

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0175.13
(437 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
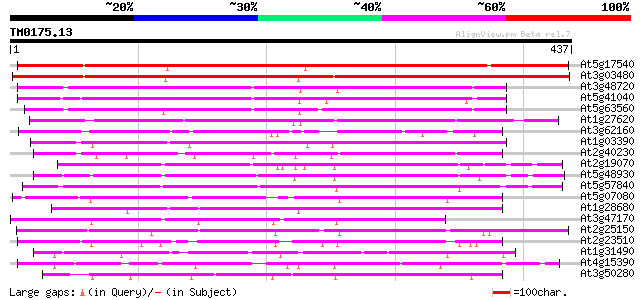
Score E
Sequences producing significant alignments: (bits) Value
At5g17540 unknown protein 417 e-117
At3g03480 putative hypersensitivity-related gene 410 e-115
At3g48720 unknown protein 233 2e-61
At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein 230 1e-60
At5g63560 acyltransferase-like protein 229 2e-60
At1g27620 putative hypersensitivity-related protein 197 7e-51
At3g62160 unknown protein 184 1e-46
At1g03390 hypothetical protein 181 5e-46
At2g40230 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 176 2e-44
At2g19070 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 160 2e-39
At5g48930 anthranilate N-benzoyltransferase 159 2e-39
At5g57840 N-hydroxycinnamoyl/benzoyltransferase 156 2e-38
At5g07080 unknown protein 144 7e-35
At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase lik... 144 7e-35
At3g47170 hypersensitivity-related protein-like protein 142 5e-34
At2g25150 unknown protein 134 8e-32
At2g23510 putative anthranilate N-hydroxycinnamoyl/benzoyltransf... 129 3e-30
At1g31490 putative protein 116 3e-26
At4g15390 HSR201 like protein 110 2e-24
At3g50280 anthranilate N-hydroxycinnamoyl/benzoyltransferase - l... 110 2e-24
>At5g17540 unknown protein
Length = 461
Score = 417 bits (1071), Expect = e-117
Identities = 221/439 (50%), Positives = 298/439 (67%), Gaps = 13/439 (2%)
Query: 7 SLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQA 66
SLTFK+ R KPELV PA TP E+K LSDIDDQ+GLRF+IP I YRH P+ + DPV
Sbjct: 4 SLTFKIYRQKPELVSPAKPTPRELKPLSDIDDQEGLRFHIPTIFFYRHNPT-TNSDPVAV 62
Query: 67 IRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPL--PPF 124
IR+AL+ TLVYYYPFAGRL+EGP RKL VDCTGEGV+FIEA+ADV+LVEF E PPF
Sbjct: 63 IRRALAETLVYYYPFAGRLREGPNRKLAVDCTGEGVLFIEADADVTLVEFEEKDALKPPF 122
Query: 125 PCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEM 184
PCFEELL++V GS ++L++PL+L+QVTRLKCGGFI A+R NH MSD GL FL + E
Sbjct: 123 PCFEELLFNVEGSCEMLNTPLMLMQVTRLKCGGFIFAVRINHAMSDAGGLTLFLKTMCEF 182
Query: 185 AQGASQPLTPPVWCRELLMARDPPRITCNHYEFEQVPSDSTEEGA-----ITRSFFFGSN 239
+G P PVW R LL AR R+T H E++++P+ TE G+ + RS FFG
Sbjct: 183 VRGYHAPTVAPVWERHLLSARVLLRVTHAHREYDEMPAIGTELGSRRDNLVGRSLFFGPC 242
Query: 240 EIAALRRLVPLDLRHCST-FDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRF-NPPI 297
E++A+RRL+P +L + ST +++T+ W RT AL+ ++RL+ IVN R++ NPP+
Sbjct: 243 EMSAIRRLLPPNLVNSSTNMEMLTSFLWRYRTIALRPDQDKEMRLILIVNARSKLKNPPL 302
Query: 298 PVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLF 357
P GYYGN F +P + T +L +A+ L+++AK+ TEEYM S+AD +V R F
Sbjct: 303 PRGYYGNAFAFPVAIATANELTKKPLEFALRLIKEAKSSVTEEYMRSLADLMVIKGRPSF 362
Query: 358 TTVRSCIVSDLTRFKLHETDFG-WGEPVCGGVAKGGAGLYGGASYIIACKNAKGEDGRVL 416
++ + +VSD+ F + DFG WG+PV GG+ G GAS+ ++ + GE G V+
Sbjct: 363 SSDGAYLVSDVRIFA--DIDFGIWGKPVYGGIGTAGVEDLPGASFYVSFEKRNGEIGIVV 420
Query: 417 VICLPVENMKRFAKELNNM 435
+CLP + M+RF +EL +
Sbjct: 421 PVCLPEKAMQRFVEELEGV 439
>At3g03480 putative hypersensitivity-related gene
Length = 454
Score = 410 bits (1054), Expect = e-115
Identities = 223/442 (50%), Positives = 291/442 (65%), Gaps = 10/442 (2%)
Query: 3 SSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKD 62
S+ L+FKV R + ELV PA TP E+K LSDIDDQ GLRF IP I YR S +D D
Sbjct: 10 STTTGLSFKVHRQQRELVTPAKPTPRELKPLSDIDDQQGLRFQIPVIFFYRPNLS-SDLD 68
Query: 63 PVQAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETP-- 120
PVQ I++AL+ LVYYYPFAGRL+E RKL VDCTGEGV+FIEA ADV+L E E
Sbjct: 69 PVQVIKKALADALVYYYPFAGRLRELSNRKLAVDCTGEGVLFIEAEADVALAELEEADAL 128
Query: 121 LPPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSA 180
LPPFP EELL+DV GS VL++PLLL+QVTRLKC GFI ALRFNHTM+DGAGL FL +
Sbjct: 129 LPPFPFLEELLFDVEGSSDVLNTPLLLVQVTRLKCCGFIFALRFNHTMTDGAGLSLFLKS 188
Query: 181 LAEMAQGASQPLTPPVWCRELL-MARDPPRITCNHYEF-EQVPSD--STEEGAITRSFFF 236
L E+A G P PPVW R LL ++ R+T H E+ +QV D +T ++RSFFF
Sbjct: 189 LCELACGLHAPSVPPVWNRHLLTVSASEARVTHTHREYDDQVGIDVVATGHPLVSRSFFF 248
Query: 237 GSNEIAALRRLVPLDLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRF-NP 295
+ EI+A+R+L+P DL H ++F+ +++ W CRT AL P+ ++RL I+N+R++ NP
Sbjct: 249 RAEEISAIRKLLPPDL-HNTSFEALSSFLWRCRTIALNPDPNTEMRLTCIINSRSKLRNP 307
Query: 296 PIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRC 355
P+ GYYGN F PA + T L +A+ L+++ K+ TE+Y+ S+ + R
Sbjct: 308 PLEPGYYGNVFVIPAAIATARDLIEKPLEFALRLIQETKSSVTEDYVRSVTALMATRGRP 367
Query: 356 LFTTVRSCIVSDLTRFKLHETDFG-WGEPVCGGVAKGGAGLYGGASYIIACKNAKGEDGR 414
+F + I+SDL F L + DFG WG+PV GG AK G L+ G S+ + KN KGE G
Sbjct: 368 MFVASGNYIISDLRHFDLGKIDFGPWGKPVYGGTAKAGIALFPGVSFYVPFKNKKGETGT 427
Query: 415 VLVICLPVENMKRFAKELNNMI 436
V+ I LPV M+ F ELN ++
Sbjct: 428 VVAISLPVRAMETFVAELNGVL 449
>At3g48720 unknown protein
Length = 430
Score = 233 bits (593), Expect = 2e-61
Identities = 137/391 (35%), Positives = 212/391 (54%), Gaps = 16/391 (4%)
Query: 7 SLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQA 66
S F V R PEL+PP S TP+ LS++D + I + Y S +++
Sbjct: 4 SSEFIVTRKNPELIPPVSETPNGHYYLSNLDQNIAI---IVKTLYYYKSESRTNQESYNV 60
Query: 67 IRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPP-FP 125
I+++LS LV+YYP AGRL P K+ V+CTGEGV+ +EA A+ + E
Sbjct: 61 IKKSLSEVLVHYYPVAGRLTISPEGKIAVNCTGEGVVVVEAEANCGIDTIKEAISENRME 120
Query: 126 CFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMA 185
E+L+YDVPG+ +L+ P +++QVT KCGGF++ L +H M DG +FL++ EMA
Sbjct: 121 TLEKLVYDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMSHNMFDGVAAAEFLNSWCEMA 180
Query: 186 QGASQPLT-PPVWCRELLMARDPPRITCNHYEFEQVPSDST------EEGAITRSFFFGS 238
+G PL+ PP R +L +R+PP+I H EF+++ S EE I +SF F
Sbjct: 181 KGL--PLSVPPFLDRTILRSRNPPKIEFPHNEFDEIEDISDTGKIYDEEKLIYKSFLFEP 238
Query: 239 NEIAALRRLVPLDLRH--CSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPP 296
++ L+ + + + STF +T W R +AL+ P ++L+ + R+RF P
Sbjct: 239 EKLEKLKIMAIEENNNNKVSTFQALTGFLWKSRCEALRFKPDQRVKLLFAADGRSRFIPR 298
Query: 297 IPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCL 356
+P GY GN +VT+ G+L GN ++V LV++ T+ +M S D+ NR
Sbjct: 299 LPQGYCGNGIVLTGLVTSSGELVGNPLSHSVGLVKRLVELVTDGFMRSAMDYFEVNRTRP 358
Query: 357 FTTVRSCIVSDLTRFKLHETDFGWGEPVCGG 387
+ +++ ++ LH+ DFGWGEPV G
Sbjct: 359 SMNA-TLLITSWSKLTLHKLDFGWGEPVFSG 388
>At5g41040 N-hydroxycinnamoyl/benzoyltransferase-like protein
Length = 457
Score = 230 bits (587), Expect = 1e-60
Identities = 139/395 (35%), Positives = 216/395 (54%), Gaps = 22/395 (5%)
Query: 7 SLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQA 66
++ +V + +P LV P S T + LS++D + + I ++ E +++ VQ
Sbjct: 28 NIHLEVHQKEPALVKPESETRKGLYFLSNLDQN--IAVIVRTIYCFKSE-ERGNEEAVQV 84
Query: 67 IRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPC 126
I++ALS+ LV+YYP AGRL P KL VDCT EGV+F+EA A+ + E G+ P
Sbjct: 85 IKKALSQVLVHYYPLAGRLTISPEGKLTVDCTEEGVVFVEAEANCKMDEIGDITKPDPET 144
Query: 127 FEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQ 186
+L+YDV ++ +L+ P + QVT+ KCGGF++ L NH M DG G +F+++ ++A+
Sbjct: 145 LGKLVYDVVDAKNILEIPPVTAQVTKFKCGGFVLGLCMNHCMFDGIGAMEFVNSWGQVAR 204
Query: 187 GASQPL-TPPVWCRELLMARDPPRITCNHYEFEQVPSDS------TEEGAITRSFFFGSN 239
G PL TPP R +L AR+PP+I H EFE++ S T+E + RSF F
Sbjct: 205 GL--PLTTPPFSDRTILNARNPPKIENLHQEFEEIEDKSNINSLYTKEPTLYRSFCFDPE 262
Query: 240 EIAALR-----RLVPLDLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFN 294
+I L+ L C++F+ ++A W RTK+L++ +L+ V+ R +F
Sbjct: 263 KIKKLKLQATENSESLLGNSCTSFEALSAFVWRARTKSLKMLSDQKTKLLFAVDGRAKFE 322
Query: 295 PPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRR 354
P +P GY+GN + G+L +AV LVR+A T+ YM S D+ R
Sbjct: 323 PQLPKGYFGNGIVLTNSICEAGELIEKPLSFAVGLVREAIKMVTDGYMRSAIDYFEVTRA 382
Query: 355 --CLFTTVRSCIVSDLTRFKLHETDFGWGEPVCGG 387
L +T+ +++ +R H TDFGWGEP+ G
Sbjct: 383 RPSLSSTL---LITTWSRLGFHTTDFGWGEPILSG 414
>At5g63560 acyltransferase-like protein
Length = 426
Score = 229 bits (585), Expect = 2e-60
Identities = 138/385 (35%), Positives = 207/385 (52%), Gaps = 18/385 (4%)
Query: 12 VQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQAL 71
V R +P LV PAS TP + LS++D + I Y S ++++ + I+++L
Sbjct: 9 VTRKEPVLVSPASETPKGLHYLSNLDQNIAI---IVKTFYYFKSNSRSNEESYEVIKKSL 65
Query: 72 SRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGE--TPLPPFPCFEE 129
S LV+YYP AGRL P K+ VDCTGEGV+ +EA A+ + + + + + E+
Sbjct: 66 SEVLVHYYPAAGRLTISPEGKIAVDCTGEGVVVVEAEANCGIEKIKKAISEIDQPETLEK 125
Query: 130 LLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGAS 189
L+YDVPG+ +L+ P +++QVT KCGGF++ L NH M DG +FL++ AE A+G
Sbjct: 126 LVYDVPGARNILEIPPVVVQVTNFKCGGFVLGLGMNHNMFDGIAAMEFLNSWAETARGL- 184
Query: 190 QPLT-PPVWCRELLMARDPPRITCNHYEFEQVPSDS------TEEGAITRSFFFGSNEIA 242
PL+ PP R LL R PP+I H EFE + S ++E + +SF FG +
Sbjct: 185 -PLSVPPFLDRTLLRPRTPPKIEFPHNEFEDLEDISGTGKLYSDEKLVYKSFLFGPEK-- 241
Query: 243 ALRRLVPLDLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYY 302
L RL + +TF +T W R +AL L P I+L+ + R+RF P +P GY
Sbjct: 242 -LERLKIMAETRSTTFQTLTGFLWRARCQALGLKPDQRIKLLFAADGRSRFVPELPKGYS 300
Query: 303 GNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRS 362
GN + VTT G++ N ++V LV++A + +M S D+ R T +
Sbjct: 301 GNGIVFTYCVTTAGEVTLNPLSHSVCLVKRAVEMVNDGFMRSAIDYFEVTRARPSLTA-T 359
Query: 363 CIVSDLTRFKLHETDFGWGEPVCGG 387
+++ + H DFGWGEPV G
Sbjct: 360 LLITSWAKLSFHTKDFGWGEPVVSG 384
>At1g27620 putative hypersensitivity-related protein
Length = 442
Score = 197 bits (502), Expect = 7e-51
Identities = 134/424 (31%), Positives = 209/424 (48%), Gaps = 28/424 (6%)
Query: 16 KPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTL 75
+P L+ P S TP+ LS++DD LRF+I ++ +++ S ++ +LSR L
Sbjct: 13 QPTLITPLSPTPNHSLYLSNLDDHHFLRFSIKYLYLFQKSISPL------TLKDSLSRVL 66
Query: 76 VYYYPFAGRLK-EGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDV 134
V YYPFAGR++ G KL VDC GEG +F EA D++ +F + P + +LL+ V
Sbjct: 67 VDYYPFAGRIRVSDEGSKLEVDCNGEGAVFAEAFMDITCQDFVQLSPKPNKSWRKLLFKV 126
Query: 135 PGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTP 194
++ LD P L+IQVT L+CGG I+ NH + DG G QFL A A + T
Sbjct: 127 Q-AQSFLDIPPLVIQVTYLRCGGMILCTAINHCLCDGIGTSQFLHAWAHATTSQAHLPTR 185
Query: 195 PVWCRELLMARDPPRITCNHYEFEQ---VPSDST--------EEGAITRSFFFGSNEIAA 243
P R +L R+PPR+T +H F + V ST + + F + +
Sbjct: 186 PFHSRHVLDPRNPPRVTHSHPGFTRTTTVDKSSTFDISKYLQSQPLAPATLTFNQSHLLR 245
Query: 244 LRRLVPLDLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYG 303
L++ L+ C+TF+ + A W ++L L ++L+ VN R R P +P GYYG
Sbjct: 246 LKKTCAPSLK-CTTFEALAANTWRSWAQSLDLPMTMLVKLLFSVNMRKRLTPELPQGYYG 304
Query: 304 NCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRSC 363
N F + V L + +AV+ +++AK++ T+EY+ S D L+ ++ S
Sbjct: 305 NGFVLACAESKVQDLVNGNIYHAVKSIQEAKSRITDEYVRSTID-LLEDKTVKTDVSCSL 363
Query: 364 IVSDLTRFKLHETDFGWGEPVCGGVAKGGAGLYGGASYIIACKNAKGEDGRVLVICLPVE 423
++S + L E D G G+P+ G Y + A D + + LP E
Sbjct: 364 VISQWAKLGLEELDLGGGKPMYMGPLTSDI-------YCLFLPVASDNDAIRVQMSLPEE 416
Query: 424 NMKR 427
+KR
Sbjct: 417 VVKR 420
>At3g62160 unknown protein
Length = 428
Score = 184 bits (466), Expect = 1e-46
Identities = 132/402 (32%), Positives = 199/402 (48%), Gaps = 54/402 (13%)
Query: 8 LTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAI 67
+ F + R L+ +TP V LS ID+ LR I ++ H P D +AI
Sbjct: 1 MAFSLTRVNRVLIQACETTPSVVLDLSLIDNIPVLRCFARTIHVFTHGP-----DAARAI 55
Query: 68 RQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGE-GVMFIEANADVSLVEFGETP-LPPFP 125
++AL++ LV YYP +GRLKE KL +DCTG+ G+ F++A DV L G L P
Sbjct: 56 QEALAKALVSYYPLSGRLKELNQGKLQIDCTGKTGIWFVDAVTDVKLESVGYFDNLMEIP 115
Query: 126 CFEELLYD-VPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEM 184
+++LL D +P + PL+ +QVT+ CGGF+M LRF H + DG G QFL+A+ E+
Sbjct: 116 -YDDLLPDQIPKDDDA--EPLVQMQVTQFSCGGFVMGLRFCHAICDGLGAAQFLTAVGEI 172
Query: 185 AQGASQPLTPPVWCRELL----MARDP-------------PRITCNHYEFEQVPSDSTEE 227
A G + PVW R+ + A D P H F+ +PSD E
Sbjct: 173 ACGQTNLGVSPVWHRDFIPQQHRANDVISPAIPPPFPPAFPEYKLEHLSFD-IPSDLIER 231
Query: 228 GAITRSFFFGSNEIAALRRLVPLDLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIV 287
R F + EI CS F+VI ACFW RT+A+ L +++L+
Sbjct: 232 --FKREFRASTGEI-------------CSAFEVIAACFWKLRTEAINLREDTEVKLVFFA 276
Query: 288 NTRNRFNPPIPVGYYGNCFTYPAVVTTVGKLCG--NSFGYAVELVRKAKAQATEEYMHSM 345
N R+ NPP+P+G+YGNCF +P ++ S V+L+++AK + EE+
Sbjct: 277 NCRHLVNPPLPIGFYGNCF-FPVKISAESHEISKEKSVFDVVKLIKQAKTKLPEEF---- 331
Query: 346 ADFLVANRRCLFTTV---RSCIVSDLTRFKLHETDFGWGEPV 384
F+ + F + +S+ + ++ D+GWG P+
Sbjct: 332 EKFVSGDGDDPFAPAVGYNTLFLSEWGKLGFNQVDYGWGPPL 373
>At1g03390 hypothetical protein
Length = 461
Score = 181 bits (460), Expect = 5e-46
Identities = 125/388 (32%), Positives = 193/388 (49%), Gaps = 21/388 (5%)
Query: 17 PELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDP--VQAIRQALSRT 74
P PA+ +PH LS++DD G R P + Y PS +++ ++ ++ ALS
Sbjct: 27 PSSPTPANQSPHHSLYLSNLDDIIGARVFTPSVYFY---PSTNNRESFVLKRLQDALSEV 83
Query: 75 LVYYYPFAGRLKEGPGRKLMVDCTGE-GVMFIEANADVSLVEFGETPLPPFPCFEELLYD 133
LV YYP +GRL+E KL V E GV+ + AN+ + L + G+ +P P + L++
Sbjct: 84 LVPYYPLSGRLREVENGKLEVFFGEEQGVLMVSANSSMDLADLGDLTVPN-PAWLPLIFR 142
Query: 134 VPGSE--QVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQP 191
PG E ++L+ PLL+ QVT CGGF + +R H + DG G QFL + A A+
Sbjct: 143 NPGEEAYKILEMPLLIAQVTFFTCGGFSLGIRLCHCICDGFGAMQFLGSWAATAKTGKLI 202
Query: 192 LTP-PVWCRELLMARDPPRITCNHYEFEQVPSDSTEEGAI-----TRSFFFGSNEIAALR 245
P PVW RE R+PP + H+E+ + S ++ + + S E
Sbjct: 203 ADPEPVWDRETFKPRNPPMVKYPHHEYLPIEERSNLTNSLWDTKPLQKCYRISKEFQCRV 262
Query: 246 RLVPL---DLRHCSTFDVITACFWYCRTKALQLAPHD-DIRLMTIVNTRNRFNP-PIPVG 300
+ + CSTFD + A W KAL + P D ++RL VN R R + G
Sbjct: 263 KSIAQGEDPTLVCSTFDAMAAHIWRSWVKALDVKPLDYNLRLTFSVNVRTRLETLKLRKG 322
Query: 301 YYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTV 360
+YGN +++V L +S LV+ A+ + +E+Y+ SM D++ R
Sbjct: 323 FYGNVVCLACAMSSVESLINDSLSKTTRLVQDARLRVSEDYLRSMVDYVDVKRPKRLEFG 382
Query: 361 RSCIVSDLTRFKLHET-DFGWGEPVCGG 387
++ TRF+++ET DFGWG+PV G
Sbjct: 383 GKLTITQWTRFEMYETADFGWGKPVYAG 410
>At2g40230 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 433
Score = 176 bits (446), Expect = 2e-44
Identities = 125/387 (32%), Positives = 207/387 (53%), Gaps = 32/387 (8%)
Query: 19 LVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQA--IRQALSRTLV 76
++ P+ TP V LS +D Q LRF I ++++Y P ++D + + ++ ALSR LV
Sbjct: 12 VITPSDQTPSSVLSLSALDSQLFLRFTIEYLLVY---PPVSDPEYSLSGRLKSALSRALV 68
Query: 77 YYYPFAGRLKEGP--GRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDV 134
Y+PF+GR++E P G L V+C G+G +F+EA +D+ + P + +LL
Sbjct: 69 PYFPFSGRVREKPDGGGGLEVNCRGQGALFLEAVSDILTCLDFQKPPRHVTSWRKLL--- 125
Query: 135 PGSEQVLD----SPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGA-- 188
S V+D +P L++Q+T L+ GG +A+ NH +SDG G +FL+ AE+++ +
Sbjct: 126 --SLHVIDVLAGAPPLVVQLTWLRDGGAALAVGVNHCVSDGIGSAEFLTLFAELSKDSLS 183
Query: 189 -SQPLTPPVWCRELLMARDPPRITCNHYEFEQVP------SDSTEEGAITRSFFFGSNEI 241
++ +W R+LLM P R + +H EF +VP + E + S F ++
Sbjct: 184 QTELKRKHLWDRQLLMP-SPIRDSLSHPEFNRVPDLCGFVNRFNAERLVPTSVVFERQKL 242
Query: 242 AALRRLVP----LDLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPI 297
L++L + +H S F+V++A W ++L L + ++L+ VN R+R P +
Sbjct: 243 NELKKLASRLGEFNSKHTS-FEVLSAHVWRSWARSLNLPSNQVLKLLFSVNIRDRVKPSL 301
Query: 298 PVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLF 357
P G+YGN F TTV L YA LV++AK + +EY+ S+ + V+ R
Sbjct: 302 PSGFYGNAFVVGCAQTTVKDLTEKGLSYATMLVKQAKERVGDEYVRSVVE-AVSKERASP 360
Query: 358 TTVRSCIVSDLTRFKLHETDFGWGEPV 384
+V I+S +R L + DFG G+PV
Sbjct: 361 DSVGVLILSQWSRLGLEKLDFGLGKPV 387
>At2g19070 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 160 bits (404), Expect = 2e-39
Identities = 129/429 (30%), Positives = 201/429 (46%), Gaps = 48/429 (11%)
Query: 38 DQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDC 97
DQ G +IP + Y + V+ ++ +LSR LV++YP AGRL+ P + ++C
Sbjct: 29 DQVGTITHIPTLYFYDKPSESFQGNVVEILKTSLSRVLVHFYPMAGRLRWLPRGRFELNC 88
Query: 98 TGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGG 157
EGV FIEA ++ L +F + P P FE L+ V + PL L QVT+ KCGG
Sbjct: 89 NAEGVEFIEAESEGKLSDFKD--FSPTPEFENLMPQVNYKNPIETIPLFLAQVTKFKCGG 146
Query: 158 FIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPL-TPPVWCRELLMARDP--PRIT--- 211
+++ +H + DG +S +A+G +PL T P R++L A +P P ++
Sbjct: 147 ISLSVNVSHAIVDGQSALHLISEWGRLARG--EPLETVPFLDRKILWAGEPLPPFVSPPK 204
Query: 212 CNHYEFEQVP-----SDSTEEG---AITRSFFFGSNEIAALRRLVPLD-----LRHCSTF 258
+H EF+Q P +D+ EE I ++++ LR + + +
Sbjct: 205 FDHKEFDQPPFLIGETDNVEERKKKTIVVMLPLSTSQLQKLRSKANGSKHSDPAKGFTRY 264
Query: 259 DVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTTVGKL 318
+ +T W C KA +P L ++TR+R PP+P GY+GN +T G+L
Sbjct: 265 ETVTGHVWRCACKARGHSPEQPTALGICIDTRSRMEPPLPRGYFGNATLDVVAASTSGEL 324
Query: 319 CGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCL----------------FTTVRS 362
N G+A L+ KA T EY+ ++L N++ L +
Sbjct: 325 ISNELGFAASLISKAIKNVTNEYVMIGIEYL-KNQKDLKKFQDLHALGSTEGPFYGNPNL 383
Query: 363 CIVSDLTRFKLHETDFGWGEPVCGGVAKGGAGLYGGASYIIACKNAKGEDGRV-LVICLP 421
+VS LT ++ DFGWG+ G G + G S I+ +N EDG V L CL
Sbjct: 384 GVVSWLT-LPMYGLDFGWGKEFYTG---PGTHDFDGDSLILPDQN---EDGSVILATCLQ 436
Query: 422 VENMKRFAK 430
V +M+ F K
Sbjct: 437 VAHMEAFKK 445
>At5g48930 anthranilate N-benzoyltransferase
Length = 433
Score = 159 bits (403), Expect = 2e-39
Identities = 125/433 (28%), Positives = 199/433 (45%), Gaps = 31/433 (7%)
Query: 19 LVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTLVYY 78
+V PA+ TP S++D RF+ P + YR + DP Q +++ALS+ LV +
Sbjct: 10 MVRPATETPITNLWNSNVDLVIP-RFHTPSVYFYRPTGASNFFDP-QVMKEALSKALVPF 67
Query: 79 YPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPGSE 138
YP AGRLK ++ +DC G GV+F+ A+ + +FG+ P +L+ +V S
Sbjct: 68 YPMAGRLKRDDDGRIEIDCNGAGVLFVVADTPSVIDDFGD--FAPTLNLRQLIPEVDHSA 125
Query: 139 QVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTPPVWC 198
+ PLL++QVT KCGG + + H +DG F++ ++MA+G + PP
Sbjct: 126 GIHSFPLLVLQVTFFKCGGASLGVGMQHHAADGFSGLHFINTWSDMARGLDLTI-PPFID 184
Query: 199 RELLMARDPPRITCNHYEFEQV--------PSDSTEEGAITRSFFFGSNEIAALRRLVPL 250
R LL ARDPP+ +H E++ PS S E F +++ AL+
Sbjct: 185 RTLLRARDPPQPAFHHVEYQPAPSMKIPLDPSKSGPENTTVSIFKLTRDQLVALKAKSKE 244
Query: 251 D--LRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTY 308
D S+++++ W KA L + +L + R+R P +P GY+GN
Sbjct: 245 DGNTVSYSSYEMLAGHVWRSVGKARGLPNDQETKLYIATDGRSRLRPQLPPGYFGNVIFT 304
Query: 309 PAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRSCI---- 364
+ G L YA + + + Y+ S D+L + L VR
Sbjct: 305 ATPLAVAGDLLSKPTWYAAGQIHDFLVRMDDNYLRSALDYL-EMQPDLSALVRGAHTYKC 363
Query: 365 ----VSDLTRFKLHETDFGWGEPVCGGVAKGGAGLYGGASYIIACKNAKGEDGRVLV-IC 419
++ R +++ DFGWG P+ G G Y G S+++ + DG + V I
Sbjct: 364 PNLGITSWVRLPIYDADFGWGRPIFMG---PGGIPYEGLSFVLP---SPTNDGSLSVAIA 417
Query: 420 LPVENMKRFAKEL 432
L E+MK F K L
Sbjct: 418 LQSEHMKLFEKFL 430
>At5g57840 N-hydroxycinnamoyl/benzoyltransferase
Length = 443
Score = 156 bits (395), Expect = 2e-38
Identities = 122/443 (27%), Positives = 201/443 (44%), Gaps = 35/443 (7%)
Query: 11 KVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEP-SMADKDPVQAIRQ 69
K++ + +V PA TP LS++D ++ + +Y ++P S +D+ Q++ +
Sbjct: 2 KIRVKQATIVKPAEETPTHNLWLSNLDL---IQVRLHMGTLYFYKPCSSSDRPNTQSLIE 58
Query: 70 ALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEE 129
ALS+ LV++YP AGRL++ +L V C GEGV+F+EA +D ++ + G L +
Sbjct: 59 ALSKVLVFFYPAAGRLQKNTNGRLEVQCNGEGVLFVEAESDSTVQDIG--LLTQSLDLSQ 116
Query: 130 LLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGAS 189
L+ V + + PLLL QVT KCG + +HT + L + A + A+G
Sbjct: 117 LVPTVDYAGDISSYPLLLFQVTYFKCGTICVGSSIHHTFGEATSLGYIMEAWSLTARGLL 176
Query: 190 QPLTPPVWCRELLMARDPPRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRRLVP 249
LT P R +L AR+PP H E++ P + ++ S+ A +L
Sbjct: 177 VKLT-PFLDRTVLHARNPPSSVFPHTEYQSPPFHNHPMKSLAYRSNPESDSAIATLKLTR 235
Query: 250 LDLR-----------HCSTFDVITACFWYCRTKALQ-LAPHDDIRLMTIVNTRNRFNPPI 297
L L+ ST++V+ A W C + A + L+ RL I++ R R P +
Sbjct: 236 LQLKALKARAEIADSKFSTYEVLVAHTWRCASFANEDLSEEHSTRLHIIIDGRPRLQPKL 295
Query: 298 PVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFL-------- 349
P GY GN + V+ +G SF VE V + + EY+ S D+L
Sbjct: 296 PQGYIGNTLFHARPVSPLGAFHRESFSETVERVHREIRKMDNEYLRSAIDYLERHPDLDQ 355
Query: 350 --VANRRCLFTTVRSCIVSDLTRFKLHETDFGWGEPVCGGVAKGGAGLYGGASYIIACKN 407
+F+ + + L + +E DFGWG+ K + L G ++ N
Sbjct: 356 LVPGEGNTIFSCAANFCIVSLIKQTAYEMDFGWGK----AYYKRASHLNEGKGFVTG--N 409
Query: 408 AKGEDGRVLVICLPVENMKRFAK 430
E +L +CL + +F K
Sbjct: 410 PDEEGSMMLTMCLKKTQLSKFRK 432
>At5g07080 unknown protein
Length = 450
Score = 144 bits (364), Expect = 7e-35
Identities = 123/414 (29%), Positives = 177/414 (42%), Gaps = 49/414 (11%)
Query: 3 SSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADK- 61
SSP L V++ + +V P+ +TP LS +D+ L I +Y PS D
Sbjct: 7 SSPSPLV--VKKSQVVIVKPSKATPDVSLSLSTLDNDPYLETLAKTIYVYA-PPSPNDHV 63
Query: 62 -DPVQAIRQALSRTLVYYYPFAGRLKEGP-GRKLMVDCT-GEGVMFIEANADVSLVEFGE 118
DP +QALS LVYYYP AG+L G +L + C+ EGV F+ A AD +L
Sbjct: 64 HDPASLFQQALSDALVYYYPLAGKLHRGSHDHRLELRCSPAEGVPFVRATADCTLSSLNY 123
Query: 119 TPLPPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFL 178
++ + DV D L +Q+T CGG +A +H++ DG G QF
Sbjct: 124 LKDMDTDLYQLVPCDVAAPSG--DYNPLALQITLFACGGLTLATALSHSLCDGFGASQFF 181
Query: 179 SALAEMAQGASQPLTPPVWCRELLMARDPPRITCNHYEFEQVPSDSTEEGAITRSFFFGS 238
L E+A G +QP PVW R R+T N++ +D EEG + FG
Sbjct: 182 KTLTELAAGKTQPSIIPVWDRH--------RLTSNNFSL----NDQVEEGQAPKLVDFGE 229
Query: 239 N-EIAALRRLVPLDLRHC---------------------STFDVITACFWYCRTKALQLA 276
AA P + C +T +++ A W R +AL+L+
Sbjct: 230 ACSSAATSPYTPSNDMVCEILNVTSEDITQLKEKVAGVVTTLEILAAHVWRARCRALKLS 289
Query: 277 PHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQ 336
P V R PP+P GYYGN F V G+L + + V+L+++AK
Sbjct: 290 PDGTSLFGMAVGIRRIVEPPLPEGYYGNAFVKANVAMKAGELSNSPLSHVVQLIKEAKKA 349
Query: 337 ATE-----EYMHSMADFLVANRRCLFTTVRSCIVSDLTRF-KLHETDFGWGEPV 384
A E E + L N C +++D + L E DFG+G V
Sbjct: 350 AQEKRYVLEQLRETEKTLKMNVACEGGKGAFMLLTDWRQLGLLDEIDFGYGGSV 403
>At1g28680 anthranilate N-hydroxycinnamoyl/benzoyltransferase like
protein
Length = 451
Score = 144 bits (364), Expect = 7e-35
Identities = 101/363 (27%), Positives = 176/363 (47%), Gaps = 13/363 (3%)
Query: 33 LSDIDDQDGLRFNIPFIMMYRHEPS-MADKDPVQAIRQALSRTLVYYYPFAGRLKEGPG- 90
LS +D+ + L + ++ +Y S +A + P + +L+ LV+YYP AG L+
Sbjct: 27 LSHLDNDNNLHVSFRYLRVYSSSSSTVAGESPSAVVSASLATALVHYYPLAGSLRRSASD 86
Query: 91 -RKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPGSEQVLDSPLLLIQ 149
R ++ G+ V + A + +L G P P F E L P E+ + +P +L Q
Sbjct: 87 NRFELLCSAGQSVPLVNATVNCTLESVGYLDGPD-PGFVERLVPDPTREEGMVNPCIL-Q 144
Query: 150 VTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTPPVWCRE-LLMARDPP 208
VT +CGG+++ +H + DG G F +A+AE+A+GA++ PVW RE LL R+ P
Sbjct: 145 VTMFQCGGWVLGASIHHAICDGLGASLFFNAMAELARGATKISIEPVWDRERLLGPREKP 204
Query: 209 RITCNHYEFEQVPSD----STEEGAITRSFFFGSNE-IAALR-RLVPLDLRHCSTFDVIT 262
+ +F + D G + R FF +++ + L+ +L+ + +TF+ +
Sbjct: 205 WVGAPVRDFLSLDKDFDPYGQAIGDVKRDCFFVTDDSLDQLKAQLLEKSGLNFTTFEALG 264
Query: 263 ACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTTVGKLCGNS 322
A W + +A + ++++ + +N R NPP+P GY+GN G+L
Sbjct: 265 AYIWRAKVRAAKTEEKENVKFVYSINIRRLMNPPLPKGYWGNGCVPMYAQIKAGELIEQP 324
Query: 323 FGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRSCIVSDLTRFKLHET-DFGWG 381
EL++++K+ ++EY+ S DF + + R+ H T DFGWG
Sbjct: 325 IWKTAELIKQSKSNTSDEYVRSFIDFQELHHKDGINAGTGVTGFTDWRYLGHSTIDFGWG 384
Query: 382 EPV 384
PV
Sbjct: 385 GPV 387
>At3g47170 hypersensitivity-related protein-like protein
Length = 447
Score = 142 bits (357), Expect = 5e-34
Identities = 106/369 (28%), Positives = 166/369 (44%), Gaps = 34/369 (9%)
Query: 1 MDSSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMAD 60
M S S F V++ + LV P+ TP LS ID+ + + ++ P + D
Sbjct: 1 MPRSDNSCPFVVEKSEVVLVKPSKPTPDVTLSLSTIDNDPNNEVILDILCVFSPNPYVRD 60
Query: 61 KD---PVQAIRQALSRTLVYYYPFAGRL-KEGPGRKLMVDCT-GEGVMFIEANADVSLVE 115
P I+ ALS LVYYYP AG+L + + L +DCT G+GV I+A A +L
Sbjct: 61 HTNYHPASFIQLALSNALVYYYPLAGKLHRRTSDQTLQLDCTQGDGVPLIKATASCTLSS 120
Query: 116 FGETPLPPFPCFEELLYDVPGSEQVLDSPL----LLIQVTRLKCGGFIMALRFNHTMSDG 171
L E VP + + L L +Q+T+ CGG + +HT+ DG
Sbjct: 121 LNY--LESGNHLEATYQLVPSHDTLKGCNLGYRPLALQITKFTCGGMTIGSVHSHTVCDG 178
Query: 172 AGLKQFLSALAEMAQGASQPLTPPVWCRELLMA------------RDPPRITCNHYEFEQ 219
G+ QF A+ E+A G +QP PVW R ++ + ++ P++ E +
Sbjct: 179 IGMAQFSQAILELAAGRAQPTVIPVWDRHMITSNQISTLCKLGNDKNNPKLV--DVEKDC 236
Query: 220 VPSDSTEEGAITRSFFFGSNEIAALRRLVPLDLR---------HCSTFDVITACFWYCRT 270
D+ E + S +I L+ ++ D +T +V+ A W R
Sbjct: 237 SSPDTPTEDMVREILNITSEDITKLKNIIIEDENLTNEKEKNMKITTVEVLAAYVWRARC 296
Query: 271 KALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELV 330
+A++L P L+ V+ R+ PP+P GYYGN FT+ +V T +L V L+
Sbjct: 297 RAMKLNPDTITDLVISVSIRSSIEPPLPEGYYGNAFTHASVALTAKELSKTPISRLVRLI 356
Query: 331 RKAKAQATE 339
+ AK A +
Sbjct: 357 KDAKRAALD 365
>At2g25150 unknown protein
Length = 461
Score = 134 bits (338), Expect = 8e-32
Identities = 126/453 (27%), Positives = 204/453 (44%), Gaps = 30/453 (6%)
Query: 6 PSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQ 65
P L +++ ELV P+ T E LS +D+ I +++ D DPV
Sbjct: 7 PILPLLLEKKPVELVKPSKHTHCETLSLSTLDNDPFNEVMYATIYVFKANGKNLD-DPVS 65
Query: 66 AIRQALSRTLVYYYPFAGRLKEGPGR-KLMVDCTGEGVMFIEANA--DVSLVEFGETPLP 122
+R+ALS LV+YYP +G+L KL + GEGV F A + D+S + + E
Sbjct: 66 LLRKALSELLVHYYPLSGKLMRSESNGKLQLVYLGEGVPFEVATSTLDLSSLNYIENLDD 125
Query: 123 PFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALA 182
++ V PL L QVT+ CGGF + H + DG G+ Q + AL
Sbjct: 126 QVALRLVPEIEIDYESNVCYHPLAL-QVTKFACGGFTIGTALTHAVCDGYGVAQIIHALT 184
Query: 183 EMAQGASQPLTPPVWCRELLMAR---DPPRITCNHYE-FEQVPSDSTEEGAITRSFFFGS 238
E+A G ++P VW RE L+ + P ++ +H + F + +T + +
Sbjct: 185 ELAAGKTEPSVKSVWQRERLVGKIDNKPGKVPGSHIDGFLATSAYLPTTDVVTETINIRA 244
Query: 239 NEIAALRRLVPLDLRHC-------STFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRN 291
+I +RL ++ C +T++V+++ W R++AL+L P L V R+
Sbjct: 245 GDI---KRLKDSMMKECEYLKESFTTYEVLSSYIWKLRSRALKLNPDGITVLGVAVGIRH 301
Query: 292 RFNPPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVA 351
+PP+P GYYGN + V TV +L +S V+KAK A E+ + + L
Sbjct: 302 VLDPPLPKGYYGNAYIDVYVELTVRELEESSISNIANRVKKAKKTAYEK--GYIEEELKN 359
Query: 352 NRRCLFTTVRSCIVSD----LTRFK----LHETDFGWGEPV-CGGVAKGGAGLYGGASYI 402
R + VSD LT ++ DFGW EPV + + + ++ G
Sbjct: 360 TERLMRDDSMFEGVSDGLFFLTDWRNIGWFGSMDFGWNEPVNLRPLTQRESTVHVGMILK 419
Query: 403 IACKNAKGEDGRVLVICLPVENMKRFAKELNNM 435
+ + E G +++ LP + M F +E+ M
Sbjct: 420 PSKSDPSMEGGVKVIMKLPRDAMVEFKREMATM 452
>At2g23510 putative anthranilate
N-hydroxycinnamoyl/benzoyltransferase
Length = 451
Score = 129 bits (324), Expect = 3e-30
Identities = 120/416 (28%), Positives = 184/416 (43%), Gaps = 63/416 (15%)
Query: 7 SLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKD--PV 64
S+ V++ E+V P+ P + LS +D+ +++ + ++AD D P
Sbjct: 8 SIPLMVEKMLTEMVKPSKHIPQQTLNLSTLDNDPYNEVIYKACYVFKAK-NVADDDNRPE 66
Query: 65 QAIRQALSRTLVYYYPFAGRLK-EGPGRKLMVDCTGEG--VMFIEANADVSLVEF----- 116
+R+ALS L YYYP +G LK + RKL + C G+G V F A A+V L
Sbjct: 67 ALLREALSDLLGYYYPLSGSLKRQESDRKLQLSCGGDGGGVPFTVATANVELSSLKNLEN 126
Query: 117 --GETPLPPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGL 174
+T L P L D+ G +QVT+ +CGGFI+ + +H M DG G
Sbjct: 127 IDSDTALNFLPV---LHVDIDGYRP------FALQVTKFECGGFILGMAMSHAMCDGYGE 177
Query: 175 KQFLSALAEMAQGASQPLTPPVWCRELLMAR---DPPRITCNHYEFEQVPSDSTEEGAIT 231
+ AL ++A G +P+ P+W RE L+ + D P VP D T
Sbjct: 178 GHIMCALTDLAGGKKKPMVTPIWERERLVGKPEDDQPPF---------VPGDDTAASPYL 228
Query: 232 RSFFFGSNEIA----ALRRLVPLDLRH-------CSTFDVITACFWYCRTKALQLAPHDD 280
+ + + +I ++RRL L+ +TF+VI A W R KAL L
Sbjct: 229 PTDDWVTEKITIRADSIRRLKEATLKEYDFSNETITTFEVIGAYLWKSRVKALNLDRDGV 288
Query: 281 IRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEE 340
L V RN +PP+P GYYGN + V T ++ + V+L+++AK A ++
Sbjct: 289 TVLGLSVGIRNVVDPPLPDGYYGNAYIDMYVPLTAREVEEFTISDIVKLIKEAKRNAHDK 348
Query: 341 YMHSMADFL---VANRRCLF---TTVRS------CIVSDLTRFKLHETDFGWGEPV 384
D+L +AN + T++ C+ DFGW EPV
Sbjct: 349 ------DYLQEELANTEKIIKMNLTIKGKKDGLFCLTDWRNIGIFGSMDFGWDEPV 398
>At1g31490 putative protein
Length = 444
Score = 116 bits (290), Expect = 3e-26
Identities = 103/402 (25%), Positives = 180/402 (44%), Gaps = 42/402 (10%)
Query: 19 LVPPASSTPHEVKLL--SDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTLV 76
+V + P +V L S+ D G RF + + Y +P ++ + V++++ +LS+TL
Sbjct: 13 IVKASGDQPEKVNSLNLSNFDLLSG-RFPVTYFYFYPKQPQLSFETIVKSLQSSLSQTLS 71
Query: 77 YYYPFAGRL--KEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDV 134
Y+YPFAG++ E + M+ C G +F+EA A V L L F + L
Sbjct: 72 YFYPFAGQIVPNETSQEEPMIICNNNGALFVEARAHVDLKSLDFYNLDAF--LQSKLV-- 127
Query: 135 PGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLT- 193
QV L IQ T +CGG + F+H + D + +FL+ +E+++ ++P++
Sbjct: 128 ----QVNPDFSLQIQATEFECGGLALTFTFDHALGDASSFGKFLTLWSEISR--NKPVSC 181
Query: 194 PPVWCRELLMARDPPR---------ITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAAL 244
P R LL AR PPR I C+ + + +P T I R + G++ + AL
Sbjct: 182 VPDHRRNLLRARSPPRYDPHLDKTFIKCSEEDIKNIPMSKT---LIKRLYHIGASSLDAL 238
Query: 245 RRLVPLDLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGN 304
+ L ++ + + +A W +++ + H ++ +V+ R R + Y GN
Sbjct: 239 QALATVNGESRTKIEAFSAYVWKKMVDSIE-SGHKTCKMGWLVDGRGRLE-TVTSSYIGN 296
Query: 305 CFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATE--------EYMHSMADFLVANRRCL 356
+ ++ L N +V K+ + T +++ S L+ R L
Sbjct: 297 VLSIAVGEASIENLKKNHVSDIANIVHKSITEVTNDTHFTDLIDWIESHRPGLMLARVVL 356
Query: 357 FTTVRSCIVSDLTRFKLHETDFGWGEP----VCGGVAKGGAG 394
+ ++S RF + E DFG+G P VC V K G G
Sbjct: 357 GQEGPALVLSSGRRFPVAELDFGFGAPFLGTVCSTVEKIGVG 398
>At4g15390 HSR201 like protein
Length = 446
Score = 110 bits (275), Expect = 2e-24
Identities = 109/450 (24%), Positives = 197/450 (43%), Gaps = 52/450 (11%)
Query: 7 SLTFKVQRCKPELVPPASSTPHEVKLL--SDIDDQDGLRFNIPFIMMYRHEPSMADKDPV 64
++T KV+ E++ P+S TP+ ++ L S D + + F+ Y ++ +
Sbjct: 3 TMTMKVETISKEIIKPSSPTPNNLQTLQLSIYDHILPPVYTVAFLF-YTKNDLISQEHTS 61
Query: 65 QAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANAD-VSLVEFGETPLPP 123
++ +LS TL +YP AGR+ + VDCT EG +F++A + L EF P
Sbjct: 62 HKLKTSLSETLTKFYPLAGRITG-----VTVDCTDEGAIFVDARVNNCPLTEF--LKCPD 114
Query: 124 FPCFEELL-YDVPGSEQVLDS--PLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSA 180
F ++LL DV + V + PLLL++ T CGG + + H ++D A + F+ +
Sbjct: 115 FDALQQLLPLDVVDNPYVAAATWPLLLVKATYFGCGGMAIGICITHKIADAASISTFIRS 174
Query: 181 LAEMAQGASQPLTPPVWCRELLMARDPPRITCNHY----EFEQVPSD--STEEGAITRSF 234
A A+G + + P N Y E ++P+D + + +IT+ F
Sbjct: 175 WAATARGEN----------DAAAMESPVFAGANFYPPANEAFKLPADEQAGKRSSITKRF 224
Query: 235 FFGSNEIAALRRLVPLD--LRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNR 292
F ++++ LR + + + + +TA W C + + D L+ + N R++
Sbjct: 225 VFEASKVEDLRTKAASEETVDQPTRVESVTALIWKCFVASSKTTTCDHKVLVQLANLRSK 284
Query: 293 FNPPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAK-------------AQATE 339
+ GN + +VV ++G+ AV +RK K + ++
Sbjct: 285 IPSLLQESSIGN-LMFSSVVLSIGRGGEVKIEEAVRDLRKKKEELGTVILDEGGSSDSSS 343
Query: 340 EYMHSMADFLVAN-RRCLFTTVRSCIVSDLTRFKLHETDFGWGEPVCGGVAKGGAGLYGG 398
+A+ ++ N R + T VS + L+E FGW PV V + + G
Sbjct: 344 MIGSKLANLMLTNYSRLSYETHEPYTVSSWCKLPLYEASFGWDSPV--WVVGNVSPVLGN 401
Query: 399 ASYIIACKNAKGEDGRVLVICLPVENMKRF 428
+ +I K+ +G + + LP ENM F
Sbjct: 402 LAMLIDSKDGQGIEA---FVTLPEENMSSF 428
>At3g50280 anthranilate N-hydroxycinnamoyl/benzoyltransferase - like
protein
Length = 443
Score = 110 bits (274), Expect = 2e-24
Identities = 102/382 (26%), Positives = 157/382 (40%), Gaps = 39/382 (10%)
Query: 26 TPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDP----VQAIRQALSRTLVYYYPF 81
TP ++ LL Q GL F P DP + +R +LS L Y+PF
Sbjct: 28 TPFDLNLLYVDYTQRGLLFPKP--------------DPETHFISRLRTSLSSALDIYFPF 73
Query: 82 AGRLKEGPGRK-----LMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPG 136
AGRL + + ++C G G FI A +D V P P F + Y + G
Sbjct: 74 AGRLNKVENHEDETVSFYINCDGSGAKFIHAVSDSVSVSDLLRPDGSVPDFFRIFYPMNG 133
Query: 137 SEQV--LDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTP 194
+ + L PLL +QVT ++ G FI +NH ++DGA + F +++ +
Sbjct: 134 VKSIDGLSEPLLALQVTEMRDGVFI-GFGYNHMVADGASIWNFFRTWSKICSNGQRENLQ 192
Query: 195 PVWCRELLM--ARDPPRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRRLVPLDL 252
P+ + L + P I + E PS R F F I+ L+ V ++
Sbjct: 193 PLALKGLFVDGMDFPIHIPVSDTETSP-PSRELSPTFKERVFHFTKRNISDLKAKVNGEI 251
Query: 253 ----RHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTY 308
S+ ++A W + L + R V+ R R NPP+ +G+
Sbjct: 252 GLRDHKVSSLQAVSAHMWRSIIRHSGLNQEEKTRCFVAVDLRQRLNPPLDKECFGHVIYN 311
Query: 309 PAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFT------TVRS 362
V TTVG+L G+A + T E A+ V N + + T S
Sbjct: 312 SVVTTTVGELHDQGLGWAFLQINNMLRSLTNEDYRIYAENWVRNMKIQKSGLGSKMTRDS 371
Query: 363 CIVSDLTRFKLHETDFGWGEPV 384
IVS RF++++ DFGWG+P+
Sbjct: 372 VIVSSSPRFEVYDNDFGWGKPI 393
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.140 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,181,847
Number of Sequences: 26719
Number of extensions: 437677
Number of successful extensions: 1119
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 866
Number of HSP's gapped (non-prelim): 85
length of query: 437
length of database: 11,318,596
effective HSP length: 102
effective length of query: 335
effective length of database: 8,593,258
effective search space: 2878741430
effective search space used: 2878741430
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0175.13


