjct: 7 DLINEMFLRLRATTLVKCRVLSKPCFSLIDSPE-FVSSHLRRRLETGEHLMILLR----- 60
Query: 90 DHREFCVTHL-SGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSI 148
R L S ++S + +P L+ G V G NG+I L VD
Sbjct: 61 GPRLLRTVELDSPENVSDIPHP-----------LQAGGFTEVFGSFNGVIGLCNSPVD-- 107
Query: 149 FQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSF-----GYDNSTDTYKVVFFEIEKI 203
L+ +NP+TR + +L + D+ + GYD+ D +KVV K+
Sbjct: 108 -----LAIFNPSTRKI-HRLPIEPIDFPERDITREYVFYGLGYDSVGDDFKVVRIVQCKL 161
Query: 204 RRASL-------VSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGV---NDGVHLSGTINW 253
+ V VF+L N W + F +L SY + GV ++ ++W
Sbjct: 162 KEGKKKFPCPVEVKVFSLKK-NSWKRVCLMFEFQILWISYYYHLLPRRGYGVVVNNHLHW 220
Query: 254 MTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCLC 313
+ R + +N I+ DL ++ +LS P+ L ++ +GVL C+C
Sbjct: 221 ILPRRQGVIAFN---------AIIKYDLASDDIGVLSFPQELYIEDNM--DIGVLDGCVC 269
Query: 314 FSHDLEGTHFVIWQMKKFGDAKSWTQLLKV 343
E +H +W +K++ D KSWT+L +V
Sbjct: 270 LMCYDEYSHVDVWVLKEYEDYKSWTKLYRV 299
>At1g71320 hypothetical protein
Length = 392
Score = 60.8 bits (146), Expect = 1e-09
Identities = 88/367 (23%), Positives = 147/367 (39%), Gaps = 51/367 (13%)
Query: 21 NLPRTQLHNDLIAE-VLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPH 79
N P+T D IAE + LP+KSL R K + K W S+I S F L R+ L P+
Sbjct: 7 NNPKTIFIPDDIAEGIFHHLPIKSLARFKVLSKKWTSMIESTY-FSHKRLIRTGLP-TPN 64
Query: 80 LLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPR-YRLKKGDRYHVVGCCNGLI 138
+ + H V S S L+E +N + + + V+G C+GL+
Sbjct: 65 MKFLHISQHFTAN--FVEEYSNSITFLLETFSRDDQNNRKTFDESQNKTIQVLGSCDGLV 122
Query: 139 CLHGYS-----------------VDSIFQQGW---LSFWNPAT-----RLMSEKLGCFCV 173
L + + F Q W LS+ PA R +S+ + + +
Sbjct: 123 LLRIHDDFRSIYLINPTTKEHMKLSPEFMQ-WPFTLSYLTPAMANKPWRQLSQSVLDYDI 181
Query: 174 KTKNFDVKLSFGYDNSTDTYKVV--FFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPL 231
K FG D +YKVV + E V +LD NG RD+ + +
Sbjct: 182 SVKRMPSLAGFGKDIVKKSYKVVLIYTRYEIHDHEFKAKVLSLD--NGEQ--RDVGFYDI 237
Query: 232 LPFSYDDWGVNDGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSP 291
+ D V+ +G++ W+T + S Y +L++DL TE + +
Sbjct: 238 HHCIFCD--EQTSVYANGSLFWLTLKKLSQTSYQ----------LLAIDLHTEEFRWILL 285
Query: 292 PKGLDEVPSVVPSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQRE 351
P+ D + + L LC S LE ++ V+W + + + W ++ ++ ++
Sbjct: 286 PE-CDTKYATNIEMWNLNERLCLSDVLESSNLVVWSLHQEYPTEKWEKIYSIKIDVIRTN 344
Query: 352 PLHSMFY 358
LH F+
Sbjct: 345 QLHEKFW 351
>At5g10340 putative protein
Length = 445
Score = 58.5 bits (140), Expect = 7e-09
Identities = 109/423 (25%), Positives = 171/423 (39%), Gaps = 99/423 (23%)
Query: 8 TVTVLSMNTPHPDNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKL 67
++T+ T + LP H+ + ++ L VK+L + K V K W S I S +F +
Sbjct: 55 SITLRDSETSMEELLP----HDVIEYHIMVRLDVKTLLKFKSVSKQWMSTIQSP-SFQER 109
Query: 68 HL--HRSALHGKPHLLL--------QRENSHHDHREFCVTHLSGSSLSLVE--NPWISLA 115
L H S G PH+LL ++++ E T L SS + V+ PW
Sbjct: 110 QLIHHLSQSSGDPHVLLVSLYDPCARQQDPSISSFEALRTFLVESSAASVQIPTPW---- 165
Query: 116 HNPRYRLKKGDRYHVV--GCCNGLICLHG-YSVDSIFQQGWLSFWNPATRL--------- 163
D+ + V C+GLICL Y + SI NP TR
Sbjct: 166 ---------EDKLYFVCNTSCDGLICLFSFYELPSIVV-------NPTTRWHRTFPKCNY 209
Query: 164 ------MSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFF----EIEKIRRASLVSVFT 213
E+ CF V FG D + TYK V+ E++ + + VF
Sbjct: 210 QLVAADKGERHECFKVACPT----PGFGKDKISGTYKPVWLYNSAELDLNDKPTTCEVFD 265
Query: 214 LDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPYYNPVTITVEQ 273
N WR + FP P D V++ G+++W T+ +
Sbjct: 266 F----ATNAWRYV--FPASPHLI--LHTQDPVYVDGSLHWFTALSHEG-----------E 306
Query: 274 LGILSLDLRTETYTLLSPPKGL---DEVPSVVPSVGVLMNCLCFSHDLEGTHFVIWQMKK 330
+LSLDL +ET+ ++S L DE V+ ++G + LC S + + + VIW +
Sbjct: 307 TMVLSLDLHSETFQVISKAPFLNVSDEYYIVMCNLG---DRLCVS-EQKWPNQVIWSLDD 362
Query: 331 FGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQLF-PLCLHDGDTLILASYSDRYDGNEAI 389
D K+W Q+ + DL S+F+ F PL + D D L+ + D G+ +
Sbjct: 363 -SDHKTWKQIYSI---DL--IITSSLFFSAIFAFTPLAVLDKDKLL---FYDSTHGDAFL 413
Query: 390 LYD 392
+D
Sbjct: 414 THD 416
>At3g21410 hypothetical protein
Length = 374
Score = 57.8 bits (138), Expect = 1e-08
Identities = 82/336 (24%), Positives = 124/336 (36%), Gaps = 79/336 (23%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
+L E+L +P KSL RLK CK W +L +D+ FI HL L RE+
Sbjct: 14 ELFEEILCRVPTKSLLRLKLTCKRWLAL-FNDKRFI-----------YKHLALVREHI-- 59
Query: 90 DHREFCVTHLSGSSLSLVENPWISLAHN---PRYRLKKGDRYHVVGCCNGLICLHGYSVD 146
+ + + + NP + + P KG+ Y +V C L+C
Sbjct: 60 ---------IRTNQMVKIINPVVGACSSFSLPNKFQVKGEIYTMVPCDGLLLC------- 103
Query: 147 SIFQQGWLSFWNPATR-------LMSEKLGCFCVKTKNFDVKLSFGYDN-STDTYKVVFF 198
IF+ G ++ WNP L GC C GYD S D+YK++ F
Sbjct: 104 -IFETGSMAVWNPCLNQVRWIFLLNPSFRGCSC---------YGIGYDGLSRDSYKILRF 153
Query: 199 EIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGV---NDGVHLSGTINWMT 255
V ++ L N W+ + DW V G+ L G + W+
Sbjct: 154 YANTGSYKPEVDIYELKS----NSWKTFK-------VSLDWHVVLRCKGLSLKGNMYWIA 202
Query: 256 SRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCLCFS 315
++ + I S + TET+ L +V +VV + L
Sbjct: 203 KWNRKPDIF-----------IQSFNFSTETFEPLCSLPVRYDVHNVVALSAFKGDNLSLL 251
Query: 316 HDLEGTHFV-IWQMKKFGDAKS--WTQLLKVRYQDL 348
H + T + +W K + S WT+L V DL
Sbjct: 252 HQSKETSKIDVWVTNKVKNGVSILWTKLFSVTRPDL 287
>At3g20690 hypothetical protein
Length = 370
Score = 57.4 bits (137), Expect = 2e-08
Identities = 66/251 (26%), Positives = 110/251 (43%), Gaps = 41/251 (16%)
Query: 25 TQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQR 84
+ L +DL+ E+LS LP+ SL ++ CK+WN ++ ++F H+ GK L++ +
Sbjct: 4 SDLPHDLVEEILSRLPLISLKAMRSTCKTWN-VLSKHRSFANKHIGNVTASGKRDLIMIK 62
Query: 85 ENSHHDHREFCVTHLSGSSLSLVENP----WISLAHNPRYRLKKGD----RYHVVGCCNG 136
+ C + G +L ++N +S+ + L+ D V CNG
Sbjct: 63 D---------CKVYSIGVNLHGIQNNNNIIDLSIKNKGILHLENLDLIFKEIFNVFHCNG 113
Query: 137 LICLHGYSVDSIFQQGWLSFWNP--ATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYK 194
L+ L+G D + L NP R +++ F +FD KL+FGYD S +K
Sbjct: 114 LLLLYGSITDDSIR---LVVCNPYWGKREWVKRINNFA----SFD-KLAFGYDKSCGCHK 165
Query: 195 VV-FFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINW 253
++ F+ R L+ N + R + +FPL +D + DGV L G W
Sbjct: 166 ILRVFDSYPNR---------LEIYNLRSNSRMVSSFPL---KWDIAFMQDGVSLKGNTYW 213
Query: 254 MTSRDKSAPYY 264
+S Y
Sbjct: 214 YAKDRRSEDCY 224
>At1g09650 hypothetical protein
Length = 382
Score = 57.0 bits (136), Expect = 2e-08
Identities = 95/384 (24%), Positives = 147/384 (37%), Gaps = 77/384 (20%)
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLL-QRE 85
L ++++ +L L L R K V K W S I S F + + G P +L+ R
Sbjct: 13 LPHEVVECILERLDADPLLRFKAVSKQWKSTIESP-FFQRRQFTQRQQSGNPDVLMVSRC 71
Query: 86 NSHHDHREFCVTHLSGSSLSL-VENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYS 144
+ E T + GSS S+ + PW + D C+GL+CL
Sbjct: 72 ADINSEIEALTTLVLGSSSSVKIPTPWEEEEEEDKEYSVSQDS------CDGLVCLFDK- 124
Query: 145 VDSIFQQGWLSFWNPATRLMSEKLGC------FCVKTKNFDV-----KLSFGYDNSTDTY 193
F+ G++ +NPATR C + +D+ +L FG D T TY
Sbjct: 125 ----FKSGFV--FNPATRWYHPLPLCQLQQLLIAIGDGFYDLGYRVSRLGFGKDKLTGTY 178
Query: 194 KVVFF--EIE-KIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGT 250
K V+ IE + A+ VF N WR + P P Y G V + G+
Sbjct: 179 KPVWLYNSIEIGLENATTCEVFDFST----NAWRYVS--PAAP--YRIVGCPAPVCVDGS 230
Query: 251 INWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLS--PPKGLDEVPSVVPSVGVL 308
++W T E+ ILS DL TET+ ++S P +D V+ + L
Sbjct: 231 LHWFTE--------------CEETKILSFDLHTETFQVVSKAPFANVDGFDIVMCN---L 273
Query: 309 MNCLCFSHDLEGTHFVIWQMKK-------------------FGDAKSWTQLLKVRYQDLQ 349
N LC S +++ + VIW FG + L +
Sbjct: 274 DNRLCVS-EMKLPNQVIWSFNSGNKTWHKMCSINLDITSRWFGPTQVCAVLPLALLDGKK 332
Query: 350 REPLHSMFYCHYQLFPLCLHDGDT 373
++ +FYC + + +HD +T
Sbjct: 333 KKKKKLLFYCRVRKRTMVVHDDET 356
>At3g18910 unknown protein
Length = 388
Score = 55.8 bits (133), Expect = 5e-08
Identities = 68/274 (24%), Positives = 115/274 (41%), Gaps = 45/274 (16%)
Query: 26 QLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRE 85
QL NDL+ E+L +P SL RL+ CK+WN L D+ H +SA + L
Sbjct: 7 QLPNDLVEEILCRVPATSLRRLRSTCKAWNRLFKGDRILASKHFEKSAKQFRSLSL---- 62
Query: 86 NSHHDHREFCVT-HLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYS 144
+D+R F ++ +L G+S SL + H+ + + V+ C L+C
Sbjct: 63 --RNDYRIFPISFNLHGNSPSLELKSELIDPHS-KNSAAPFEISRVIHCEGLLLC----- 114
Query: 145 VDSIFQQGWLSFWNPAT-RLMSEKLGCFCVKTKNFDVKLSFGYDNST--DTYKVVFFEIE 201
S + + WNP T + G F K ++FDV + D + +YK++ +
Sbjct: 115 -SSQLDESRVVVWNPLTGETRWIRTGDFRQKGRSFDVGYYYQKDKRSWIKSYKLLCY--- 170
Query: 202 KIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGV-NDGVHLSGTINW--MTSRD 258
+R + F + F D W + +D + G+I + ++
Sbjct: 171 ---------------------YRGTKYFEIYDFDSDSWRILDDIIAPRGSIGYSELSVSL 209
Query: 259 KSAPYYNPVTITVEQLGILSLDLRTETYTLLSPP 292
K Y+ +T E+ +SL L+ + YT S P
Sbjct: 210 KGNTYWFAKGVTEERPRTISL-LKFDFYTEKSVP 242
>At2g07140 pseudogene
Length = 384
Score = 55.8 bits (133), Expect = 5e-08
Identities = 80/329 (24%), Positives = 133/329 (40%), Gaps = 47/329 (14%)
Query: 26 QLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRE 85
+L DL+ E+LS++P SL RL+ CK WN L D+ F ++H ++A +P L +
Sbjct: 5 ELPKDLVEEILSFVPATSLKRLRSTCKGWNRLFKDDKRFTRIHTEKAAKQFQPLTLTK-- 62
Query: 86 NSHHDHREFCV--THLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGY 143
+ C +L G++ SL +SL V C+GL+ L
Sbjct: 63 -----NYRICPINVNLHGTTPSLEVKNEVSLLDPHSKNSAAQFNIDRVFHCDGLL-LCTS 116
Query: 144 SVDSIFQQGWLSFWNPATRLMS-EKLGCFCVKTKNFDVKLSFGYDNST--DTYKVVFFEI 200
DS F WNP T + +LG N + GYDN + +YK + F
Sbjct: 117 QKDSRF-----VVWNPLTGVTKWIELG----DRYNEGMAFILGYDNKSCNKSYKAMSFNY 167
Query: 201 EKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKS 260
+ + S + F+ D WR I + + P Y D+ + L G W+ +
Sbjct: 168 --LDKDSEIYEFSSD------SWRVIDDI-IKPPHYMDY-FRECFSLKGNTYWLGIDRRR 217
Query: 261 APYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVL----MNCLCFSH 316
P +T ++ D TE + +S P ++ V+ ++ L +
Sbjct: 218 RPPDLRIT-------LIKFDFGTEKFGYVSLPPPCQVHGFEASNLSVVGDEKLSVLVQAG 270
Query: 317 DLEGTHFVIWQMKKFGDAK--SWTQLLKV 343
T +W K G+A SW+++L +
Sbjct: 271 STSKTE--VWVTSKIGEANVVSWSKVLSL 297
>At3g07870 unknown protein
Length = 417
Score = 55.5 bits (132), Expect = 6e-08
Identities = 81/331 (24%), Positives = 126/331 (37%), Gaps = 56/331 (16%)
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQREN 86
L D+IA++ S LP+ S+ RL VC+SW S++ S+ KP LLL ++
Sbjct: 28 LPEDIIADIFSRLPISSIARLMFVCRSWRSVLTQHGRLSS----SSSSPTKPCLLLHCDS 83
Query: 87 SHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVD 146
+ F LS E + R+ + + VVG CNGL+CL D
Sbjct: 84 PIRNGLHFL-------DLSEEEKRIKTKKFTLRFASSMPE-FDVVGSCNGLLCLS----D 131
Query: 147 SIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKV---VFFEIEKI 203
S++ L +NP T E C K + ++ FG+ T YKV V+F
Sbjct: 132 SLYNDS-LYLYNPFTTNSLELPEC-SNKYHDQELVFGFGFHEMTKEYKVLKIVYFRGSSS 189
Query: 204 R-------------RASLVSVFTLDG--CNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLS 248
+ S V + TL + WR + P Y + ++
Sbjct: 190 NNNGIYRGRGRIQYKQSEVQILTLSSKTTDQSLSWRSLGKAP-----YKFVKRSSEALVN 244
Query: 249 GTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPK--GLDEVPSVVPSVG 306
G ++++T + P +S DL E + + P GL+ + +
Sbjct: 245 GRLHFVTRPRRHVP----------DRKFVSFDLEDEEFKEIPKPDCGGLNRTNHRLVN-- 292
Query: 307 VLMNCLCFSHDLEGTHFVIWQMKKFGDAKSW 337
L CLC IW MK +G +SW
Sbjct: 293 -LKGCLCAVVYGNYGKLDIWVMKTYGVKESW 322
>At5g62660 putative protein
Length = 379
Score = 54.7 bits (130), Expect = 1e-07
Identities = 70/283 (24%), Positives = 111/283 (38%), Gaps = 45/283 (15%)
Query: 20 DNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPH 79
D L ++ DL+ E+L+ LP KSL R KCV K W+SLI S + F +L P
Sbjct: 34 DALVAPEIPLDLLIEILTKLPAKSLMRFKCVSKLWSSLIRS-RFFSNCYLTVKTPRRPPR 92
Query: 80 LLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNG--L 137
L + V HL +SL + P S+ + + ++ + G +
Sbjct: 93 LYMS-----------LVDHLLCNSLMVCHYPCESVLLSSSSSAESLEQNLTIAGMGGRNM 141
Query: 138 ICLHGYSVDSIFQQGWLSFWNPATR----LMSEKLGCFCVKTKNFDVKLSFGYDNSTDTY 193
+ L G + + + S +NP TR L + K K+ + FGYD D Y
Sbjct: 142 VVLRGLILYVVCRTA--SIYNPTTRQSVTLPAVKSNILAQKSHWNSLLYFFGYDPVLDQY 199
Query: 194 KVV----FFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSG 249
KVV F R S VF L+ W + F G+ ++G
Sbjct: 200 KVVCTVALFSKRLKRITSEHWVFVLEPGGSWK---------RIEFDQPHLPTRLGLCVNG 250
Query: 250 TINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPP 292
I ++ S K ++S D+R+E ++++ P
Sbjct: 251 VIYYLASTWKRRDI------------VVSFDVRSEEFSMIQGP 281
>At3g62380 putative protein
Length = 325
Score = 54.7 bits (130), Expect = 1e-07
Identities = 57/226 (25%), Positives = 99/226 (43%), Gaps = 41/226 (18%)
Query: 130 VVGCCNGLICLHGYSVDSIFQQGWLSFWNPATR----LMSEKLGCFCVKT---KNFDVKL 182
++G CNG++C I+ G++ NPATR L E L + +T + + +
Sbjct: 74 IMGYCNGIVC--------IYDLGYIYLINPATRKLRILSPEFLREYTGRTGWSNDLPINV 125
Query: 183 SFGYDNSTDTYKVV---FFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDW 239
FG D T TYKV+ F+ ++ +L N NG R FP+ SY
Sbjct: 126 GFGRDMVTGTYKVILMYLFDRNFVKTEAL---------NLKNGERRYVYFPI---SYGQL 173
Query: 240 GVND--GVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDE 297
G ND + +G++ W+ D +Y T + + + ++DL TET+ + P +
Sbjct: 174 G-NDKRSIFANGSLYWLPKPD--VDFY-----TNQLIKLAAIDLHTETFRYVPLPSWYTK 225
Query: 298 VPSVVPSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKV 343
V + L + LC S L+ +W +++ + W ++ V
Sbjct: 226 YCKSV-YLWSLKDSLCVSDVLQNPSVDVWSLQQEEPSVKWEKMFSV 270
>At3g19410 hypothetical protein
Length = 373
Score = 53.1 bits (126), Expect = 3e-07
Identities = 49/183 (26%), Positives = 80/183 (42%), Gaps = 28/183 (15%)
Query: 26 QLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRE 85
+L DLI E+L ++P L RL+ CK WN L D+ F K H ++A K L L
Sbjct: 5 ELPKDLIEEILCYVPATYLKRLRSTCKGWNRLFKDDRRFAKKHYDKAA---KQFLPLMST 61
Query: 86 NSHHDHREFCV--THLSGS--SLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLH 141
N E C +L G+ SL + + PW+ ++ + ++ +H G L+C
Sbjct: 62 N-----EELCAMSVNLHGTIPSLEVKDKPWLFVSDSKHCDIEISRIFHSGGL---LLCFS 113
Query: 142 GYSVDSIFQQGWLSFWNP---ATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFF 198
SI WNP TRL+ + + K + L + ++ YK++ F
Sbjct: 114 RDGEISII------VWNPLTSETRLIRTRNR----RDKGRNFVLGYYQEDKKTYYKILSF 163
Query: 199 EIE 201
++
Sbjct: 164 YLD 166
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,749,944
Number of Sequences: 26719
Number of extensions: 474795
Number of successful extensions: 1503
Number of sequences better than 10.0: 261
Number of HSP's better than 10.0 without gapping: 227
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 1181
Number of HSP's gapped (non-prelim): 309
length of query: 440
length of database: 11,318,596
effective HSP length: 102
effective length of query: 338
effective length of database: 8,593,258
effective search space: 2904521204
effective search space used: 2904521204
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0176a.1


