

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0175.11
(923 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
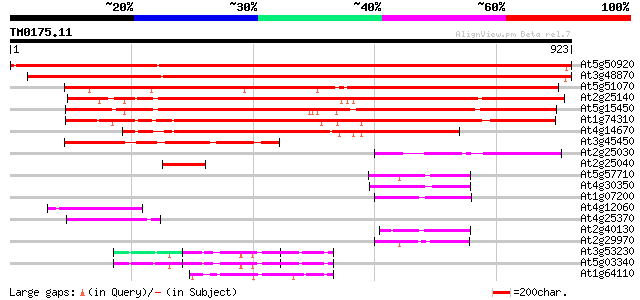
Score E
Sequences producing significant alignments: (bits) Value
At5g50920 ATP-dependent Clp protease, ATP-binding subunit 1511 0.0
At3g48870 AtClpC 1469 0.0
At5g51070 Erd1 protein precursor (sp|P42762) 749 0.0
At2g25140 putative ATP-dependent CLPB protein 703 0.0
At5g15450 clpB heat shock protein-like 702 0.0
At1g74310 heat shock protein 101 640 0.0
At4g14670 heat shock protein like 493 e-139
At3g45450 clpC-like protein 347 1e-95
At2g25030 putative ATP-dependent CLPB protein 194 1e-49
At2g25040 hypothetical protein 92 1e-18
At5g57710 101 kDa heat shock protein; HSP101-like protein 69 9e-12
At4g30350 unknown protein 69 2e-11
At1g07200 unknown protein 64 5e-10
At4g12060 unknown protein 61 3e-09
At4g25370 unknown protein 59 1e-08
At2g40130 unknown protein 59 2e-08
At2g29970 unknown protein 53 7e-07
At3g53230 CDC48 - like protein 49 1e-05
At5g03340 transitional endoplasmic reticulum ATPase 49 2e-05
At1g64110 unknown protein 48 2e-05
>At5g50920 ATP-dependent Clp protease, ATP-binding subunit
Length = 929
Score = 1511 bits (3913), Expect = 0.0
Identities = 787/927 (84%), Positives = 847/927 (90%), Gaps = 8/927 (0%)
Query: 2 SRVLAQSITIPGLVCGRSHGHNNRSTMSRRSLKMM-STLQAPALRMSGFSGLRTYNNLDT 60
+RVLAQS T P L C + + + S SRRS+KMM S LQ LRM GF GLR N LDT
Sbjct: 5 TRVLAQS-TPPSLACYQRNVPSRGSGRSRRSVKMMCSQLQVSGLRMQGFMGLRGNNALDT 63
Query: 61 MLRPGLDFRSKVFGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGT 120
+ + DF SKV K +ASR KAMFERFTEKAIKVIML+QEEARRLGHNFVGT
Sbjct: 64 LGKSRQDFHSKVRQAMNVPKGKASRFTVKAMFERFTEKAIKVIMLAQEEARRLGHNFVGT 123
Query: 121 EQILLGLVGEGTGIAARVLKAMGISLKDARVEVEKTIGRGSGFVAVEIPFTPRAKRVLEL 180
EQILLGL+GEGTGIAA+VLK+MGI+LKDARVEVEK IGRGSGFVAVEIPFTPRAKRVLEL
Sbjct: 124 EQILLGLIGEGTGIAAKVLKSMGINLKDARVEVEKIIGRGSGFVAVEIPFTPRAKRVLEL 183
Query: 181 SLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRMVGESNNETA 240
SLEEARQLGHNYIGSEHLLLGLLREGEGVAARVL++LGADP+NIR QVIRMVGE+N TA
Sbjct: 184 SLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLENLGADPSNIRTQVIRMVGENNEVTA 243
Query: 241 GVAVGRGGSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLV 300
V G G SSNKMPTLEEYGTNLTKLA EGKLDPVVGRQ QIERV+QILGRRTKNNPCL+
Sbjct: 244 NV--GGGSSSNKMPTLEEYGTNLTKLAEEGKLDPVVGRQPQIERVVQILGRRTKNNPCLI 301
Query: 301 GEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEE 360
GEPGVGKTAIAEGLAQRIA+GDVPETIEGK+VITLDMGLLVAGTKYRGEFEERLKKLMEE
Sbjct: 302 GEPGVGKTAIAEGLAQRIASGDVPETIEGKKVITLDMGLLVAGTKYRGEFEERLKKLMEE 361
Query: 361 IKQNDNIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKD 420
I+Q+D IILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKD
Sbjct: 362 IRQSDEIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKD 421
Query: 421 PALERRFQPVRVPEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLSHQYISDRFL 480
PALERRFQPV+VPEPTVDE+IQIL+GLRERYE HHKL YTD++LVAA+QLS+QYISDRFL
Sbjct: 422 PALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDESLVAAAQLSYQYISDRFL 481
Query: 481 PDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEKAGELRDREM 540
PDKAIDLIDEAGSRVRLRHAQ+PEEAREL+KE+RQI KEK+EAVR QDFEKAG LRDRE+
Sbjct: 482 PDKAIDLIDEAGSRVRLRHAQVPEEARELEKELRQITKEKNEAVRGQDFEKAGTLRDREI 541
Query: 541 DLKTQISALIEKGKEMSKAESEADGAGPVVTEVDIQHIVASWTGVPVDKVSSDESDRLLK 600
+L+ ++SA+ KGKEMSKAESE GP+VTE DIQHIV+SWTG+PV+KVS+DESDRLLK
Sbjct: 542 ELRAEVSAIQAKGKEMSKAESETGEEGPMVTESDIQHIVSSWTGIPVEKVSTDESDRLLK 601
Query: 601 MEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASY 660
MEETLHKR+IGQDEAVKAI RAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAK LA+Y
Sbjct: 602 MEETLHKRIIGQDEAVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAY 661
Query: 661 YFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEK 720
YFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEK
Sbjct: 662 YFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEK 721
Query: 721 AHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEK 780
AHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGR+IGFDLDYDEK
Sbjct: 722 AHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRRIGFDLDYDEK 781
Query: 781 DSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKE 840
DSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADI+LKEVF+RLK KE
Sbjct: 782 DSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADILLKEVFERLKKKE 841
Query: 841 IDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADS 900
I+L VTERF++RVV+EGY+PSYGARPLRRAIMRLLEDSMAEKMLA EIKEGDSVI+D D+
Sbjct: 842 IELQVTERFKERVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSVIVDVDA 901
Query: 901 DGKVIVLNGSSGAP----ESLPEALPV 923
+G V VLNG SG P E ++LPV
Sbjct: 902 EGNVTVLNGGSGTPTTSLEEQEDSLPV 928
>At3g48870 AtClpC
Length = 952
Score = 1469 bits (3803), Expect = 0.0
Identities = 752/900 (83%), Positives = 829/900 (91%), Gaps = 7/900 (0%)
Query: 30 RRSLKMMSTLQAPALRMSGFSGLRTYNNLDTMLRPGLDFRSKVFGVTTSRKARASRCIPK 89
R +KMMS+LQAP L + FSGLR + LD + RP F K +S + +ASRC+PK
Sbjct: 53 RGRVKMMSSLQAPLLTIQSFSGLRAPSALDYLGRPSPGFLVKYKLAKSSGREKASRCVPK 112
Query: 90 AMFERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDA 149
AMFERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGL+GEGTGIAA+VLK+MGI+LKD+
Sbjct: 113 AMFERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDS 172
Query: 150 RVEVEKTIGRGSGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGV 209
RVEVEK IGRGSGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGV
Sbjct: 173 RVEVEKIIGRGSGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGV 232
Query: 210 AARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKLANE 269
AARVL++LGADP+NIR QVIRMVGE+N TA V G G+S KMPTLEEYGTNLTKLA E
Sbjct: 233 AARVLENLGADPSNIRTQVIRMVGENNEVTASVGGGSSGNS-KMPTLEEYGTNLTKLAEE 291
Query: 270 GKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEG 329
GKLDPVVGRQ QIER++QIL RRTKNNPCL+GEPGVGKTAIAEGLAQRIA+GDVPETIEG
Sbjct: 292 GKLDPVVGRQPQIERMVQILARRTKNNPCLIGEPGVGKTAIAEGLAQRIASGDVPETIEG 351
Query: 330 KEVITLDMGLLVAGTKYRGEFEERLKKLMEEIKQNDNIILFIDEVHTLIGAGAAEGAIDA 389
K VITLDMGLLVAGTKYRGEFEERLKKLMEEI+Q+D IILFIDEVHTLIGAGAAEGAIDA
Sbjct: 352 KTVITLDMGLLVAGTKYRGEFEERLKKLMEEIRQSDEIILFIDEVHTLIGAGAAEGAIDA 411
Query: 390 ANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQILRGLRE 449
ANILKPALARGELQCIGATT+DEYRKHIEKDPALERRFQPV+VPEPTV+E+IQIL+GLRE
Sbjct: 412 ANILKPALARGELQCIGATTIDEYRKHIEKDPALERRFQPVKVPEPTVEEAIQILQGLRE 471
Query: 450 RYERHHKLSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEAREL 509
RYE HHKL YTD+ALVAA+QLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEAREL
Sbjct: 472 RYEIHHKLRYTDEALVAAAQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEAREL 531
Query: 510 DKEVRQIVKEKDEAVRNQDFEKAGELRDREMDLKTQISALIEKGKEMSKAESEADGAGPV 569
+K++RQI KEK+EAVR+QDFE AG RDRE++LK +I+ ++ +GKE++KAE+EA+ GP
Sbjct: 532 EKQLRQITKEKNEAVRSQDFEMAGSHRDREIELKAEIANVLSRGKEVAKAENEAEEGGPT 591
Query: 570 VTEVDIQHIVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVG 629
VTE DIQHIVA+WTG+PV+KVSSDES RLL+ME+TLH RVIGQDEAVKAI RAIRRARVG
Sbjct: 592 VTESDIQHIVATWTGIPVEKVSSDESSRLLQMEQTLHTRVIGQDEAVKAISRAIRRARVG 651
Query: 630 LKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSP 689
LKNPNRPIASFIFSGPTGVGKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSP
Sbjct: 652 LKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSP 711
Query: 690 PGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFK 749
PGYVGYTEGGQLTEAVRRRPYT+VLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFK
Sbjct: 712 PGYVGYTEGGQLTEAVRRRPYTLVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFK 771
Query: 750 NTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDE 809
NTLLIMTSNVGSSVIEKGGR+IGFDLD+DEKDSSYNRIKSLVTEELKQYFRPEFLNRLDE
Sbjct: 772 NTLLIMTSNVGSSVIEKGGRRIGFDLDHDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDE 831
Query: 810 MIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRR 869
MIVFRQLTKLEVKEIADIMLKEV RL+ KEI+L VTERF++RVV+EG+DPSYGARPLRR
Sbjct: 832 MIVFRQLTKLEVKEIADIMLKEVVARLEVKEIELQVTERFKERVVDEGFDPSYGARPLRR 891
Query: 870 AIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLNGSSG------APESLPEALPV 923
AIMRLLEDSMAEKML+ +IKEGDSVI+D D++G V+VL+G++G A E++ + +P+
Sbjct: 892 AIMRLLEDSMAEKMLSRDIKEGDSVIVDVDAEGSVVVLSGTTGRVGGFAAEEAMEDPIPI 951
>At5g51070 Erd1 protein precursor (sp|P42762)
Length = 945
Score = 749 bits (1934), Expect = 0.0
Identities = 405/855 (47%), Positives = 567/855 (65%), Gaps = 51/855 (5%)
Query: 90 AMFERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGE--------GTGIAA-RVLK 140
A+FERFTE+AI+ I+ SQ+EA+ LG + V T+ +LLGL+ E G+GI + +
Sbjct: 78 AVFERFTERAIRAIIFSQKEAKSLGKDMVYTQHLLLGLIAEDRDPQGFLGSGITIDKARE 137
Query: 141 AMGISLKDARVEVEKTIGRGSGFV-AVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLL 199
A+ +A + ++ + + + ++PF+ KRV E ++E +R + YI EH+
Sbjct: 138 AVWSIWDEANSDSKQEEASSTSYSKSTDMPFSISTKRVFEAAVEYSRTMDCQYIAPEHIA 197
Query: 200 LGLLREGEGVAARVLQDLGADPNNIRAQVIRMV----------------GESNNETAGVA 243
+GL +G A RVL+ LGA+ N + A + + G + +G
Sbjct: 198 VGLFTVDDGSAGRVLKRLGANMNLLTAAALTRLKGEIAKDGREPSSSSKGSFESPPSGRI 257
Query: 244 VGRG-GSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGE 302
G G G LE++ +LT A+EG +DPV+GR+++++RVIQIL RRTKNNP L+GE
Sbjct: 258 AGSGPGGKKAKNVLEQFCVDLTARASEGLIDPVIGREKEVQRVIQILCRRTKNNPILLGE 317
Query: 303 PGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEEIK 362
GVGKTAIAEGLA IA P + K +++LD+GLL+AG K RGE E R+ L+ E+K
Sbjct: 318 AGVGKTAIAEGLAISIAEASAPGFLLTKRIMSLDIGLLMAGAKERGELEARVTALISEVK 377
Query: 363 QNDNIILFIDEVHTLIGAGAAE-----GAIDAANILKPALARGELQCIGATTLDEYRKHI 417
++ +ILFIDEVHTLIG+G +D AN+LKP+L RGELQCI +TTLDE+R
Sbjct: 378 KSGKVILFIDEVHTLIGSGTVGRGNKGSGLDIANLLKPSLGRGELQCIASTTLDEFRSQF 437
Query: 418 EKDPALERRFQPVRVPEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLSHQYISD 477
EKD AL RRFQPV + EP+ +++++IL GLRE+YE HH YT +A+ AA LS +YI+D
Sbjct: 438 EKDKALARRFQPVLINEPSEEDAVKILLGLREKYEAHHNCKYTMEAIDAAVYLSSRYIAD 497
Query: 478 RFLPDKAIDLIDEAGSRVRLRHAQLPEE---------ARELDKEVRQIVKEKDEAVRNQD 528
RFLPDKAIDLIDEAGSR R+ + +E + +E++ + + + ++
Sbjct: 498 RFLPDKAIDLIDEAGSRARIEAFRKKKEDAICILSKPPNDYWQEIKTVQAMHEVVLSSRQ 557
Query: 529 FEKAGELRDREMDLKTQISALIEKGKEMSKAESEADGAGPVVTEVDIQHIVASWTGVPVD 588
+ G+ + + L+E E S + D +V DI + + W+G+PV
Sbjct: 558 KQDDGDA------ISDESGELVE---ESSLPPAAGDDEPILVGPDDIAAVASVWSGIPVQ 608
Query: 589 KVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGV 648
++++DE L+ +E+ L RV+GQDEAV AI RA++R+RVGLK+P+RPIA+ +F GPTGV
Sbjct: 609 QITADERMLLMSLEDQLRGRVVGQDEAVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGV 668
Query: 649 GKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRR 708
GK+EL K LA+ YFGSEE+M+RLDMSE+MERHTVSKLIGSPPGYVG+ EGG LTEA+RRR
Sbjct: 669 GKTELTKALAANYFGSEESMLRLDMSEYMERHTVSKLIGSPPGYVGFEEGGMLTEAIRRR 728
Query: 709 PYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGG 768
P+TVVLFDEIEKAHPD+FN++LQ+ EDG LTDS+GR V FKN L+IMTSNVGS I KG
Sbjct: 729 PFTVVLFDEIEKAHPDIFNILLQLFEDGHLTDSQGRRVSFKNALIIMTSNVGSLAIAKGR 788
Query: 769 R-KIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADI 827
IGF LD DE+ +SY +K+LV EELK YFRPE LNR+DE+++FRQL K ++ EI ++
Sbjct: 789 HGSIGFILDDDEEAASYTGMKALVVEELKNYFRPELLNRIDEIVIFRQLEKAQMMEILNL 848
Query: 828 MLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGE 887
ML+++ RL + L V+E ++ + ++GYDP+YGARPLRR + ++ED ++E LAG
Sbjct: 849 MLQDLKSRLVALGVGLEVSEPVKELICKQGYDPAYGARPLRRTVTEIVEDPLSEAFLAGS 908
Query: 888 IKEGDSVIIDADSDG 902
K GD+ + D G
Sbjct: 909 FKPGDTAFVVLDDTG 923
>At2g25140 putative ATP-dependent CLPB protein
Length = 964
Score = 703 bits (1815), Expect = 0.0
Identities = 395/892 (44%), Positives = 558/892 (62%), Gaps = 95/892 (10%)
Query: 95 FTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGIS------LKD 148
FTE A + ++ + + AR V +E ++ L+ + G+A ++ GI D
Sbjct: 88 FTEMAWEGLINAFDAARESKQQIVESEHLMKALLEQKDGMARKIFTKAGIDNSSVLQATD 147
Query: 149 ARVEVEKTIGRGSGFVAVEIPFTPRAKRVLELSLEEARQ----LGHNYIGSEHLLLGLLR 204
+ + T+ SG R L + LE A++ + +Y+ EH LL
Sbjct: 148 LFISKQPTVSDASG---------QRLGSSLSVILENAKRHKKDMLDSYVSVEHFLLAYYS 198
Query: 205 EGEGVAARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLT 264
+ +D+ D ++ + + G+ V +K LE+YG +LT
Sbjct: 199 DTR-FGQEFFRDMKLDIQVLKDAIKDVRGDQR-------VTDRNPESKYQALEKYGNDLT 250
Query: 265 KLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVP 324
++A GKLDPV+GR +I R IQIL RRTKNNP ++GEPGVGKTAIAEGLAQRI GDVP
Sbjct: 251 EMARRGKLDPVIGRDDEIRRCIQILCRRTKNNPVIIGEPGVGKTAIAEGLAQRIVRGDVP 310
Query: 325 ETIEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEEIK-QNDNIILFIDEVHTLIGAGAA 383
E + +++I+LDMG L+AG K+RG+FEERLK +M+E+ N ILFIDE+HT++GAGA
Sbjct: 311 EPLMNRKLISLDMGSLLAGAKFRGDFEERLKAVMKEVSASNGQTILFIDEIHTVVGAGAM 370
Query: 384 EGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQI 443
+GA+DA+N+LKP L RGEL+CIGATTL EYRK+IEKDPALERRFQ V +P+V+++I I
Sbjct: 371 DGAMDASNLLKPMLGRGELRCIGATTLTEYRKYIEKDPALERRFQQVLCVQPSVEDTISI 430
Query: 444 LRGLRERYERHHKLSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLP 503
LRGLRERYE HH ++ +D ALV+A+ L+ +YI++RFLPDKAIDL+DEAG+++++ P
Sbjct: 431 LRGLRERYELHHGVTISDSALVSAAVLADRYITERFLPDKAIDLVDEAGAKLKMEITSKP 490
Query: 504 EEARELDKEVRQIVKEKDEAVRNQDFEKAGELRDREMDLKT------QISALIEKGK--- 554
E +D+ V ++ EK + D L+ E DL T +++ EK K
Sbjct: 491 TELDGIDRAVIKLEMEKLSLKNDTDKASKERLQKIENDLSTLKQKQKELNVQWEKEKSLM 550
Query: 555 ---------------EMSKAESEAD--------------------------------GAG 567
E+ AE E D G
Sbjct: 551 TKIRSFKEEIDRVNLEIESAEREYDLNRAAELKYGTLLSLQRQLEEAEKNLTNFRQFGQS 610
Query: 568 ---PVVTEVDIQHIVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIR 624
VVT++DI IV+ WTG+P+ + E ++L+ +EE LH RVIGQD AVK++ AIR
Sbjct: 611 LLREVVTDLDIAEIVSKWTGIPLSNLQQSEREKLVMLEEVLHHRVIGQDMAVKSVADAIR 670
Query: 625 RARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSK 684
R+R GL +PNRPIASF+F GPTGVGK+ELAK LA Y F +E A++R+DMSE+ME+H+VS+
Sbjct: 671 RSRAGLSDPNRPIASFMFMGPTGVGKTELAKALAGYLFNTENAIVRVDMSEYMEKHSVSR 730
Query: 685 LIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGR 744
L+G+PPGYVGY EGGQLTE VRRRPY+VVLFDEIEKAHPDVFN++LQ+L+DGR+TDS+GR
Sbjct: 731 LVGAPPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHPDVFNILLQLLDDGRITDSQGR 790
Query: 745 TVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFL 804
TV FKN ++IMTSN+GS I + R + D K++ Y +K V E +Q FRPEF+
Sbjct: 791 TVSFKNCVVIMTSNIGSHHILETLRN-----NEDSKEAVYEIMKRQVVELARQNFRPEFM 845
Query: 805 NRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGA 864
NR+DE IVF+ L E+ +I ++ ++ V + L+ K+I L T+ D + + G+DP+YGA
Sbjct: 846 NRIDEYIVFQPLDSNEISKIVELQMRRVKNSLEQKKIKLQYTKEAVDLLAQLGFDPNYGA 905
Query: 865 RPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD---SDGKVIVLNGSSGA 913
RP++R I +++E+ +A +L G+ E D+V++D D SD K+++ S A
Sbjct: 906 RPVKRVIQQMVENEIAVGILKGDFAEEDTVLVDVDHLASDNKLVIKKLESNA 957
>At5g15450 clpB heat shock protein-like
Length = 968
Score = 702 bits (1813), Expect = 0.0
Identities = 397/883 (44%), Positives = 547/883 (60%), Gaps = 105/883 (11%)
Query: 93 ERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVE 152
+ FTE A + I+ S + A+ V TE ++ L+ + G+A R+ +G+
Sbjct: 80 QEFTEMAWQSIVSSPDVAKENKQQIVETEHLMKALLEQKNGLARRIFSKIGVDNTKVLEA 139
Query: 153 VEKTIGRGS---GFVAVEIPFTPRAKRVLELSLEEARQ----LGHNYIGSEHLLLGLLRE 205
EK I R G A + R LE + ARQ L +Y+ EHL+L +
Sbjct: 140 TEKFIQRQPKVYGDAAGSM-----LGRDLEALFQRARQFKKDLKDSYVSVEHLVLAFA-D 193
Query: 206 GEGVAARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTK 265
+ ++ +D ++++ + + G+ + V K LE+YG +LT
Sbjct: 194 DKRFGKQLFKDFQISERSLKSAIESIRGKQS-------VIDQDPEGKYEALEKYGKDLTA 246
Query: 266 LANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPE 325
+A EGKLDPV+GR +I R IQIL RRTKNNP L+GEPGVGKTAI+EGLAQRI GDVP+
Sbjct: 247 MAREGKLDPVIGRDDEIRRCIQILSRRTKNNPVLIGEPGVGKTAISEGLAQRIVQGDVPQ 306
Query: 326 TIEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEEIKQNDN-IILFIDEVHTLIGAGAAE 384
+ +++I+LDMG L+AG KYRGEFE+RLK +++E+ ++ IILFIDE+HT++GAGA
Sbjct: 307 ALMNRKLISLDMGALIAGAKYRGEFEDRLKAVLKEVTDSEGQIILFIDEIHTVVGAGATN 366
Query: 385 GAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQIL 444
GA+DA N+LKP L RGEL+CIGATTLDEYRK+IEKDPALERRFQ V V +PTV+++I IL
Sbjct: 367 GAMDAGNLLKPMLGRGELRCIGATTLDEYRKYIEKDPALERRFQQVYVDQPTVEDTISIL 426
Query: 445 RGLRERYERHHKLSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAG------------ 492
RGLRERYE HH + +D ALV A+ LS +YIS RFLPDKAIDL+DEA
Sbjct: 427 RGLRERYELHHGVRISDSALVEAAILSDRYISGRFLPDKAIDLVDEAAAKLKMEITSKPT 486
Query: 493 --------------------------SRVRL------------RHAQLPEE--------- 505
SR RL + A+L E+
Sbjct: 487 ALDELDRSVIKLEMERLSLTNDTDKASRERLNRIETELVLLKEKQAELTEQWEHERSVMS 546
Query: 506 -ARELDKEVRQIVKEKDEAVRNQDFEKAGELR-------DREM-DLKTQISALIEKGKEM 556
+ + +E+ ++ E +A R D +A EL+ R++ + + +++ + GK M
Sbjct: 547 RLQSIKEEIDRVNLEIQQAEREYDLNRAAELKYGSLNSLQRQLNEAEKELNEYLSSGKSM 606
Query: 557 SKAESEADGAGPVVTEVDIQHIVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAV 616
+ E V DI IV+ WTG+PV K+ E D+LL +EE LHKRV+GQ+ AV
Sbjct: 607 FREE---------VLGSDIAEIVSKWTGIPVSKLQQSERDKLLHLEEELHKRVVGQNPAV 657
Query: 617 KAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEF 676
A+ AI+R+R GL +P RPIASF+F GPTGVGK+ELAK LASY F +EEA++R+DMSE+
Sbjct: 658 TAVAEAIQRSRAGLSDPGRPIASFMFMGPTGVGKTELAKALASYMFNTEEALVRIDMSEY 717
Query: 677 MERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDG 736
ME+H VS+LIG+PPGYVGY EGGQLTE VRRRPY+V+LFDEIEKAH DVFN+ LQIL+DG
Sbjct: 718 MEKHAVSRLIGAPPGYVGYEEGGQLTETVRRRPYSVILFDEIEKAHGDVFNVFLQILDDG 777
Query: 737 RLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELK 796
R+TDS+GRTV F NT++IMTSNVGS I + D D + SY IK V +
Sbjct: 778 RVTDSQGRTVSFTNTVIIMTSNVGSQFILN-------NTDDDANELSYETIKERVMNAAR 830
Query: 797 QYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEE 856
FRPEF+NR+DE IVF+ L + ++ I + L V R+ +++ +++T+ D +
Sbjct: 831 SIFRPEFMNRVDEYIVFKPLDREQINRIVRLQLARVQKRIADRKMKINITDAAVDLLGSL 890
Query: 857 GYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 899
GYDP+YGARP++R I + +E+ +A+ +L G+ KE D ++ID +
Sbjct: 891 GYDPNYGARPVKRVIQQNIENELAKGILRGDFKEEDGILIDTE 933
>At1g74310 heat shock protein 101
Length = 911
Score = 640 bits (1651), Expect = 0.0
Identities = 358/869 (41%), Positives = 535/869 (61%), Gaps = 85/869 (9%)
Query: 93 ERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVE 152
E+FT K + I + E A GH + L+ + TGI + + + G ++A
Sbjct: 4 EKFTHKTNETIATAHELAVNAGHAQFTPLHLAGALISDPTGIFPQAISSAGG--ENAAQS 61
Query: 153 VEKTIGRGSGFVAVE------IPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREG 206
E+ I + + + IP + +V+ + + G ++ + L++GLL +
Sbjct: 62 AERVINQALKKLPSQSPPPDDIPASSSLIKVIRRAQAAQKSRGDTHLAVDQLIMGLLEDS 121
Query: 207 EGVAARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKL 266
+ +L ++G ++++V ++ G+ G V L+ YG +L +
Sbjct: 122 Q--IRDLLNEVGVATARVKSEVEKLRGKE-----GKKVESASGDTNFQALKTYGRDLVEQ 174
Query: 267 ANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPET 326
A GKLDPV+GR ++I RV++IL RRTKNNP L+GEPGVGKTA+ EGLAQRI GDVP +
Sbjct: 175 A--GKLDPVIGRDEEIRRVVRILSRRTKNNPVLIGEPGVGKTAVVEGLAQRIVKGDVPNS 232
Query: 327 IEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEEIKQNDN-IILFIDEVHTLIGAGAAEG 385
+ +I+LDMG LVAG KYRGEFEERLK +++E++ + +ILFIDE+H ++GAG EG
Sbjct: 233 LTDVRLISLDMGALVAGAKYRGEFEERLKSVLKEVEDAEGKVILFIDEIHLVLGAGKTEG 292
Query: 386 AIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQILR 445
++DAAN+ KP LARG+L+CIGATTL+EYRK++EKD A ERRFQ V V EP+V ++I ILR
Sbjct: 293 SMDAANLFKPMLARGQLRCIGATTLEEYRKYVEKDAAFERRFQQVYVAEPSVPDTISILR 352
Query: 446 GLRERYERHHKLSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEE 505
GL+E+YE HH + D AL+ A+QLS +YI+ R LPDKAIDL+DEA + VR++ PEE
Sbjct: 353 GLKEKYEGHHGVRIQDRALINAAQLSARYITGRHLPDKAIDLVDEACANVRVQLDSQPEE 412
Query: 506 ARELDK-------EVRQIVKEKDEAVRNQDFEKAGELRD-------------REMDLKTQ 545
L++ E+ + +EKD+A + + E EL D +E + +
Sbjct: 413 IDNLERKRMQLEIELHALEREKDKASKARLIEVRKELDDLRDKLQPLTMKYRKEKERIDE 472
Query: 546 ISALIEKGKEMSKAESEADGAGPVVTEVDIQH---------------------------- 577
I L +K +E+ + EA+ + D+++
Sbjct: 473 IRRLKQKREELMFSLQEAERRYDLARAADLRYGAIQEVESAIAQLEGTSSEENVMLTENV 532
Query: 578 -------IVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGL 630
+V+ WTG+PV ++ +E +RL+ + + LHKRV+GQ++AV A+ AI R+R GL
Sbjct: 533 GPEHIAEVVSRWTGIPVTRLGQNEKERLIGLADRLHKRVVGQNQAVNAVSEAILRSRAGL 592
Query: 631 KNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPP 690
P +P SF+F GPTGVGK+ELAK LA F E ++R+DMSE+ME+H+VS+LIG+PP
Sbjct: 593 GRPQQPTGSFLFLGPTGVGKTELAKALAEQLFDDENLLVRIDMSEYMEQHSVSRLIGAPP 652
Query: 691 GYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKN 750
GYVG+ EGGQLTEAVRRRPY V+LFDE+EKAH VFN +LQ+L+DGRLTD +GRTVDF+N
Sbjct: 653 GYVGHEEGGQLTEAVRRRPYCVILFDEVEKAHVAVFNTLLQVLDDGRLTDGQGRTVDFRN 712
Query: 751 TLLIMTSNVGSSVIEKG-GRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDE 809
+++IMTSN+G+ + G K+ ++ D V E++++FRPE LNRLDE
Sbjct: 713 SVIIMTSNLGAEHLLAGLTGKVTMEVARD-----------CVMREVRKHFRPELLNRLDE 761
Query: 810 MIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRR 869
++VF L+ +++++A + +K+V RL + + L+VT+ D ++ E YDP YGARP+RR
Sbjct: 762 IVVFDPLSHDQLRKVARLQMKDVAVRLAERGVALAVTDAALDYILAESYDPVYGARPIRR 821
Query: 870 AIMRLLEDSMAEKMLAGEIKEGDSVIIDA 898
+ + + +++ ++ EI E +V IDA
Sbjct: 822 WMEKKVVTELSKMVVREEIDENSTVYIDA 850
>At4g14670 heat shock protein like
Length = 668
Score = 493 bits (1268), Expect = e-139
Identities = 278/613 (45%), Positives = 385/613 (62%), Gaps = 79/613 (12%)
Query: 186 RQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVG 245
R LG +G L++ LL + + + VL++ G P ++++V ++ GE
Sbjct: 77 RNLGDTKVGVAVLVISLLEDSQ--ISDVLKEAGVVPEKVKSEVEKLRGEVI--------- 125
Query: 246 RGGSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGV 305
+ L+ YGT+L + A GKLDPV+GR ++I RVI++L RRTKNNP L+GEPGV
Sbjct: 126 -------LRALKTYGTDLVEQA--GKLDPVIGRHREIRRVIEVLSRRTKNNPVLIGEPGV 176
Query: 306 GKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEEIKQ-N 364
GKTA+ EGLAQRI GDVP + G ++I+L+ G +VAGT RG+FEERLK +++ +++
Sbjct: 177 GKTAVVEGLAQRILKGDVPINLTGVKLISLEFGAMVAGTTLRGQFEERLKSVLKAVEEAQ 236
Query: 365 DNIILFIDEVHTLIGAGAAEGAIDAANILKPALARGELQCIGATTLDEYRKHIEKDPALE 424
++LFIDE+H +GA A G+ DAA +LKP LARG+L+ IGATTL+EYR H+EKD A E
Sbjct: 237 GKVVLFIDEIHMALGACKASGSTDAAKLLKPMLARGQLRFIGATTLEEYRTHVEKDAAFE 296
Query: 425 RRFQPVRVPEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLSHQYISDRFLPDKA 484
RRFQ V V EP+V ++I ILRGL+E+YE HH + D ALV ++QLS +YI+ R LPDKA
Sbjct: 297 RRFQQVFVAEPSVPDTISILRGLKEKYEGHHGVRIQDRALVLSAQLSERYITGRRLPDKA 356
Query: 485 IDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEKAGELRDREM---- 540
IDL+DE+ + V+ + PEE L+++V Q+ E + +D +KA E R E+
Sbjct: 357 IDLVDESCAHVKAQLDIQPEEIDSLERKVMQLEIEIHALEKEKD-DKASEARLSEVRKEL 415
Query: 541 -DLKTQISALIEKGKEMSKAESEA-------DGAGPVVTEVDIQH--------------- 577
DL+ ++ L K K+ K +E D + E + QH
Sbjct: 416 DDLRDKLEPLTIKYKKEKKIINETRRLKQNRDDLMIALQEAERQHDVPKAAVLKYGAIQE 475
Query: 578 -----------------------------IVASWTGVPVDKVSSDESDRLLKMEETLHKR 608
+V+ WTG+PV ++ +E RL+ + + LH+R
Sbjct: 476 VESAIAKLEKSAKDNVMLTETVGPENIAEVVSRWTGIPVTRLDQNEKKRLISLADKLHER 535
Query: 609 VIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAM 668
V+GQDEAVKA+ AI R+RVGL P +P SF+F GPTGVGK+ELAK LA F SE +
Sbjct: 536 VVGQDEAVKAVAAAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLL 595
Query: 669 IRLDMSEFMERHTVSKLIGSPPG-YVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFN 727
+RLDMSE+ ++ +V+KLIG+PPG Y+G+ EGGQLTE VRRRPY VVLFDE+EK H VFN
Sbjct: 596 VRLDMSEYNDKFSVNKLIGAPPGYYIGHEEGGQLTEPVRRRPYCVVLFDEVEKTHVTVFN 655
Query: 728 MMLQILEDGRLTD 740
+LQ+LEDGRLTD
Sbjct: 656 TLLQVLEDGRLTD 668
>At3g45450 clpC-like protein
Length = 341
Score = 347 bits (891), Expect = 1e-95
Identities = 211/355 (59%), Positives = 244/355 (68%), Gaps = 48/355 (13%)
Query: 91 MFERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDAR 150
MFERFTEKAIKVI L+QEEARRLG+NF GTE ILL L+GEGTGIAA+VLK+MGI+LKDAR
Sbjct: 1 MFERFTEKAIKVITLAQEEARRLGYNFFGTEHILLSLIGEGTGIAAKVLKSMGINLKDAR 60
Query: 151 VEVEKTIGRGSGFVAVEIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVA 210
VEVEK IGRGSGFV VEIPFTPR K + H +G+A
Sbjct: 61 VEVEKIIGRGSGFVVVEIPFTPR-KACAGFVTRGSSTTRH----------------KGIA 103
Query: 211 ARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKLANEG 270
RVL+ LGADP+NIR QV + N++ A VA + E T G
Sbjct: 104 VRVLEILGADPSNIRTQVNAPI-LVNSQQALVA-----------EVMETATWQVGNTRRG 151
Query: 271 KLDPVVGRQQQIERVIQILGRRT-KNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEG 329
KLDPVVGRQ QI+RV+QIL RRT +NN CL+G+PGVGK AIAEG+AQRIA+GDVPETI+G
Sbjct: 152 KLDPVVGRQPQIKRVVQILARRTCRNNACLIGKPGVGKRAIAEGIAQRIASGDVPETIKG 211
Query: 330 KEVITLDMGLLVAGTKYRGEFEERLKKLMEEI-KQNDNIILFIDEVHTLIGAGAAEGAID 388
K + VAG E R + +EE+ Q+D+IILFIDE+H LIGAGA EGAID
Sbjct: 212 K--------MNVAGNCGWNEIRWRSRGKIEEVYGQSDDIILFIDEMHLLIGAGAVEGAID 263
Query: 389 AANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQI 443
AANILKPAL R ELQ YRKHIE DPALERRFQPV+VPEPTV+E+IQI
Sbjct: 264 AANILKPALERCELQ---------YRKHIENDPALERRFQPVKVPEPTVEEAIQI 309
Score = 35.8 bits (81), Expect = 0.11
Identities = 15/30 (50%), Positives = 23/30 (76%)
Query: 545 QISALIEKGKEMSKAESEADGAGPVVTEVD 574
QI+ ++ KGKE++KA++EA+ P VTE D
Sbjct: 308 QITTVLAKGKEVNKADNEAEEGRPTVTEAD 337
>At2g25030 putative ATP-dependent CLPB protein
Length = 265
Score = 194 bits (494), Expect = 1e-49
Identities = 110/307 (35%), Positives = 176/307 (56%), Gaps = 55/307 (17%)
Query: 601 MEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASY 660
+E+ LH+R+I QD V+++ AIR ++ G+ +PNR IASF+F G V
Sbjct: 2 LEQILHERIIAQDLDVESVADAIRCSKAGISDPNRLIASFMFMGQPSV------------ 49
Query: 661 YFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEK 720
VS+L+G+ PGYVGY +GG+LTE VRRRPY+VV FDEIEK
Sbjct: 50 ---------------------VSQLVGASPGYVGYGDGGKLTEVVRRRPYSVVQFDEIEK 88
Query: 721 AHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNVGSSVIEKGGRKIGFDLDYDEK 780
H DVF+++LQ+L+DGR+T+S G + L + +N D K
Sbjct: 89 PHLDVFSILLQLLDDGRITNSHGSL----HILETIRNNE------------------DIK 126
Query: 781 DSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKE 840
++ Y +K V E ++ FRP+F+NR+DE IV + L E+ +I ++ +++V RL+ +
Sbjct: 127 EAFYEMMKQQVVELARKTFRPKFMNRIDEYIVSQPLNSSEISKIVELQMRQVKKRLEQNK 186
Query: 841 IDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADS 900
I+L T+ D + + G+DP+ GARP+++ I +L++ + K+L G+ E +++IDAD
Sbjct: 187 INLEYTKEAVDLLAQLGFDPNNGARPVKQMIEKLVKKEITLKVLKGDFAEDGTILIDADQ 246
Query: 901 DGKVIVL 907
+V+
Sbjct: 247 PNNKLVI 253
>At2g25040 hypothetical protein
Length = 135
Score = 92.0 bits (227), Expect = 1e-18
Identities = 42/71 (59%), Positives = 55/71 (77%)
Query: 252 KMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIA 311
K LE YG++LTK+A +GKL P++GR ++ R IQIL R T++NP ++GEPGVGKTAI
Sbjct: 65 KHEALETYGSDLTKMARQGKLPPLIGRDDEVNRCIQILCRMTESNPVIIGEPGVGKTAIV 124
Query: 312 EGLAQRIATGD 322
EGLA+RI GD
Sbjct: 125 EGLAERIVKGD 135
>At5g57710 101 kDa heat shock protein; HSP101-like protein
Length = 990
Score = 69.3 bits (168), Expect = 9e-12
Identities = 45/173 (26%), Positives = 88/173 (50%), Gaps = 14/173 (8%)
Query: 591 SSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIAS-----FIFSGP 645
+S + D K+ + + ++V Q++A A+ + + ++G R + S +FSGP
Sbjct: 602 NSLDIDLFKKLLKGMTEKVWWQNDAAAAVAATVSQCKLG-NGKRRGVLSKGDVWLLFSGP 660
Query: 646 TGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAV 705
VGK ++ L+S +G+ MI+L + S + G T ++ E V
Sbjct: 661 DRVGKRKMVSALSSLVYGTNPIMIQLGSRQDAGDGNSS--------FRGKTALDKIAETV 712
Query: 706 RRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 758
+R P++V+L ++I++A V + Q ++ GR+ DS GR + N + +MT++
Sbjct: 713 KRSPFSVILLEDIDEADMLVRGSIKQAMDRGRIRDSHGREISLGNVIFVMTAS 765
Score = 40.0 bits (92), Expect = 0.006
Identities = 33/146 (22%), Positives = 66/146 (44%), Gaps = 9/146 (6%)
Query: 196 EHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKM-- 253
E L++ +L + +RV+++ ++A + + + S T +V G + +
Sbjct: 131 EQLIISILDDPS--VSRVMREASFSSPAVKATIEQSLNNSVTPTPIPSVSSVGLNFRPGG 188
Query: 254 --PTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIA 311
P N N + V + +ERV+ ILGR K NP LVG+ G+ +
Sbjct: 189 GGPMTRNSYLNPRLQQNASSVQSGVSKNDDVERVMDILGRAKKKNPVLVGDSEPGR--VI 246
Query: 312 EGLAQRIATGDVPE-TIEGKEVITLD 336
+ ++I G+V ++ +V++L+
Sbjct: 247 REILKKIEVGEVGNLAVKNSKVVSLE 272
>At4g30350 unknown protein
Length = 924
Score = 68.6 bits (166), Expect = 2e-11
Identities = 49/167 (29%), Positives = 82/167 (48%), Gaps = 10/167 (5%)
Query: 592 SDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKS 651
S + D K+ + L K V Q +A ++ AI + G ++ +F+GP GKS
Sbjct: 564 SFDIDLFKKLLKGLAKSVWWQHDAASSVAAAITECKHG-NGKSKGDIWLMFTGPDRAGKS 622
Query: 652 ELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYT 711
++A L+ GS+ I L S M+ + G T + EAVRR P+
Sbjct: 623 KMASALSDLVSGSQPITISLGSSSRMDDGLNIR---------GKTALDRFAEAVRRNPFA 673
Query: 712 VVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 758
V++ ++I++A + N + +E GR+ DS GR V N ++I+T+N
Sbjct: 674 VIVLEDIDEADILLRNNVKIAIERGRICDSYGREVSLGNVIIILTAN 720
Score = 37.7 bits (86), Expect = 0.030
Identities = 39/164 (23%), Positives = 76/164 (45%), Gaps = 15/164 (9%)
Query: 275 VVGRQQQIERVIQILGRRTKNNPCLVG--EPGVGKTAIAEGLAQRIATGDVPE-TIEGKE 331
++ R + +RVI+I+ R K NP LVG EP + + + + ++I G+ + + +
Sbjct: 233 MIQRTDEAKRVIEIMIRTRKRNPVLVGDSEPHI----LVKEILEKIENGEFSDGALRNFQ 288
Query: 332 VITLDMGLLVAGTKYRGEFEERLKKLMEEIKQNDNIILFIDEVHTLIGAGAAEG--AIDA 389
VI L+ L+ GE + L+E ++L + ++ L+ AA G ++
Sbjct: 289 VIRLEKELVSQLATRLGE----ISGLVETRIGGGGVVLDLGDLKWLVEHPAANGGAVVEM 344
Query: 390 ANILKPALARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVP 433
+L+ +G L IG T + Y + P++E + +P
Sbjct: 345 RKLLE--RYKGRLCFIGTATCETYLRCQVYYPSMENDWDLQAIP 386
>At1g07200 unknown protein
Length = 979
Score = 63.5 bits (153), Expect = 5e-10
Identities = 44/161 (27%), Positives = 79/161 (48%), Gaps = 11/161 (6%)
Query: 601 MEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIASFI-FSGPTGVGKSELAKTLAS 659
+ E L ++V Q EAV AI + I + N+ ++ GP VGK ++A TL+
Sbjct: 621 LREILSRKVAWQTEAVNAISQIICGCKTDSTRRNQASGIWLALLGPDKVGKKKVAMTLSE 680
Query: 660 YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIE 719
+FG + I +D ++ + G T +T + R+P++VVL + +E
Sbjct: 681 VFFGGKVNYICVDFG--------AEHCSLDDKFRGKTVVDYVTGELSRKPHSVVLLENVE 732
Query: 720 KAH-PDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNV 759
KA PD + + + G++ D GR + KN ++++TS +
Sbjct: 733 KAEFPDQMRLS-EAVSTGKIRDLHGRVISMKNVIVVVTSGI 772
Score = 33.5 bits (75), Expect = 0.57
Identities = 37/159 (23%), Positives = 68/159 (42%), Gaps = 12/159 (7%)
Query: 277 GRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLD 336
G + R+ ++LGR+ K NP L+G G + I G +I+++
Sbjct: 218 GFDENSRRIGEVLGRKDKKNPLLIGNCANEALKTFTDSINSGKLGFLQMDISGLSLISIE 277
Query: 337 ---MGLLVAGTKYRGEFEERLKKLMEEIKQN---DNIILFIDEVHTLIGAGAAEGAI--- 387
+L G+K E ++ L ++Q+ I+L + E+ L A I
Sbjct: 278 KEISEILADGSKNEEEIRMKVDDLGRTVEQSGSKSGIVLNLGELKVLTSEANAALEILVS 337
Query: 388 DAANILKPALARGELQCIGATTLDE-YRKHIEKDPALER 425
+++LK +L IG + +E Y K I++ P +E+
Sbjct: 338 KLSDLLKH--ESKQLSFIGCVSSNETYTKLIDRFPTIEK 374
>At4g12060 unknown protein
Length = 241
Score = 61.2 bits (147), Expect = 3e-09
Identities = 46/159 (28%), Positives = 78/159 (48%), Gaps = 6/159 (3%)
Query: 63 RPGLDFRSKVFGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGTEQ 122
RP +S + + T A + A +++ +AIK + + EAR+L + GTE
Sbjct: 65 RPRRIHKSAISSLPT---ANPDLVVSDAKKPKWSWRAIKSFAMGELEARKLKYPNTGTEA 121
Query: 123 ILLGLVGEGTGIAARVLKAMGISLKDARVEVEKTIGRGS--GFVAVEIPFTPRAKRVLEL 180
+L+G++ EGT ++ L+A I L R E K +G+ F P T A+R L+
Sbjct: 122 LLMGILIEGTSFTSKFLRANKIMLYKVREETVKLLGKADMYFFSPEHPPLTEDAQRALDS 181
Query: 181 SLEEARQLGH-NYIGSEHLLLGLLREGEGVAARVLQDLG 218
+L++ + G + H+LLG+ E E ++L LG
Sbjct: 182 ALDQNLKAGGIGEVMPAHILLGIWSEVESPGHKILATLG 220
>At4g25370 unknown protein
Length = 238
Score = 58.9 bits (141), Expect = 1e-08
Identities = 40/158 (25%), Positives = 81/158 (50%), Gaps = 8/158 (5%)
Query: 94 RFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVEV 153
+++ +AIK + + + EAR+L + GTE IL+G++ EGT A+ L+ G++L R E
Sbjct: 85 KWSARAIKSLAMGELEARKLKYPSTGTEAILMGILVEGTSTVAKFLRGNGVTLFKVRDET 144
Query: 154 EKTIGRGS--GFVAVEIPFTPRAKRVLELSLEEARQLG-HNYIGSEHLLLGLLREGEGVA 210
+G+ F P T A++ + +++E + + + +LLLG+ + +
Sbjct: 145 LSLLGKSDMYFFSPEHPPLTEPAQKAIAWAIDEKNKSDVDGELTTAYLLLGVWSQKDSAG 204
Query: 211 ARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGG 248
++L+ LG + + + V +S NE ++ + G
Sbjct: 205 RQILEKLGFNEDKAKE-----VEKSMNEDVDLSFKKQG 237
>At2g40130 unknown protein
Length = 907
Score = 58.5 bits (140), Expect = 2e-08
Identities = 45/153 (29%), Positives = 69/153 (44%), Gaps = 12/153 (7%)
Query: 609 VIGQDEAVKAICRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKTLASYYFGSEEAM 668
V GQDEA + I A+ + K+ R GP VGK ++ LA + SE
Sbjct: 545 VSGQDEAARVISCALSQPP---KSVTRRDVWLNLVGPDTVGKRRMSLVLAEIVYQSEHRF 601
Query: 669 IRLDMSEFMERHTVSKLIGS---PPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDV 725
+ +D+ + +G P G T + E + R P+ VV + IEKA +
Sbjct: 602 MAVDLG------AAEQGMGGCDDPMRLRGKTMVDHIFEVMCRNPFCVVFLENIEKADEKL 655
Query: 726 FNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSN 758
+ + +E G+ DS GR V NT+ +MTS+
Sbjct: 656 QMSLSKAIETGKFMDSHGREVGIGNTIFVMTSS 688
>At2g29970 unknown protein
Length = 1002
Score = 53.1 bits (126), Expect = 7e-07
Identities = 44/162 (27%), Positives = 74/162 (45%), Gaps = 16/162 (9%)
Query: 601 MEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRPIAS-----FIFSGPTGVGKSELAK 655
+ E L ++V Q+EAV AI + R + N +A+ GP GK ++A
Sbjct: 631 LRELLSRKVGFQNEAVNAISEIVCGYRDESRRRNNHVATTSNVWLALLGPDKAGKKKVAL 690
Query: 656 TLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLF 715
LA + G ++ I +D F + ++ + G T + V RR +VV
Sbjct: 691 ALAEVFCGGQDNFICVD---FKSQDSLDDR------FRGKTVVDYIAGEVARRADSVVFI 741
Query: 716 DEIEKAH-PDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMT 756
+ +EKA PD + + + G+L DS GR + KN +++ T
Sbjct: 742 ENVEKAEFPDQIRLS-EAMRTGKLRDSHGREISMKNVIVVAT 782
Score = 42.0 bits (97), Expect = 0.002
Identities = 42/154 (27%), Positives = 75/154 (48%), Gaps = 18/154 (11%)
Query: 280 QQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMG- 338
+ R+ ++L R+ K NP LVG GV R G +P I G V+++ +
Sbjct: 223 ENCRRIGEVLARKDKKNPLLVGVCGVEALKTFTDSINRGKFGFLPLEISGLSVVSIKISE 282
Query: 339 LLVAGTKYRGEFEE--RLKKLM----EEIKQNDNIILFIDEVHTLIGAGAAEGAIDAANI 392
+LV G++ +F++ RLK M E+K + + +D + + + A++
Sbjct: 283 VLVDGSRIDIKFDDLGRLKSGMVLNLGELKVLASDVFSVDVIEKFV--------LKLADL 334
Query: 393 LKPALARGELQCIGATTLDE-YRKHIEKDPALER 425
LK L R +L IG+ + +E Y K IE+ P +++
Sbjct: 335 LK--LHREKLWFIGSVSSNETYLKLIERFPTIDK 366
>At3g53230 CDC48 - like protein
Length = 815
Score = 49.3 bits (116), Expect = 1e-05
Identities = 69/296 (23%), Positives = 119/296 (39%), Gaps = 43/296 (14%)
Query: 171 TPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQD-LGADPNNIRAQVI 229
T K ++ LE + H Y+G++ L L E R D + D I A+++
Sbjct: 389 TKNMKLAEDVDLERVSKDTHGYVGAD--LAALCTEAALQCIREKMDVIDLDDEEIDAEIL 446
Query: 230 RMVGESNNETAGVAVGRGGSSNKMPTLEEYGT----NLTKLAN-EGKLDPVVGRQQQIER 284
+ SN+ A+G S T+ E ++ L N + +L V +
Sbjct: 447 NSMAVSNDHFQ-TALGNSNPSALRETVVEVPNVSWEDIGGLENVKRELQETVQYPVEHPE 505
Query: 285 VIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGT 344
+ G G PG GKT +A+ +A + +I+G E++T+
Sbjct: 506 KFEKFGMSPSKGVLFYGPPGCGKTLLAKAIANECQANFI--SIKGPELLTM--------- 554
Query: 345 KYRGEFEERLKKLMEEIKQNDNIILFIDEVHTLI-----GAGAAEGAIDAANILKPAL-- 397
+ GE E ++++ ++ +Q+ +LF DE+ ++ G A GA D +L L
Sbjct: 555 -WFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSVGDAGGAAD--RVLNQLLTE 611
Query: 398 -----ARGELQCIGATTLDEYRKHIEKDPAL---ERRFQPVRVPEPTVDESIQILR 445
A+ + IGAT + DPAL R Q + +P P + QI +
Sbjct: 612 MDGMNAKKTVFIIGATNRPDI-----IDPALLRPGRLDQLIYIPLPDEESRYQIFK 662
Score = 47.0 bits (110), Expect = 5e-05
Identities = 67/256 (26%), Positives = 108/256 (42%), Gaps = 35/256 (13%)
Query: 284 RVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAG 343
++ + +G + L G PG GKT IA +A TG I G E+++
Sbjct: 232 QLFKSIGVKPPKGILLYGPPGSGKTLIARAVANE--TGAFFFCINGPEIMS--------- 280
Query: 344 TKYRGEFEERLKKLMEEIKQNDNIILFIDEVHTLIGA-----GAAEGAI--DAANILKPA 396
K GE E L+K EE ++N I+FIDE+ ++ G E I ++
Sbjct: 281 -KLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKTHGEVERRIVSQLLTLMDGL 339
Query: 397 LARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQILRGLRERYERHHK 456
+R + +GAT + DPAL R + R + V + I L LR + ++ K
Sbjct: 340 KSRAHVIVMGAT-----NRPNSIDPALRRFGRFDREIDIGVPDEIGRLEVLR-IHTKNMK 393
Query: 457 LSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQI 516
L+ D L S+ +H Y+ D A + A +R + + + E+D E+
Sbjct: 394 LA-EDVDLERVSKDTHGYVG----ADLAALCTEAALQCIREKMDVIDLDDEEIDAEILNS 448
Query: 517 VKEKDEAVRNQDFEKA 532
+ AV N F+ A
Sbjct: 449 M-----AVSNDHFQTA 459
>At5g03340 transitional endoplasmic reticulum ATPase
Length = 810
Score = 48.5 bits (114), Expect = 2e-05
Identities = 66/293 (22%), Positives = 119/293 (40%), Gaps = 37/293 (12%)
Query: 171 TPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQD-LGADPNNIRAQVI 229
T K ++ LE + H Y+G++ L L E R D + + ++I A+++
Sbjct: 388 TKNMKLAEDVDLERISKDTHGYVGAD--LAALCTEAALQCIREKMDVIDLEDDSIDAEIL 445
Query: 230 RMVGESNNETAGVAVGRGGSSNKMPTLEEYGT----NLTKLAN-EGKLDPVVGRQQQIER 284
+ SN E A+G S T+ E ++ L N + +L V +
Sbjct: 446 NSMAVSN-EHFHTALGNSNPSALRETVVEVPNVSWEDIGGLENVKRELQETVQYPVEHPE 504
Query: 285 VIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGT 344
+ G G PG GKT +A+ +A + +++G E++T+
Sbjct: 505 KFEKFGMSPSKGVLFYGPPGCGKTLLAKAIANECQANFI--SVKGPELLTM--------- 553
Query: 345 KYRGEFEERLKKLMEEIKQNDNIILFIDEVHTLI-----GAGAAEGAIDAANILKPAL-- 397
+ GE E ++++ ++ +Q+ +LF DE+ ++ AG A GA D +L L
Sbjct: 554 -WFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSAGDAGGAAD--RVLNQLLTE 610
Query: 398 -----ARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQILR 445
A+ + IGAT + P R Q + +P P D + I +
Sbjct: 611 MDGMNAKKTVFIIGATNRPDIIDSALLRPG--RLDQLIYIPLPDEDSRLNIFK 661
Score = 45.1 bits (105), Expect = 2e-04
Identities = 67/256 (26%), Positives = 107/256 (41%), Gaps = 35/256 (13%)
Query: 284 RVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAG 343
++ + +G + L G PG GKT IA +A TG I G E+++
Sbjct: 231 QLFKSIGVKPPKGILLYGPPGSGKTLIARAVANE--TGAFFFCINGPEIMS--------- 279
Query: 344 TKYRGEFEERLKKLMEEIKQNDNIILFIDEVHTLIGA-----GAAEGAI--DAANILKPA 396
K GE E L+K EE ++N I+FIDE+ ++ G E I ++
Sbjct: 280 -KLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKTNGEVERRIVSQLLTLMDGL 338
Query: 397 LARGELQCIGATTLDEYRKHIEKDPALERRFQPVRVPEPTVDESIQILRGLRERYERHHK 456
+R + +GAT + DPAL R + R + V + I L LR + ++ K
Sbjct: 339 KSRAHVIVMGAT-----NRPNSIDPALRRFGRFDREIDIGVPDEIGRLEVLR-IHTKNMK 392
Query: 457 LSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQI 516
L+ D L S+ +H Y+ D A + A +R + + E +D E+
Sbjct: 393 LA-EDVDLERISKDTHGYVG----ADLAALCTEAALQCIREKMDVIDLEDDSIDAEILNS 447
Query: 517 VKEKDEAVRNQDFEKA 532
+ AV N+ F A
Sbjct: 448 M-----AVSNEHFHTA 458
>At1g64110 unknown protein
Length = 824
Score = 48.1 bits (113), Expect = 2e-05
Identities = 63/256 (24%), Positives = 107/256 (41%), Gaps = 40/256 (15%)
Query: 297 PC----LVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEE 352
PC L G PG GKT +A+ +A+ G I + M + + K+ GE E+
Sbjct: 549 PCRGILLFGPPGTGKTMLAKAIAKEA----------GASFINVSMSTITS--KWFGEDEK 596
Query: 353 RLKKLMEEIKQNDNIILFIDEVHTLIGAGAAEGAIDAANILKPALAR---------GELQ 403
++ L + I+F+DEV +++G G +A +K GE
Sbjct: 597 NVRALFTLASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLMTKPGERI 656
Query: 404 CIGATTLDEYRKHIEKDPALERRFQ-PVRVPEPTVDESIQILRGLRERYERHHKLSYTDD 462
+ A T + + D A+ RRF+ + V P V+ +ILR L + + L Y +
Sbjct: 657 LVLAAT----NRPFDLDEAIIRRFERRIMVGLPAVENREKILRTLLAKEKVDENLDYKEL 712
Query: 463 ALV----AASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELD--KEVRQI 516
A++ S L + + + P + + + E + + P +A E D KE R I
Sbjct: 713 AMMTEGYTGSDLKNLCTTAAYRPVREL-IQQERIKDTEKKKQREPTKAGEEDEGKEERVI 771
Query: 517 VKEKDEAVRNQDFEKA 532
+ QDF++A
Sbjct: 772 TL---RPLNRQDFKEA 784
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,404,592
Number of Sequences: 26719
Number of extensions: 841373
Number of successful extensions: 3573
Number of sequences better than 10.0: 162
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 132
Number of HSP's that attempted gapping in prelim test: 3297
Number of HSP's gapped (non-prelim): 320
length of query: 923
length of database: 11,318,596
effective HSP length: 108
effective length of query: 815
effective length of database: 8,432,944
effective search space: 6872849360
effective search space used: 6872849360
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0175.11


