

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0175.7
(402 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
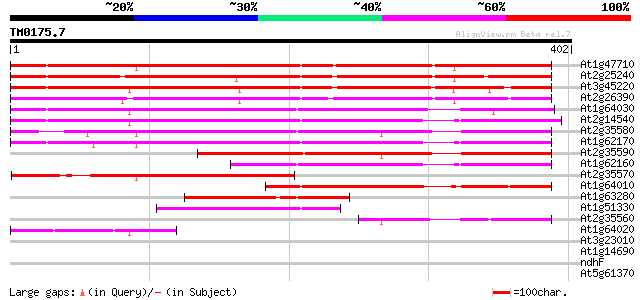
Score E
Sequences producing significant alignments: (bits) Value
At1g47710 serpin, putative 369 e-102
At2g25240 putative serpin 320 7e-88
At3g45220 serpin-like protein 304 5e-83
At2g26390 putative serpin 302 2e-82
At1g64030 hypothetical protein 297 6e-81
At2g14540 putative serpin 283 1e-76
At2g35580 putative serpin 280 1e-75
At1g62170 hypothetical protein 277 8e-75
At2g35590 putative serpin 220 1e-57
At1g62160 hypothetical protein 185 3e-47
At2g35570 putative serpin 166 2e-41
At1g64010 hypothetical protein 166 2e-41
At1g63280 hypothetical protein 99 5e-21
At1g51330 hypothetical protein 92 5e-19
At2g35560 putative serpin 86 3e-17
At1g64020 hypothetical protein 65 7e-11
At3g23010 disease resistance protein, putative 33 0.28
At1g14690 hypothetical protein 33 0.36
ndhF -chloroplast genome- NADH dehydrogenase ND5 32 0.81
At5g61370 unknown protein 31 1.1
>At1g47710 serpin, putative
Length = 391
Score = 369 bits (948), Expect = e-102
Identities = 195/393 (49%), Positives = 269/393 (67%), Gaps = 9/393 (2%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MD+++SI Q +V++++ KH+ + Q N++FSP S++ +LS++A GSAG+T D++ S
Sbjct: 1 MDVRESISLQNQVSMNLAKHVITTVS-QNSNVIFSPASINVVLSIIAAGSAGATKDQILS 59
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FL+F S D LN+F S+++S VL+D + + LS N W DKSL+ SFKQL+ YK
Sbjct: 60 FLKFSSTDQLNSFSSEIVSAVLADGSANGGPKLSVANGAWIDKSLSFKPSFKQLLEDSYK 119
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKH 178
A DF++K +V EVNSW EKETNGLITE+LP + +TKLIFANALYFKG W
Sbjct: 120 AASNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKLIFANALYFKGTWNE 179
Query: 179 TFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMC 238
FD S+T +FHLL+G + PFM+S+KK Q++ +DGF VL L Y QG+DK+ +FSM
Sbjct: 180 KFDESLTQEGEFHLLDGNKVTAPFMTSKKK-QYVSAYDGFKVLGLPYLQGQDKR-QFSMY 237
Query: 239 IFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGV 298
+LP+A +GL L++K+ S FL +PR++V+VR+F+IPKFK F +ASNVLK LG+
Sbjct: 238 FYLPDANNGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFSFGFDASNVLKGLGL 297
Query: 299 VSPFSKSNANFTKMVVSPL--DELCVESIHHKASIEVNEEGTEAGVA-CSMCDVKLCVSA 355
SPFS T+MV SP LCV +I HKA IEVNEEGTEA A + ++ +
Sbjct: 298 TSPFS-GEEGLTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAAASAGVIKLRGLLME 356
Query: 356 RTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
IDFVADHPFL ++ E++TG +LFIGQV+ P
Sbjct: 357 EDEIDFVADHPFLLVVTENITGVVLFIGQVVDP 389
>At2g25240 putative serpin
Length = 385
Score = 320 bits (821), Expect = 7e-88
Identities = 184/393 (46%), Positives = 249/393 (62%), Gaps = 16/393 (4%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
M+L KSI +V + +TKH+ + NLVFSP S++ +LS++A GS T +++ S
Sbjct: 1 MELGKSIENHNDVVVRLTKHVIATVA-NGSNLVFSPISINVLLSLIAAGSCSVTKEQILS 59
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSFL-LSFVNEMWADKSLALSHSFKQLMTTHYKA 119
FL S DHLN +Q++ T S L LS N +W DK +L SFK L+ YKA
Sbjct: 60 FLMLPSTDHLNLVLAQIID---GGTEKSDLRLSIANGVWIDKFFSLKLSFKDLLENSYKA 116
Query: 120 TLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKL--TKLIFANALYFKGVWK 177
T + VDF +K +V EVN+W E TNGLI ++L +++ + + L+ ANA+YFKG W
Sbjct: 117 TCSQVDFASKPSEVIDEVNTWAEVHTNGLIKQILSRDSIDTIRSSTLVLANAVYFKGAWS 176
Query: 178 HTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSM 237
FD +MT +DFHLL+GTS++VPFM++ + Q++R++DGF VLRL Y + + + FSM
Sbjct: 177 SKFDANMTKKNDFHLLDGTSVKVPFMTNYED-QYLRSYDGFKVLRLPYIEDQRQ---FSM 232
Query: 238 CIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELG 297
I+LPN K+GL L+EK+ SE F +P + V FRIPKFK FE AS VLK++G
Sbjct: 233 YIYLPNDKEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVLKDMG 292
Query: 298 VVSPFSKSNANFTKMVVSPL--DELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSA 355
+ SPF+ + T+MV SP D+L V SI HKA IEV+EEGTEA A V C S
Sbjct: 293 LTSPFN-NGGGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEA--AAVSVGVVSCTSF 349
Query: 356 RTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
R DFVAD PFLF +RED +G ILF+GQVL P
Sbjct: 350 RRNPDFVADRPFLFTVREDKSGVILFMGQVLDP 382
>At3g45220 serpin-like protein
Length = 393
Score = 304 bits (779), Expect = 5e-83
Identities = 175/402 (43%), Positives = 249/402 (61%), Gaps = 26/402 (6%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
M+L KS+ QT+V + + KH+ NLVFSP S++ +L ++A GS T +++ S
Sbjct: 1 MELGKSMENQTDVMVLLAKHVIPTVA-NGSNLVFSPMSINVLLCLIAAGSNCVTKEQILS 59
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYK 118
F+ S D+LN ++ +S L+D LS +W DKSL+ SFK L+ Y
Sbjct: 60 FIMLPSSDYLNAVLAKTVSVALNDGMERSDLHLSTAYGVWIDKSLSFKPSFKDLLENSYN 119
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTK--LIFANALYFKGVW 176
AT VDF TK +V EVN+W E TNGLI E+L +++ + + LI ANA+YFKG W
Sbjct: 120 ATCNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFKGAW 179
Query: 177 KHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFS 236
FD +T DFHLL+GT ++VPFM++ KK Q++ +DGF VLRL Y + + + F+
Sbjct: 180 SKKFDAKLTKSYDFHLLDGTMVKVPFMTNYKK-QYLEYYDGFKVLRLPYVEDQRQ---FA 235
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M I+LPN +DGLP+L+E+++S+ FL +PRQ++ F+IPKFK FE +AS+VLKE+
Sbjct: 236 MYIYLPNDRDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVLKEM 295
Query: 297 GVVSPFSKSNANFTKMVVSP--------LDELCVESIHHKASIEVNEEGTEAGV--ACSM 346
G+ PF ++ + T+MV SP + L V ++ HKA IEV+EEGTEA SM
Sbjct: 296 GLTLPF--THGSLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAVSVASM 353
Query: 347 CDVKLCVSARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
L + DFVADHPFLF +RE+ +G ILF+GQVL P
Sbjct: 354 TKDMLLMG-----DFVADHPFLFTVREEKSGVILFMGQVLDP 390
>At2g26390 putative serpin
Length = 389
Score = 302 bits (773), Expect = 2e-82
Identities = 179/397 (45%), Positives = 238/397 (59%), Gaps = 20/397 (5%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
M+L KSI Q V + K + + N+VFSP S++ +LS++A GS T +E+ S
Sbjct: 1 MELGKSIENQNNVVARLAKKVIETDVANGSNVVFSPMSINVLLSLIAAGSNPVTKEEILS 60
Query: 61 FLRFDSVDHLNTFFSQVLS--TVLSDTTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FL S DHLN +++ T SD LS + +W DKS L SFK+L+ YK
Sbjct: 61 FLMSPSTDHLNAVLAKIADGGTERSD----LCLSTAHGVWIDKSSYLKPSFKELLENSYK 116
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTK-----LIFANALYFK 173
A+ + VDF TK +V EVN W + TNGLI ++L + + + LI ANA+YFK
Sbjct: 117 ASCSQVDFATKPVEVIDEVNIWADVHTNGLIKQILSRDCTDTIKEIRNSTLILANAVYFK 176
Query: 174 GVWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKL 233
W FD +T +DFHLL+G +++VPFM S K Q++R +DGF VLRL Y + K
Sbjct: 177 AAWSRKFDAKLTKDNDFHLLDGNTVKVPFMMSYKD-QYLRGYDGFQVLRLPYVED---KR 232
Query: 234 RFSMCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVL 293
FSM I+LPN KDGL +L+EK+++E FL +P + V RIPK FE +AS VL
Sbjct: 233 HFSMYIYLPNDKDGLAALLEKISTEPGFLDSHIPLHRTPVDALRIPKLNFSFEFKASEVL 292
Query: 294 KELGVVSPFSKSNANFTKMVVSPL--DELCVESIHHKASIEVNEEGTEAGVACSMCDVKL 351
K++G+ SPF+ S N T+MV SP D+L V SI HKA IEV+EEGTEA +
Sbjct: 293 KDMGLTSPFT-SKGNLTEMVDSPSNGDKLHVSSIIHKACIEVDEEGTEAAAVSVAIMMPQ 351
Query: 352 CVSARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
C+ DFVADHPFLF +RED +G ILFIGQVL P
Sbjct: 352 CLMRNP--DFVADHPFLFTVREDNSGVILFIGQVLDP 386
>At1g64030 hypothetical protein
Length = 385
Score = 297 bits (761), Expect = 6e-81
Identities = 170/397 (42%), Positives = 240/397 (59%), Gaps = 31/397 (7%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLD-ELF 59
MD++++++ QT VA+ ++ H+ S ++ N++FSP S+++ +++ A G G + ++
Sbjct: 1 MDVREAMKNQTHVAMILSGHVLSSAP-KDSNVIFSPASINSAITMHAAGPGGDLVSGQIL 59
Query: 60 SFLRFDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTHY 117
SFLR S+D L T F ++ S V +D T ++ N +W DKSL FK L +
Sbjct: 60 SFLRSSSIDELKTVFRELASVVYADRSATGGPKITAANGLWIDKSLPTDPKFKDLFENFF 119
Query: 118 KATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWK 177
KA VDFR++ ++V +EVNSWVE TN LI +LLP +V LT I+ANAL FKG WK
Sbjct: 120 KAVYVPVDFRSEAEEVRKEVNSWVEHHTNNLIKDLLPDGSVTSLTNKIYANALSFKGAWK 179
Query: 178 HTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLR-FS 236
F+ T +DF+L+NGTS+ VPFMSS + Q++R +DGF VLRL Y++G D R FS
Sbjct: 180 RPFEKYYTRDNDFYLVNGTSVSVPFMSS-YENQYVRAYDGFKVLRLPYQRGSDDTNRKFS 238
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M +LP+ KDGL L+EK+AS FL +P + + KFRIPKFKI F ++VL L
Sbjct: 239 MYFYLPDKKDGLDDLLEKMASTPGFLDSHIPTYRDELEKFRIPKFKIEFGFSVTSVLDRL 298
Query: 297 GVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACS--MCDVKL-CV 353
G+ S S++HKA +E++EEG EA A + C L V
Sbjct: 299 GLRS----------------------MSMYHKACVEIDEEGAEAAAATADGDCGCSLDFV 336
Query: 354 SARTGIDFVADHPFLFLIREDLTGTILFIGQVLHPEG 390
IDFVADHPFLFLIRE+ TGT+LF+GQ+ P G
Sbjct: 337 EPPKKIDFVADHPFLFLIREEKTGTVLFVGQIFDPSG 373
>At2g14540 putative serpin
Length = 407
Score = 283 bits (724), Expect = 1e-76
Identities = 171/401 (42%), Positives = 237/401 (58%), Gaps = 31/401 (7%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDE-LF 59
+D+Q++++ Q EV+L + + S + N VFSP S++A+L+V A + TL +
Sbjct: 29 IDMQEAMKNQNEVSLLLVGKVISAVA-KNSNCVFSPASINAVLTVTAANTDNKTLRSFIL 87
Query: 60 SFLRFDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTHY 117
SFL+ S + N F ++ S V D T ++ VN +W ++SL+ + ++ L +
Sbjct: 88 SFLKSSSTEETNAIFHELASVVFKDGSETGGPKIAAVNGVWMEQSLSCNPDWEDLFLNFF 147
Query: 118 KATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWK 177
KA+ A VDFR K ++V +VN+W + TN LI E+LP +V LT I+ NALYFKG W+
Sbjct: 148 KASFAKVDFRHKAEEVRLDVNTWASRHTNDLIKEILPRGSVTSLTNWIYGNALYFKGAWE 207
Query: 178 HTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLR-FS 236
FD SMT FHLLNG S+ VPFM S +K QFI +DGF VLRL Y+QGRD R FS
Sbjct: 208 KAFDKSMTRDKPFHLLNGKSVSVPFMRSYEK-QFIEAYDGFKVLRLPYRQGRDDTNREFS 266
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M ++LP+ K L +L+E++ S FL +P +V V FRIPKFKI F EAS+V +
Sbjct: 267 MYLYLPDKKGELDNLLERITSNPGFLDSHIPEYRVDVGDFRIPKFKIEFGFEASSVFNDF 326
Query: 297 GVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVK-LCV-S 354
EL V S+H KA IE++EEGTEA A ++ V C+
Sbjct: 327 ----------------------ELNV-SLHQKALIEIDEEGTEAAAATTVVVVTGSCLWE 363
Query: 355 ARTGIDFVADHPFLFLIREDLTGTILFIGQVLHPEGAAMSL 395
+ IDFVADHPFLFLIRED TGT+LF GQ+ P + +L
Sbjct: 364 PKKKIDFVADHPFLFLIREDKTGTLLFAGQIFDPSELSSAL 404
>At2g35580 putative serpin
Length = 374
Score = 280 bits (715), Expect = 1e-75
Identities = 164/395 (41%), Positives = 230/395 (57%), Gaps = 45/395 (11%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGST--LDEL 58
MDL++S+ Q ++ L +T L ++ ILS++A S G T D++
Sbjct: 15 MDLKESVGNQNDIVLRLTAPL-----------------INVILSIIAASSPGDTDTADKI 57
Query: 59 FSFLRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTH 116
S L+ S D L+ S++++TVL+D+T S +S N +W +K+L + SFK L+
Sbjct: 58 VSLLQASSTDKLHAVSSEIVTTVLADSTASGGPTISAANGLWIEKTLNVEPSFKDLLLNS 117
Query: 117 YKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVH-KLTKLIFANALYFKGV 175
YKA VDFRTK D+V REVNSWVEK+TNGLIT LLP+N LT IFANAL+F G
Sbjct: 118 YKAAFNRVDFRTKADEVNREVNSWVEKQTNGLITNLLPSNPKSAPLTDHIFANALFFNGR 177
Query: 176 WKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRF 235
W FDPS+T DFHLL+GT + VPFM+ ++ ++GF V+ L Y++GR+ F
Sbjct: 178 WDSQFDPSLTKDSDFHLLDGTKVRVPFMTG-ASCRYTHVYEGFKVINLQYRRGREDSRSF 236
Query: 236 SMCIFLPNAKDGLPSLIEKLASESCFLKGK--LPRQKVRVRKFRIPKFKICFELEASNVL 293
SM I+LP+ KDGLPS++E+LAS FLK LP +++ +IP+FK F EAS L
Sbjct: 237 SMQIYLPDEKDGLPSMLERLASTRGFLKDNEVLPSHSAVIKELKIPRFKFDFAFEASEAL 296
Query: 294 KELGVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCV 353
K G+V P S I HK+ IEV+E G++A A + +
Sbjct: 297 KGFGLVVPLS--------------------MIMHKSCIEVDEVGSKAAAAAAFRGIGCRR 336
Query: 354 SARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
DFVADHPFLF+++E +G +LF+GQV+ P
Sbjct: 337 PPPEKHDFVADHPFLFIVKEYRSGLVLFLGQVMDP 371
>At1g62170 hypothetical protein
Length = 433
Score = 277 bits (708), Expect = 8e-75
Identities = 166/395 (42%), Positives = 231/395 (58%), Gaps = 32/395 (8%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDEL-- 58
+D+ ++++ Q +VA+ +T + S + N VFSP S++A L+++A S G +EL
Sbjct: 62 IDVGEAMKKQNDVAIFLTGIVISSVA-KNSNFVFSPASINAALTMVAASSGGEQGEELRS 120
Query: 59 --FSFLRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMT 114
SFL+ S D LN F ++ S VL D + ++ VN MW D+SL+++ K L
Sbjct: 121 FILSFLKSSSTDELNAIFREIASVVLVDGSKKGGPKIAVVNGMWMDQSLSVNPLSKDLFK 180
Query: 115 THYKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKG 174
+ A A VDFR+K ++V EVN+W TNGLI +LLP +V LT ++ +ALYFKG
Sbjct: 181 NFFSAAFAQVDFRSKAEEVRTEVNAWASSHTNGLIKDLLPRGSVTSLTDRVYGSALYFKG 240
Query: 175 VWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLR 234
W+ + SMT F+LLNGTS+ VPFMSS +K Q+I +DGF VLRL Y+QGRD R
Sbjct: 241 TWEEKYSKSMTKCKPFYLLNGTSVSVPFMSSFEK-QYIAAYDGFKVLRLPYRQGRDNTNR 299
Query: 235 -FSMCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVL 293
F+M I+LP+ K L L+E++ S FL P ++V+V KFRIPKFKI F EAS+
Sbjct: 300 NFAMYIYLPDKKGELDDLLERMTSTPGFLDSHNPERRVKVGKFRIPKFKIEFGFEASSAF 359
Query: 294 KELGVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCV 353
+ EL V S + K IE++E+GTEA + L
Sbjct: 360 SDF----------------------ELDV-SFYQKTLIEIDEKGTEAVTFTAFRSAYLGC 396
Query: 354 SARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+ IDFVADHPFLFLIRE+ TGT+LF GQ+ P
Sbjct: 397 ALVKPIDFVADHPFLFLIREEQTGTVLFAGQIFDP 431
>At2g35590 putative serpin
Length = 331
Score = 220 bits (560), Expect = 1e-57
Identities = 117/256 (45%), Positives = 160/256 (61%), Gaps = 23/256 (8%)
Query: 135 REVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHTFDPSMTFVDDFHLLN 194
+EVNSWVE++TNGLIT+LLP N+ LT LIFANAL+F G W F+PS+T DFHLL+
Sbjct: 94 KEVNSWVEEQTNGLITDLLPPNSASPLTDLIFANALFFNGRWDSQFNPSLTKESDFHLLD 153
Query: 195 GTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCIFLPNAKDGLPSLIEK 254
GT + VPFM+ + + ++ F VL L Y++GR+ FSM I+LP+ KDGLPS++E
Sbjct: 154 GTKVRVPFMTGAHEDS-LDVYEAFKVLNLPYREGREDSRGFSMQIYLPDEKDGLPSMLES 212
Query: 255 LASESCFLKGK--LPRQKVRVRKFRIPKFKICFELEASNVLKELGVVSPFSKSNANFTKM 312
LAS FLK LP QK V++ +IP+FK F+ EAS LK LG+ P S
Sbjct: 213 LASTRGFLKDNKVLPSQKAGVKELKIPRFKFAFDFEASKALKGLGLKVPLS--------- 263
Query: 313 VVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTGIDFVADHPFLFLIR 372
+I HK+ IEV+E G++A A ++ C DFVADHPFLF+++
Sbjct: 264 -----------TIIHKSCIEVDEVGSKAAAAAALRSCGGCYFPPKKYDFVADHPFLFIVK 312
Query: 373 EDLTGTILFIGQVLHP 388
E ++G +LF+G V+ P
Sbjct: 313 EYISGLVLFLGHVMDP 328
>At1g62160 hypothetical protein
Length = 265
Score = 185 bits (470), Expect = 3e-47
Identities = 110/232 (47%), Positives = 139/232 (59%), Gaps = 26/232 (11%)
Query: 159 HK-LTKLIFANALYFKGVWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDG 217
HK LTK I+ NALYFKG W+ F SMT F+L+NG S+ VPFMSS K Q+I +DG
Sbjct: 56 HKNLTKRIYGNALYFKGTWEKKFSKSMTKRKPFYLVNGISVSVPFMSS-SKDQYIEAYDG 114
Query: 218 FNVLRLSYKQGRDKKLR-FSMCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKF 276
F VLRL Y+QGRD R FSM +LP+ K L L++++ S FL PR++V V +F
Sbjct: 115 FKVLRLPYRQGRDNTNRNFSMYFYLPDKKGELDDLLKRMTSTPGFLDSHTPRERVEVDEF 174
Query: 277 RIPKFKICFELEASNVLKELGVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEE 336
RIPKFKI F EAS+V + E+ V S + KA IE++EE
Sbjct: 175 RIPKFKIEFGFEASSVFSDF----------------------EIDV-SFYQKALIEIDEE 211
Query: 337 GTEAGVACSMCDVKLCVSARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
GTEA A + D + +DFVADHPFLFLIRE+ TGT+LF GQ+ P
Sbjct: 212 GTEAAAATAFVDNEDGCGFVETLDFVADHPFLFLIREEQTGTVLFAGQIFDP 263
>At2g35570 putative serpin
Length = 213
Score = 166 bits (421), Expect = 2e-41
Identities = 91/205 (44%), Positives = 128/205 (62%), Gaps = 18/205 (8%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
+L+ S+ ++ L +T+H+ + + NLVFSP A+LSV + SF
Sbjct: 16 NLKGSVGNLNDIILRLTQHVIASTAGKTSNLVFSP----ALLSV------------ILSF 59
Query: 62 LRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYKA 119
+ S D LN S +++T+L+D+TPS +S N +W +K+L + SFK L+ YKA
Sbjct: 60 VAASSPDELNAVSSGIVTTILADSTPSGGPTISAANGVWIEKTLYVEPSFKDLLLNSYKA 119
Query: 120 TLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHT 179
VDFRTK D+V +EVNSWVE++TNGLIT+LLP N+ T LIFANAL+F G W
Sbjct: 120 AFNQVDFRTKADEVNKEVNSWVEEQTNGLITDLLPPNSASSWTDLIFANALFFNGRWDSQ 179
Query: 180 FDPSMTFVDDFHLLNGTSIEVPFMS 204
F+PS+T DFHLL+GT + VPFM+
Sbjct: 180 FNPSLTKESDFHLLDGTKVRVPFMT 204
>At1g64010 hypothetical protein
Length = 185
Score = 166 bits (421), Expect = 2e-41
Identities = 95/205 (46%), Positives = 124/205 (60%), Gaps = 24/205 (11%)
Query: 184 MTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCIFLPN 243
MT DFHL+NGTS+ V MSS K Q+I +DGF VL+L ++QG D FSM +LP+
Sbjct: 1 MTKDRDFHLINGTSVSVSLMSSYKD-QYIEAYDGFKVLKLPFRQGNDTSRNFSMHFYLPD 59
Query: 244 AKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVVSPFS 303
KDGL +L+EK+AS FL +P QKV+V +F IPKFKI F AS LG
Sbjct: 60 EKDGLDNLVEKMASSVGFLDSHIPSQKVKVGEFGIPKFKIEFGFSASRAFNRLG------ 113
Query: 304 KSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTGIDFVA 363
LDE+ +++ KA +E++EEG EA A ++ C + IDFVA
Sbjct: 114 -------------LDEM---ALYQKACVEIDEEGAEAIAATAVVGGFGCAFVKR-IDFVA 156
Query: 364 DHPFLFLIREDLTGTILFIGQVLHP 388
DHPFLF+IRED TGT+LF+GQ+ P
Sbjct: 157 DHPFLFMIREDKTGTVLFVGQIFDP 181
>At1g63280 hypothetical protein
Length = 120
Score = 98.6 bits (244), Expect = 5e-21
Identities = 54/119 (45%), Positives = 73/119 (60%), Gaps = 4/119 (3%)
Query: 126 FRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHTFDPSMT 185
F K QV +E+N W TNGLI +LLP +V T ++ NALYFKG W++ FD S T
Sbjct: 5 FVLKAVQVRQELNKWASDHTNGLIIDLLPRGSVKSETVQVYGNALYFKGAWENKFDKSST 64
Query: 186 FVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQG-RDKKLRFSMCIFLPN 243
++FH G + VPFM S ++Q+I DGF VL L Y+QG D K +FS+ +LP+
Sbjct: 65 KDNEFH--QGKEVHVPFMRS-YESQYIMACDGFKVLGLPYQQGLDDTKRKFSIYFYLPD 120
>At1g51330 hypothetical protein
Length = 193
Score = 92.0 bits (227), Expect = 5e-19
Identities = 56/135 (41%), Positives = 79/135 (58%), Gaps = 4/135 (2%)
Query: 106 SHSFKQLMTTHYKATLALVDFRTKG-DQVCREVNSWVEKETNGLITELLPTNAVHKLTKL 164
S S K L + K +V G ++V EVNSW + TNGLI LLP +V T
Sbjct: 7 SVSAKSLPLSLSKEAKEVVHAAVNGAEEVRMEVNSWALRHTNGLIKNLLPPGSVTNQTIK 66
Query: 165 IFANALYFKGVWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLR-L 223
I+ NALYFKG W++ F SMT FHL+NG + VPFM S ++ ++++ ++GF VLR L
Sbjct: 67 IYGNALYFKGAWENKFGKSMTIHKPFHLVNGKQVLVPFMKSYER-KYMKAYNGFKVLRIL 125
Query: 224 SYK-QGRDKKLRFSM 237
Y+ +D +FS+
Sbjct: 126 QYRVDYKDTSRQFSI 140
>At2g35560 putative serpin
Length = 121
Score = 86.3 bits (212), Expect = 3e-17
Identities = 56/140 (40%), Positives = 76/140 (54%), Gaps = 24/140 (17%)
Query: 251 LIEKLASESCFLKGK--LPRQKVRVRKFRIPKFKICFELEASNVLKELGVVSPFSKSNAN 308
++E+LAS FL GK +P V + +IP+FK F EAS LK LG+ P
Sbjct: 1 MLERLASTRGFLNGKEDIPSHWADVGELKIPRFKFDFGFEASEALKGLGLEVP------- 53
Query: 309 FTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTGIDFVADHPFL 368
+I HK+ IEV+E G++A A ++ V C A DFVADHPFL
Sbjct: 54 --------------STIIHKSCIEVDEVGSKAAAA-AVIFVGCCGPAEKRYDFVADHPFL 98
Query: 369 FLIREDLTGTILFIGQVLHP 388
FL++E +G ILF+GQV+ P
Sbjct: 99 FLVKEYRSGLILFLGQVMDP 118
>At1g64020 hypothetical protein
Length = 121
Score = 65.1 bits (157), Expect = 7e-11
Identities = 42/122 (34%), Positives = 68/122 (55%), Gaps = 5/122 (4%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTL-DELF 59
M+ + +++ Q EVA+ ++ HLFS N VFSP S+ A+ ++MA G S + D++
Sbjct: 1 MEDEGAMKKQNEVAMILSWHLFSTVAKHSNN-VFSPASITAVFTMMASGPGSSLISDQIL 59
Query: 60 SFLRFDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTHY 117
SFL S+D LN+ F +V++TV +D + N W D+S ++ S K L +
Sbjct: 60 SFLGSSSIDELNSVF-RVITTVFADGSNIGGPTIKVANGAWIDQSFSIDSSSKNLFENFF 118
Query: 118 KA 119
KA
Sbjct: 119 KA 120
>At3g23010 disease resistance protein, putative
Length = 595
Score = 33.1 bits (74), Expect = 0.28
Identities = 22/76 (28%), Positives = 35/76 (45%), Gaps = 2/76 (2%)
Query: 149 ITELLPTNAVHKLTKLIFANALYFKGVWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKK 208
I ++ N V L + FAN L VW + P ++ + + SI++ + E
Sbjct: 363 IIDISNNNFVGSLPQDYFANWLEMSLVWSGSDIPQFKYMGNVNFSTYDSIDLVYKGVE-- 420
Query: 209 TQFIRTFDGFNVLRLS 224
T F R F+GFN + S
Sbjct: 421 TDFDRIFEGFNAIDFS 436
>At1g14690 hypothetical protein
Length = 707
Score = 32.7 bits (73), Expect = 0.36
Identities = 40/159 (25%), Positives = 68/159 (42%), Gaps = 20/159 (12%)
Query: 159 HKLTKLIFANALYFKGVWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGF 218
H + KL ++ F+ V KH + + FV D+H ++ +S + K F+ +
Sbjct: 224 HMIHKLKTERSVRFQKVDKHACNAILLFVTDYHF---DALAFSPLSVQLKDVAGSLFELW 280
Query: 219 NVLRLSYKQGRDKKLRFSMCIFLPNAKDGLPSL-----IEKLASE-SCFLKGKLPRQKVR 272
N++ S ++ R K S + + P++ IE++++E CF K K R K
Sbjct: 281 NLMDTSQEE-RTKFASVSYVVRSSESDITEPNILSSETIEQVSAEVDCFNKLKASRMKEL 339
Query: 273 VRKFRIPKFKIC----------FELEASNVLKELGVVSP 301
V K R +C LE S L + G+V P
Sbjct: 340 VMKRRTELENLCRLAHIEADTSTSLEKSTALIDSGLVDP 378
>ndhF -chloroplast genome- NADH dehydrogenase ND5
Length = 746
Score = 31.6 bits (70), Expect = 0.81
Identities = 18/53 (33%), Positives = 28/53 (51%), Gaps = 5/53 (9%)
Query: 22 FSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSFLRFDSVDHLNTFF 74
+SK+E +L+FSP I +++A +AG T +F HLNT+F
Sbjct: 415 WSKDEILNDSLLFSP-----IFAIIACSTAGLTAFYMFRIYLLTFEGHLNTYF 462
>At5g61370 unknown protein
Length = 487
Score = 31.2 bits (69), Expect = 1.1
Identities = 32/128 (25%), Positives = 56/128 (43%), Gaps = 21/128 (16%)
Query: 35 SPFSLHAILSVMAVGSAGSTLDELFSFLRFDSVDHLNTFFSQVLSTVLSDTTPSFLLSFV 94
S +LH ++ +++ G LD+L L SV + +QV+ + ++T+P LL F
Sbjct: 33 SETALHEVIRIVSSPVGG--LDDLEENLNQVSVSPSSNLVTQVIESCKNETSPRRLLRFF 90
Query: 95 NEMWADKSLALSHSFKQL---------MTTHYKATLALVDFRTKGDQVCREVNSWVEKET 145
+ W+ KSL S K+ H + L D R + N ++K+T
Sbjct: 91 S--WSCKSLGSSLHDKEFNYVLRVLAEKKDHTAMQILLSDLRKE--------NRAMDKQT 140
Query: 146 NGLITELL 153
++ E L
Sbjct: 141 FSIVAETL 148
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,674,031
Number of Sequences: 26719
Number of extensions: 363914
Number of successful extensions: 899
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 817
Number of HSP's gapped (non-prelim): 21
length of query: 402
length of database: 11,318,596
effective HSP length: 102
effective length of query: 300
effective length of database: 8,593,258
effective search space: 2577977400
effective search space used: 2577977400
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0175.7


