

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0174.3
(919 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
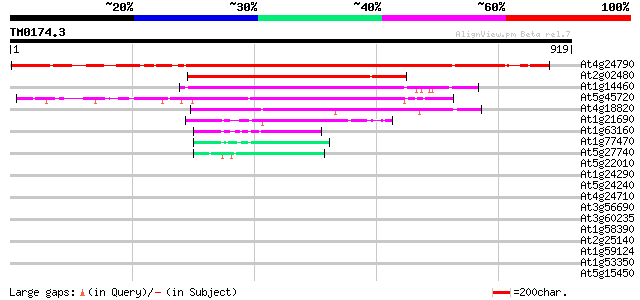
Score E
Sequences producing significant alignments: (bits) Value
At4g24790 DNA polymerase III like protein 676 0.0
At2g02480 STICHEL (STI) 362 e-100
At1g14460 hypothetical protein 355 8e-98
At5g45720 putative protein 314 1e-85
At4g18820 putative protein 311 1e-84
At1g21690 putative replication factor C subunit 73 8e-13
At1g63160 replication factor, putative 52 2e-06
At1g77470 putative replication factor C 49 2e-05
At5g27740 replication factor C - like 46 8e-05
At5g22010 replication factor C large subunit-like protein 40 0.005
At1g24290 unknown protein 37 0.051
At5g24240 ubiquitin 35 0.19
At4g24710 protein binding protein like 33 0.56
At3g56690 calmodulin-binding protein 33 0.56
At3g60235 initiation factor eIF-4 gamma like 33 0.96
At1g58390 disease resistance protein PRM1 32 1.3
At2g25140 putative ATP-dependent CLPB protein 32 2.1
At1g59124 PRM1 homolog (RF45) 31 2.8
At1g53350 hypothetical protein 31 2.8
At5g15450 clpB heat shock protein-like 31 3.7
>At4g24790 DNA polymerase III like protein
Length = 815
Score = 676 bits (1743), Expect = 0.0
Identities = 411/891 (46%), Positives = 545/891 (61%), Gaps = 94/891 (10%)
Query: 4 RRHSVDIPISKTLVALRRVRSLRDPSTNSICKRSSLIDNVQWENGSGNGISLLFPE-ASR 62
RRHSVD+PI++TLVALRRVRSLRDP T S+ K +SL+DNV+WE GS NGISL F E A
Sbjct: 5 RRHSVDVPITRTLVALRRVRSLRDPCTTSMSKFASLLDNVKWETGSNNGISLQFVEHADD 64
Query: 63 ECDSDDNVDFTSRNLGFKGQRVQDATDFKLDCGLLGSRLKPSGMSFQDSQHNDDELVYSN 122
C + D + F + + + D L S++
Sbjct: 65 ACKAA--ADAPVGLIPFGSYSIMEELESGCDLHKLSSKVI-------------------- 102
Query: 123 PNRQCSSGNKSSSESCDSNHGGEGLDLASIEPHSNNFRDGESCFIATAKSSQLGRIDHSR 182
N + + ++SS SC S+ +G DLA P + ++ +
Sbjct: 103 -NVEGDACSRSSERSC-SDLSVKGRDLACNAP-------------------SISHVEEAG 141
Query: 183 SDKKSLRANQVRPAQVEGGIVNHVGNPCLSVHDDFSPHSTSVDINQDFDVLDNNDNGCGI 242
S + + A G + +G+P S + HS D + DFD N GCGI
Sbjct: 142 SGGRYRTHYSTKLASSVGEYGSRLGSPMNSTN-----HSYYGDEDVDFDSQSNR--GCGI 194
Query: 243 SFCWSKSPRFRESNLYSKIEDRPLILHQVDDTDLHGDRNMRHDGGDISPTLET-PRSLSM 301
++CWS++PR+R SN S +E+ PL L G+ N D ++P+ E RSLS
Sbjct: 195 TYCWSRTPRYRGSNQSSDVEEYPL---------LPGNGNGESDV--VTPSHEVLSRSLSQ 243
Query: 302 KFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGKTSASRIFAAALNCLSVE 361
KFRP SF +LVGQ VVV+ LL I RG I S YLFHGPRGTGKTS S+IFAAALNCLS
Sbjct: 244 KFRPKSFDELVGQEVVVKCLLSTILRGRITSVYLFHGPRGTGKTSTSKIFAAALNCLSQA 303
Query: 362 GQ-KPCGLCRECVLFFSGRSKEVKEVDSVRINRTDQVKSLVKNACIPPISSRFKVFIIDE 420
+PCGLC EC +FSGR ++V E DS ++NR ++SL+K+A +PP+SSRFKVFIIDE
Sbjct: 304 AHSRPCGLCSECKSYFSGRGRDVMETDSGKLNRPSYLRSLIKSASLPPVSSRFKVFIIDE 363
Query: 421 CQLLKAETWASLSNNLENLSEHVVFVMITPDLDKLPRSAVSRAQTYHFPKVKDADIACRL 480
CQLL ETW +L N+L+N S+H VF+++T +L+KLPR+ +SR+Q YHF KV DADI+ +L
Sbjct: 364 CQLLCQETWGTLLNSLDNFSQHSVFILVTSELEKLPRNVLSRSQKYHFSKVCDADISTKL 423
Query: 481 EKICVEEGLDFDQAALDFIAAKSCGSVRDAEMMLDQLSLLGKKIKLSLVHELTGIVSDDE 540
KIC+EEG+DFDQ A+DFIA+KS GS+RDAE+MLDQLSLLGK+I SL ++L G+VSDDE
Sbjct: 424 AKICIEEGIDFDQGAVDFIASKSDGSLRDAEIMLDQLSLLGKRITTSLAYKLIGVVSDDE 483
Query: 541 LLNLLDLALSSDTSNTVIRARELMRSRIGPLQLVSQLANLIMDILAGKCVDGGSEVRSKF 600
LL+LLDLA+SSDTSNTVIRARELMRS+I P+QL+SQLAN+IMDI+AG + S R +F
Sbjct: 484 LLDLLDLAMSSDTSNTVIRARELMRSKIDPMQLISQLANVIMDIIAGNSQESSSATRLRF 543
Query: 601 SGRYTSEADLQKLSRAVRILSEAEKQLRISKNQTTWLTVALLQLSSVDYPS--VDVNDTK 658
R+TSE ++QKL A++ILS+AEK LR SKNQTTWLTVALLQLS+ D S D N
Sbjct: 544 LTRHTSEEEMQKLRNALKILSDAEKHLRASKNQTTWLTVALLQLSNTDSSSFATDENGRN 603
Query: 659 LCLREQSMEHLATGQCGD---KSYRLGVQEDHKGTLDAIWYKATEICQSGQLKTFLRKKG 715
++ + ++G GD G + + T++++W T++C S LK FL K+G
Sbjct: 604 QINKDVELSSTSSGCPGDVIKSDAEKGQERNCNETVESVWKTVTDLCCSDSLKRFLWKRG 663
Query: 716 KLSSLHVDRSTSWLAIAELEFRHRHHVSKAEKSWKLIASSLQFILGCNIELRITYEPRA- 774
+L+SL VD+ +AIAELEF HV++AEKSWKLIA S Q +LGCN+E+++ A
Sbjct: 664 RLTSLTVDKG---VAIAELEFYTPQHVARAEKSWKLIADSFQSVLGCNVEIQMNLVISAC 720
Query: 775 SDSKYAKLKRS-SFSIFSCSRRAQQKSLSSNGSESDYADHVSQNPMIKDKTLTSSDYHGM 833
S K AK S F +FSCSRR KS + ++SD A S+ P
Sbjct: 721 SPPKSAKAAASLFFGLFSCSRRMLHKSYLTTRTDSDCA---SEKPA-------------- 763
Query: 834 DVVTTLRSCKGNLLSSGERFLNRSFQENMGTSCSEVDSSKEKGNRCAHLVP 884
V +LRSC+GN+L + R + S + SCS H+ P
Sbjct: 764 -VTNSLRSCQGNVLRA--RSVRSSANASSRMSCSSDQGDATSAMCTPHIPP 811
>At2g02480 STICHEL (STI)
Length = 1218
Score = 362 bits (929), Expect = e-100
Identities = 195/363 (53%), Positives = 257/363 (70%), Gaps = 5/363 (1%)
Query: 292 TLETPRSLSMKFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGKTSASRIF 351
T ET RS S K+RP F +L+GQ++VV+SL+ A+ R IA YLF GPRGTGKTS +RIF
Sbjct: 444 TPETIRSFSQKYRPMFFEELIGQSIVVQSLMNAVKRSRIAPVYLFQGPRGTGKTSTARIF 503
Query: 352 AAALNCLSVEGQKPCGLCRECVLFFSGRSKEVKEVDSVRINRTDQVKSLVKN--ACIPPI 409
+AALNC++ E KPCG C+EC F SG+SK+ E+D D+V+ L+KN +P
Sbjct: 504 SAALNCVATEEMKPCGYCKECNDFMSGKSKDFWELDGANKKGADKVRYLLKNLPTILPRN 563
Query: 410 SSRFKVFIIDECQLLKAETWASLSNNLENLSEHVVFVMITPDLDKLPRSAVSRAQTYHFP 469
SS +KVF+IDEC LL ++TW S LEN + VVF+ IT DL+ +PR+ SR Q + F
Sbjct: 564 SSMYKVFVIDECHLLPSKTWLSFLKFLENPLQKVVFIFITTDLENVPRTIQSRCQKFLFD 623
Query: 470 KVKDADIACRLEKICVEEGLDFDQAALDFIAAKSCGSVRDAEMMLDQLSLLGKKIKLSLV 529
K+KD+DI RL+KI +E LD D ALD IA + GS+RDAE ML+QLSLLGK+I +LV
Sbjct: 624 KLKDSDIVVRLKKIASDENLDVDLHALDLIAMNADGSLRDAETMLEQLSLLGKRITTALV 683
Query: 530 HELTGIVSDDELLNLLDLALSSDTSNTVIRARELMRSRIGPLQLVSQLANLIMDILAG-- 587
+EL G+VSD++LL LL+LALSSDT+ TV RAREL+ P+ L+SQLA+LIMDI+AG
Sbjct: 684 NELVGVVSDEKLLELLELALSSDTAETVKRARELLDLGADPIVLMSQLASLIMDIIAGTY 743
Query: 588 KCVDGGSEVRSKFSGRYTSEADLQKLSRAVRILSEAEKQLRISKNQTTWLTVALLQLSSV 647
K VD + F GR +EAD++ L A+++LSEAEKQLR+S +++TW T LLQL S+
Sbjct: 744 KVVD-EKYSNAFFDGRNLTEADMEGLKHALKLLSEAEKQLRVSNDRSTWFTATLLQLGSM 802
Query: 648 DYP 650
P
Sbjct: 803 PSP 805
Score = 37.4 bits (85), Expect = 0.039
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 3/82 (3%)
Query: 691 LDAIWYKATEICQSGQLKTFLRKKGKLSSLHVDRSTSWLAIAELEFRHRHHVSKAEKSWK 750
L+ IW K E C S L+ L GKL S+ + +A + F +AE+
Sbjct: 912 LNDIWRKCIERCHSKTLRQLLYTHGKLISI---SEVEGILVAYIAFGENDIKLRAERFLS 968
Query: 751 LIASSLQFILGCNIELRITYEP 772
I +S++ +L ++E+RI P
Sbjct: 969 SITNSIEMVLRRSVEVRIILLP 990
>At1g14460 hypothetical protein
Length = 1102
Score = 355 bits (910), Expect = 8e-98
Identities = 221/547 (40%), Positives = 313/547 (56%), Gaps = 64/547 (11%)
Query: 278 GDRNMRHDGGDISPTLETPRSLSMKFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFH 337
G+R +GG T E+ +SLS K++P F +L+GQ++VV+SL+ A+ +G +A YLF
Sbjct: 406 GEREEEEEGGS---TPESIQSLSQKYKPMFFDELIGQSIVVQSLMNAVKKGRVAHVYLFQ 462
Query: 338 GPRGTGKTSASRIFAAALNC-LSVEGQKPCGLCRECVLFFSGRSKEVKEVDSVRINRTDQ 396
GPRGTGKTS +RI +AALNC + E KPCG C+EC + G+S+++ E+D+ + N ++
Sbjct: 463 GPRGTGKTSTARILSAALNCDVVTEEMKPCGYCKECSDYMLGKSRDLLELDAGKKNGAEK 522
Query: 397 VKSLVKN--ACIPPISSRFKVFIIDECQLLKAETWASLSNNLENLSEHVVFVMITPDLDK 454
V+ L+K P S R+KVF+IDEC LL + TW SL LEN + VFV IT DLD
Sbjct: 523 VRYLLKKLLTLAPQSSQRYKVFVIDECHLLPSRTWLSLLKFLENPLQKFVFVCITTDLDN 582
Query: 455 LPRSAVSRAQTYHFPKVKDADIACRLEKICVEEGLDFDQAALDFIAAKSCGSVRDAEMML 514
+PR+ SR Q Y F KV+D DI RL KI +E LD + ALD IA + GS+RDAE ML
Sbjct: 583 VPRTIQSRCQKYIFNKVRDGDIVVRLRKIASDENLDVESQALDLIALNADGSLRDAETML 642
Query: 515 DQLSLLGKKIKLSLVHELTGIVSDDELLNLLDLALSSDTSNTVIRARELMRSRIGPLQLV 574
+QLSL+GK+I + LV+EL G+VSDD+LL LL+LALSSDT+ TV +AREL+ P+ ++
Sbjct: 643 EQLSLMGKRITVDLVNELVGVVSDDKLLELLELALSSDTAETVKKARELLDLGADPILMM 702
Query: 575 SQLANLIMDILAGKCVDGGSEVRSKFSGRYT-SEADLQKLSRAVRILSEAEKQLRISKNQ 633
SQLA+LIMDI+AG + F R +EADL++L A+++LSEAEKQLR+S ++
Sbjct: 703 SQLASLIMDIIAGAYKALDEKYSEAFLDRRNLTEADLERLKHALKLLSEAEKQLRVSTDR 762
Query: 634 TTWLTVALLQLSSVDYPSVDVNDTKLCLREQ--------SMEHLATGQ-----CGDKSYR 680
+TW LLQL S+ PS T R+ S E +A Q C + +
Sbjct: 763 STWFIATLLQLGSM--PSPGTTHTGSSRRQSSRATEESISREVIAYKQRSGLQCSNTASP 820
Query: 681 LGVQED-------------------------HKGT--------------LDAIWYKATEI 701
+++ H T L+ IW K +
Sbjct: 821 TSIRKSGNLVREVKLSSSSSEVLESDTSMASHDDTTASTMTLTCRNSEKLNDIWIKCVDR 880
Query: 702 CQSGQLKTFLRKKGKLSSLHVDRSTSWLAIAELEFRHRHHVSKAEKSWKLIASSLQFILG 761
C S LK L GKL S+ + +A + F ++AE+ I +S++ +L
Sbjct: 881 CHSKTLKQLLYAHGKLLSI---SEVEGILVAYIAFGEGEIKARAERFVSSITNSIEMVLR 937
Query: 762 CNIELRI 768
N+E+RI
Sbjct: 938 RNVEVRI 944
>At5g45720 putative protein
Length = 900
Score = 314 bits (805), Expect = 1e-85
Identities = 251/791 (31%), Positives = 380/791 (47%), Gaps = 141/791 (17%)
Query: 12 ISKTLVALRRVRSLRDPSTNSICKRSSLIDNVQWENGSGNGISLLFP----EASRECDSD 67
+ + LV L+R RSLRDPS + K D+ + SG +S P S+ SD
Sbjct: 53 LMRDLVVLQRSRSLRDPSASPNLKEDHQ-DSREGRRRSGLRLSGSSPIVSFGTSKVTPSD 111
Query: 68 DNVDFTSRNLGFKGQRVQDATDFKLDCGLLGSRLKPSGMSFQDSQHNDDELVYSNPNRQC 127
+ D +SR K RV++ + VYS P+ +
Sbjct: 112 EKFDRSSR----KSYRVEEVNE-----------------------------VYSVPSVKS 138
Query: 128 SSG---NKSSSESCD-------SNHGGEGLDLASIEPHSNNFRDGESCFIATAKSSQLGR 177
S NK +E+ + GG+ DL S G+ C R
Sbjct: 139 VSKDRINKKVNEAIVKTLSDQLNEVGGDSDDLVSCNVRPR----GDGC-----------R 183
Query: 178 IDHSRSDKKSLRANQVRPAQVEGGIVNHVGNPC-LSVHDDFSPHSTSVDINQDFDVLDNN 236
R +++ RA VR N GN +S+ + P + + D
Sbjct: 184 RRKFRGTRRAGRAVNVRD--------NAAGNESEMSIASNSVPRGEKYEGEEGGGGRDRE 235
Query: 237 DN-GCGISFCWSK-------------------------------SPRFRESNLYSKIEDR 264
N CGI F WS+ +P F +S+ + E
Sbjct: 236 QNMSCGIPFNWSRIHHRGKTFLDIAGRSLSCGISDSKGRKGEAGTPMFSDSSSSDR-EAL 294
Query: 265 PLILHQVDDTDLHGDR---------NMRHDGGDISPTLETPR-------SLSMKFRPNSF 308
PL++ D+ + D N+ +G D ++ R S + K+ P +F
Sbjct: 295 PLLVDSADNEEWVHDYSGELGIFADNLLKNGKDSVIGKKSSRKNTRWHQSFTQKYAPRTF 354
Query: 309 SDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGKTSASRIFAAALNCLSVEGQKPCGL 368
DL+GQN+VV++L AI++ + Y+FHGP GTGKTS +R+FA ALNC S E KPCG+
Sbjct: 355 RDLLGQNLVVQALSNAIAKRRVGLLYVFHGPNGTGKTSCARVFARALNCHSTEQSKPCGV 414
Query: 369 CRECVLFFSGRSKEVKEVDSVRINRTDQVKSLVKNACIPPISSRFKVFIIDECQLLKAET 428
C CV + G+++ ++E+ V+ + ++L+ I + V I D+C + +
Sbjct: 415 CSSCVSYDDGKNRYIREMGPVK---SFDFENLLDKTNIRQQQKQQLVLIFDDCDTMSTDC 471
Query: 429 WASLSNNLENLSEHVVFVMITPDLDKLPRSAVSRAQTYHFPKVKDADIACRLEKICVEEG 488
W +LS ++ VVFV++ LD LP VSR Q + FPK+KD DI L+ I +E
Sbjct: 472 WNTLSKIVDRAPRRVVFVLVCSSLDVLPHIIVSRCQKFFFPKLKDVDIIDSLQLIASKEE 531
Query: 489 LDFDQAALDFIAAKSCGSVRDAEMMLDQLSLLGKKIKLSLVHELTGIVSDDELLNLLDLA 548
+D D+ AL +A++S GS+RDAEM L+QLSLLG +I + LV E+ G++SD++L++LLDLA
Sbjct: 532 IDIDKDALKLVASRSDGSLRDAEMTLEQLSLLGTRISVPLVQEMVGLISDEKLVDLLDLA 591
Query: 549 LSSDTSNTVIRARELMRSRIGPLQLVSQLANLIMDILAGKCVDGGSEVRSKFSGRY-TSE 607
LS+DT NTV R +M + + PL L+SQLA +I DILAG + + KF R S+
Sbjct: 592 LSADTVNTVKNLRIIMETGLEPLALMSQLATVITDILAGSYDFTKDQCKRKFFRRQPLSK 651
Query: 608 ADLQKLSRAVRILSEAEKQLRISKNQTTWLTVALLQL-----------SSVDYPSVDVND 656
D++KL +A++ LSE+EKQLR+S ++ TWLT ALLQL SS D N
Sbjct: 652 EDMEKLKQALKTLSESEKQLRVSNDKLTWLTAALLQLAPDKQYLLPHSSSAD---ASFNH 708
Query: 657 TKLCLREQSMEHLATGQCGDKSYRLGVQEDHKGTLDAIWYKATEICQSGQLKTFLRKKGK 716
T L + S H+ G D S + G ++ +++ IW E + L+ FL K+GK
Sbjct: 709 TPLTDSDPS-NHVVAGTRRDDS-KQGFSCKNRPSVEDIWLAVIENVRVNGLREFLYKEGK 766
Query: 717 LSSLHVDRSTS 727
+ S+ + + S
Sbjct: 767 IFSISIGSAKS 777
>At4g18820 putative protein
Length = 1057
Score = 311 bits (796), Expect = 1e-84
Identities = 196/501 (39%), Positives = 289/501 (57%), Gaps = 30/501 (5%)
Query: 297 RSLSMKFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGKTSASRIFAAALN 356
+SL+ K+ P +F DL+GQN+VV++L A++R + Y+FHGP GTGKTS +RIFA ALN
Sbjct: 431 QSLTEKYTPKTFRDLLGQNLVVQALSNAVARRKLGLLYVFHGPNGTGKTSCARIFARALN 490
Query: 357 CLSVEGQKPCGLCRECVLFFSGRSKEVKEVDSVRINRTDQVKSLVK-NACIPPISSRFKV 415
C S+E KPCG C CV G+S ++EV V +++ L+ N + S R V
Sbjct: 491 CHSMEQPKPCGTCSSCVSHDMGKSWNIREVGPVGNYDFEKIMDLLDGNVMVSSQSPR--V 548
Query: 416 FIIDECQLLKAETWASLSNNLENLS-EHVVFVMITPDLDKLPRSAVSRAQTYHFPKVKDA 474
FI D+C L ++ W +LS ++ + HVVF+++ LD LP +SR Q + FPK+KDA
Sbjct: 549 FIFDDCDTLSSDCWNALSKVVDRAAPRHVVFILVCSSLDVLPHVIISRCQKFFFPKLKDA 608
Query: 475 DIACRLEKICVEEGLDFDQAALDFIAAKSCGSVRDAEMMLDQLSLLGKKIKLSLVHEL-- 532
DI L+ I +E ++ D+ AL IA++S GS+RDAEM L+QLSLLG++I + LV EL
Sbjct: 609 DIVYSLQWIASKEEIEIDKDALKLIASRSDGSLRDAEMTLEQLSLLGQRISVPLVQELVS 668
Query: 533 ------------TGIVSDDELLNLLDLALSSDTSNTVIRARELMRSRIGPLQLVSQLANL 580
G+VSD++L++LLDLALS+DT NTV R +M + + PL L+SQLA +
Sbjct: 669 IVLSHHHIKQLKVGLVSDEKLVDLLDLALSADTVNTVKNLRTIMETSVEPLALMSQLATV 728
Query: 581 IMDILAGKCVDGGSEVRSKFSGRY-TSEADLQKLSRAVRILSEAEKQLRISKNQTTWLTV 639
I DILAG + + KF R + D++KL +A++ LSEAEKQLR+S ++ TWLT
Sbjct: 729 ITDILAGSYDFTKDQHKRKFFRRQPLPKEDMEKLRQALKTLSEAEKQLRVSNDKLTWLTA 788
Query: 640 ALLQLS-SVDYPSVDVNDTKLCLREQSMEHL------ATGQCGDKSYRLGVQEDHKGTLD 692
ALLQL+ +Y + RE S HL A G+ R G ++ ++
Sbjct: 789 ALLQLAPDQNYLLQRSSTADTGGRESSDHHLDPSSDAAGGRSSGLDRRRGDSRKNRPAVE 848
Query: 693 AIWYKATEICQSGQLKTFLRKKGKLSSLHVDRSTSWLAIAELEFRHRHHVSKAEKSWKLI 752
IW + E + L+ FL K+G++ SL++ + + L F S AEK I
Sbjct: 849 EIWLEVIEKLRVNGLREFLYKEGRIVSLNLGSAPT----VHLMFSSPLTKSTAEKFRSHI 904
Query: 753 ASSLQFILGCNIELRITYEPR 773
+ + +L + + I E +
Sbjct: 905 MQAFEAVLESPVTIEIRCETK 925
>At1g21690 putative replication factor C subunit
Length = 339
Score = 72.8 bits (177), Expect = 8e-13
Identities = 82/350 (23%), Positives = 145/350 (41%), Gaps = 50/350 (14%)
Query: 289 ISPTLETPRSLSMKFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGKTSAS 348
++P L++ + K+RP D+ Q VVR L + LF+GP GTGKT+ +
Sbjct: 1 MAPVLQSSQPWVEKYRPKQVKDVAHQEEVVRVLTNTLQTADCPHM-LFYGPPGTGKTTTA 59
Query: 349 RIFAAALNCLSVEGQKPCGLCRECVLFFSGRSKEVKEVDSVRINRTDQVKSLVKNACIPP 408
A A E + R E+ D IN V++ +K+
Sbjct: 60 --LAIAHQLFGPE-------------LYKSRVLELNASDDRGIN---VVRTKIKDFAAVA 101
Query: 409 ISSR----------FKVFIIDECQLLKAETWASLSNNLENLSEHVVFVMITPDLDKLPRS 458
+ S FK+ I+DE + + +L +E S+ F I + ++
Sbjct: 102 VGSNHRQSGYPCPSFKIIILDEADSMTEDAQNALRRTMETYSKVTRFFFICNYISRIIEP 161
Query: 459 AVSRAQTYHFPKVKDADIACRLEKICVEEGLDFDQAALDFIAAKSCGSVRDAEMMLDQLS 518
SR + F + + ++ R+ IC EEGL D AL +++ S G +R A L +
Sbjct: 162 LASRCAKFRFKPLSEEVMSNRILHICNEEGLSLDGEALSTLSSISQGDLRRAITYLQSAT 221
Query: 519 -LLGKKIKLSLVHELTGIVSDDELLNLLDLALSSDTSNTVIRARELMRSRIGPLQLVSQL 577
L G I + + ++G+V + + L S D A + + + + SQ+
Sbjct: 222 RLFGSTITSTDLLNVSGVVPLEVVNKLFTACKSGDFD----IANKEVDNIVAEGYPASQI 277
Query: 578 ANLIMDILAGKCVDGGSEVRSKFSGRYTSEADLQKLSRAVRILSEAEKQL 627
N + DI+A + S++ D+QK ++ + L+E +K+L
Sbjct: 278 INQLFDIVA----EADSDI-----------TDMQK-AKICKCLAETDKRL 311
>At1g63160 replication factor, putative
Length = 333
Score = 52.0 bits (123), Expect = 2e-06
Identities = 56/210 (26%), Positives = 88/210 (41%), Gaps = 19/210 (9%)
Query: 302 KFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGKTSASRIFAAALNCLSVE 361
K+RP+ D+VG V S L I+R + GP GTGKT++ I A A L
Sbjct: 20 KYRPSKVVDIVGNEDAV-SRLQVIARDGNMPNLILSGPPGTGKTTS--ILALAHELLGTN 76
Query: 362 GQKPCGLCRECVLFFSGRSKEVKEVDSVRINRTDQVKSLV-KNACIPPISSRFKVFIIDE 420
+E VL + + + + +D VR +++K K +PP R KV I+DE
Sbjct: 77 -------YKEAVLELN--ASDDRGIDVVR----NKIKMFAQKKVTLPP--GRHKVVILDE 121
Query: 421 CQLLKAETWASLSNNLENLSEHVVFVMITPDLDKLPRSAVSRAQTYHFPKVKDADIACRL 480
+ + +L +E S F + K+ SR F ++ D I RL
Sbjct: 122 ADSMTSGAQQALRRTIEIYSNSTRFALACNTSAKIIEPIQSRCALVRFSRLSDQQILGRL 181
Query: 481 EKICVEEGLDFDQAALDFIAAKSCGSVRDA 510
+ E + + L+ I + G +R A
Sbjct: 182 LVVVAAEKVPYVPEGLEAIIFTADGDMRQA 211
>At1g77470 putative replication factor C
Length = 369
Score = 48.5 bits (114), Expect = 2e-05
Identities = 48/223 (21%), Positives = 90/223 (39%), Gaps = 16/223 (7%)
Query: 302 KFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGKTSASRIFAAALNCLSVE 361
K+RP S D+ ++ ++ + + L+ GP GTGKTS I A A +
Sbjct: 44 KYRPQSLDDVAAHRDIIDTIDRLTNENKLPHLLLY-GPPGTGKTST--ILAVA---RKLY 97
Query: 362 GQKPCGLCRECVLFFSGRSKEVKEVDSVRINRTDQVKSLVKNACIPPISSRFKVFIIDEC 421
G K + E + + + +D VR Q++ S K+ ++DE
Sbjct: 98 GPKYRNMILEL------NASDDRGIDVVR----QQIQDFASTQSFSLGKSSVKLVLLDEA 147
Query: 422 QLLKAETWASLSNNLENLSEHVVFVMITPDLDKLPRSAVSRAQTYHFPKVKDADIACRLE 481
+ + +L +E ++ F +I ++K+ + SR + F + ++ RL+
Sbjct: 148 DAMTKDAQFALRRVIEKYTKSTRFALIGNHVNKIIPALQSRCTRFRFAPLDGVHMSQRLK 207
Query: 482 KICVEEGLDFDQAALDFIAAKSCGSVRDAEMMLDQLSLLGKKI 524
+ E L L + S G +R A +L + K+I
Sbjct: 208 HVIEAERLVVSDCGLAALVRLSNGDMRKALNILQSTHMASKEI 250
>At5g27740 replication factor C - like
Length = 354
Score = 46.2 bits (108), Expect = 8e-05
Identities = 54/231 (23%), Positives = 90/231 (38%), Gaps = 20/231 (8%)
Query: 302 KFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGKTS-----ASRIFAAALN 356
K+RP S ++ + + L +S LF+GP G+GK + +I+ A+
Sbjct: 6 KYRPKSLDKVIVHEDIAQKLKKLVSE-QDCPHLLFYGPSGSGKKTLIMALLKQIYGASAE 64
Query: 357 CLSVE--------GQKPCGLCRECVLFFSGRSKEVKEVDSVRINR---TDQVKSLVKNAC 405
+ VE G + L E S E+ D+ +R + +K + KN
Sbjct: 65 KVKVENRAWKVDAGSRTIDL--ELTTLSSTNHVELTPSDAGFQDRYIVQEIIKEMAKNRP 122
Query: 406 IPPISSR-FKVFIIDECQLLKAETWASLSNNLENLSEHVVFVMITPDLDKLPRSAVSRAQ 464
I + +KV +++E L E SL +E S ++ K+ + SR
Sbjct: 123 IDTKGKKGYKVLVLNEVDKLSREAQHSLRRTMEKYSSSCRLILCCNSSSKVTEAIKSRCL 182
Query: 465 TYHFPKVKDADIACRLEKICVEEGLDFDQAALDFIAAKSCGSVRDAEMMLD 515
+I LE + +E L Q IA KS S+R A + L+
Sbjct: 183 NVRINAPSQEEIVKVLEFVAKKESLQLPQGFAARIAEKSNRSLRRAILSLE 233
>At5g22010 replication factor C large subunit-like protein
Length = 956
Score = 40.4 bits (93), Expect = 0.005
Identities = 60/276 (21%), Positives = 110/276 (39%), Gaps = 38/276 (13%)
Query: 293 LETPRSLSMKFRPNSFSDLVGQNVVVRSLL-----------GAISRGMIASF-------- 333
+ET + K+RP +++VG +V L G S+G
Sbjct: 336 IETSLPWTEKYRPKVPNEIVGNQSLVTQLHNWLSHWHDQFGGTGSKGKGKKLNDAGSKKA 395
Query: 334 YLFHGPRGTGKTSASRIFAAALNCLSVEGQKPCGLCRECVLFFSGRSKEVKEV-DSVRIN 392
L G G GKT+++++ + L +VE V R K + + +
Sbjct: 396 VLLSGTPGIGKTTSAKLVSQMLGFQAVE-----------VNASDSRGKANSNIAKGIGGS 444
Query: 393 RTDQVKSLVKNACIPPISSRFK----VFIIDECQLLKAETWASLSNNLEN--LSEHVVFV 446
+ VK LV N + R K V I+DE + A +++ + + +S+ +
Sbjct: 445 NANSVKELVNNEAMAANFDRSKHPKTVLIMDEVDGMSAGDRGGVADLIASIKISKIPIIC 504
Query: 447 MITPDLDKLPRSAVSRAQTYHFPKVKDADIACRLEKICVEEGLDFDQAALDFIAAKSCGS 506
+ + +S V+ ++ K +A RL I EGL+ ++ AL+ +A + G
Sbjct: 505 ICNDRYSQKLKSLVNYCLPLNYRKPTKQQMAKRLMHIAKAEGLEINEIALEELAERVNGD 564
Query: 507 VRDAEMMLDQLSLLGKKIKLSLVHE-LTGIVSDDEL 541
+R A L +SL IK + + L D+++
Sbjct: 565 IRLAVNQLQYMSLSMSVIKYDDIRQRLLSSAKDEDI 600
>At1g24290 unknown protein
Length = 525
Score = 37.0 bits (84), Expect = 0.051
Identities = 26/74 (35%), Positives = 37/74 (49%), Gaps = 6/74 (8%)
Query: 279 DRNMRHDGGDISPTLETPRSLSMKFRPNSFSDLVGQNVVVRS---LLGAISRGMIASFYL 335
D N RH S P LS + RP + D+VGQ+ ++ L A+ + S +
Sbjct: 85 DSNKRHKLSSSSHRQHQP--LSERMRPRTLDDVVGQDHLLSPSSLLRSAVESNRLPSI-V 141
Query: 336 FHGPRGTGKTSASR 349
F GP GTGKTS ++
Sbjct: 142 FWGPPGTGKTSIAK 155
>At5g24240 ubiquitin
Length = 574
Score = 35.0 bits (79), Expect = 0.19
Identities = 32/141 (22%), Positives = 59/141 (41%), Gaps = 12/141 (8%)
Query: 611 QKLSRAVRILSEAEKQLR---ISKNQTTWLTVALLQLSSVDYPSVDVNDTKLCLREQSME 667
QKL R +S + Q+R ++ + L + L L ++ +VD + +L +
Sbjct: 72 QKLLYDGREVSRNDSQIRDYGLADGKLLHLVIRLSDLQAISVRTVDGKEFELVVERSRNV 131
Query: 668 HLATGQCGDKSYRLGVQEDHKGTLDAIWYK----ATEICQSGQ--LKTFLRKKGKLSSLH 721
Q K LG+ DH+ TLD T++CQ+G + + K K+ +
Sbjct: 132 GYVKQQIASKEKELGIPRDHELTLDGEELDDQRLITDLCQNGDNVIHLLISKSAKVRAKP 191
Query: 722 VDRSTSWLAIAELEFRHRHHV 742
V + + + H+H+V
Sbjct: 192 VGKD---FEVFIEDVNHKHNV 209
>At4g24710 protein binding protein like
Length = 467
Score = 33.5 bits (75), Expect = 0.56
Identities = 35/121 (28%), Positives = 51/121 (41%), Gaps = 9/121 (7%)
Query: 335 LFHGPRGTGKTSASRIFAAALNCLSVEGQKPCGLCRECVLFFSGRSKEVKEVDSVRINRT 394
L HGP GTGKTS + A L+ C L V S SK E +
Sbjct: 206 LLHGPPGTGKTSLCKALAQKLSIRCNSRYPHCQLIE--VNAHSLFSKWFSESGKLVAKLF 263
Query: 395 DQVKSLVKNACIPPISSRFKVFIIDECQLLKAETWASLSNNLENLSEHVVFVMITPDLDK 454
+++ +V+ +IDE + L A A+LS + + S VV ++T +DK
Sbjct: 264 QKIQEMVEE------DGNLVFVLIDEVESLAAARKAALSGSEPSDSIRVVNALLT-QMDK 316
Query: 455 L 455
L
Sbjct: 317 L 317
>At3g56690 calmodulin-binding protein
Length = 1022
Score = 33.5 bits (75), Expect = 0.56
Identities = 17/30 (56%), Positives = 20/30 (66%), Gaps = 2/30 (6%)
Query: 335 LFHGPRGTGKTSASRIFA--AALNCLSVEG 362
L HGP GTGKTS +R FA + +N SV G
Sbjct: 422 LIHGPPGTGKTSLARTFARHSGVNFFSVNG 451
>At3g60235 initiation factor eIF-4 gamma like
Length = 1544
Score = 32.7 bits (73), Expect = 0.96
Identities = 44/212 (20%), Positives = 82/212 (37%), Gaps = 5/212 (2%)
Query: 507 VRDAEMMLDQLSLLGKKIKLSLVHELT---GIVSDDELLNLLDLALSSDTSNTVIRAREL 563
+R AEM+ + +S+ + K+ H LT G D L++ + A + N + A
Sbjct: 428 IRRAEMVSESISVEDQTCKVEPPHNLTENRGQTMPDSLVSDPETATVAAKENLSLPATNG 487
Query: 564 MRSRIGPLQLVSQL-ANLIMDILAGKCVDGGSEVRSKFSGRYTSEADLQKLSRAVRILSE 622
R ++ + S + +D K +G S S+ SG E DL+ +R +
Sbjct: 488 FRKQLLKVSTTSDAPTSDSVDTSIDKSTEGSSHASSEISGSSPQEKDLKCDNRTASDKLD 547
Query: 623 AEKQLRISKNQTTWLTVALLQLSSVDYPSVDVNDTKLCLREQSMEHLA-TGQCGDKSYRL 681
+ +K++T + Q V KL + + + L + + L
Sbjct: 548 ERSVISDAKHETLSGVLEKAQNEVDGATDVCPVSEKLAVTDDTSSDLPHSTHVLSSTVPL 607
Query: 682 GVQEDHKGTLDAIWYKATEICQSGQLKTFLRK 713
G E HK ++ + T ++K L+K
Sbjct: 608 GHSETHKSAVETNTRRNTSTKGKKKIKEILQK 639
>At1g58390 disease resistance protein PRM1
Length = 907
Score = 32.3 bits (72), Expect = 1.3
Identities = 24/86 (27%), Positives = 35/86 (39%)
Query: 264 RPLILHQVDDTDLHGDRNMRHDGGDISPTLETPRSLSMKFRPNSFSDLVGQNVVVRSLLG 323
R I + D G + DGG + P + R + F + SD VG V V+ L+G
Sbjct: 116 RTRISDVIRDMQSFGVQQAIVDGGYMQPQGDRQREMRQTFSKDYESDFVGLEVNVKKLVG 175
Query: 324 AISRGMIASFYLFHGPRGTGKTSASR 349
+ G G GKT+ +R
Sbjct: 176 YLVDEENVQVVSITGMGGLGKTTLAR 201
>At2g25140 putative ATP-dependent CLPB protein
Length = 964
Score = 31.6 bits (70), Expect = 2.1
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 10/54 (18%)
Query: 311 LVGQNVVVRSLLGAISRGM---------IASFYLFHGPRGTGKTSASRIFAAAL 355
++GQ++ V+S+ AI R IASF +F GP G GKT ++ A L
Sbjct: 655 VIGQDMAVKSVADAIRRSRAGLSDPNRPIASF-MFMGPTGVGKTELAKALAGYL 707
>At1g59124 PRM1 homolog (RF45)
Length = 1155
Score = 31.2 bits (69), Expect = 2.8
Identities = 19/65 (29%), Positives = 28/65 (42%)
Query: 285 DGGDISPTLETPRSLSMKFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTGK 344
DGG P + R + KF + SD VG V+ L+G + G G GK
Sbjct: 137 DGGYKQPQGDKQREMRQKFSKDDDSDFVGLEANVKKLVGYLVDEANVQVVSITGMGGLGK 196
Query: 345 TSASR 349
T+ ++
Sbjct: 197 TTLAK 201
>At1g53350 hypothetical protein
Length = 906
Score = 31.2 bits (69), Expect = 2.8
Identities = 23/66 (34%), Positives = 30/66 (44%), Gaps = 1/66 (1%)
Query: 285 DGGDISPTL-ETPRSLSMKFRPNSFSDLVGQNVVVRSLLGAISRGMIASFYLFHGPRGTG 343
DGG S +L E R + F NS SDLVG + V L+ + G G G
Sbjct: 136 DGGGRSLSLQERQREIRQTFSRNSESDLVGLDQSVEELVDHLVENDSVQVVSVSGMGGIG 195
Query: 344 KTSASR 349
KT+ +R
Sbjct: 196 KTTLAR 201
>At5g15450 clpB heat shock protein-like
Length = 968
Score = 30.8 bits (68), Expect = 3.7
Identities = 23/77 (29%), Positives = 34/77 (43%), Gaps = 10/77 (12%)
Query: 286 GGDISPTLETPRSLSMKFRPNSFSDLVGQNVVVRSLLGAISRGM---------IASFYLF 336
G +S ++ R + +VGQN V ++ AI R IASF +F
Sbjct: 625 GIPVSKLQQSERDKLLHLEEELHKRVVGQNPAVTAVAEAIQRSRAGLSDPGRPIASF-MF 683
Query: 337 HGPRGTGKTSASRIFAA 353
GP G GKT ++ A+
Sbjct: 684 MGPTGVGKTELAKALAS 700
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,584,651
Number of Sequences: 26719
Number of extensions: 889814
Number of successful extensions: 2717
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 2673
Number of HSP's gapped (non-prelim): 38
length of query: 919
length of database: 11,318,596
effective HSP length: 108
effective length of query: 811
effective length of database: 8,432,944
effective search space: 6839117584
effective search space used: 6839117584
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0174.3


