

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0174.1
(590 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
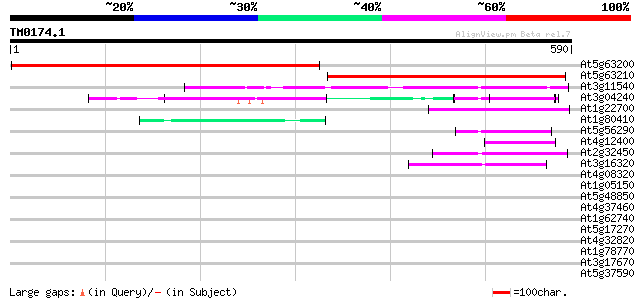
Score E
Sequences producing significant alignments: (bits) Value
At5g63200 unknown protein 416 e-116
At5g63210 putative protein 346 2e-95
At3g11540 spindly (gibberellin signal transduction protein) 72 9e-13
At3g04240 O-linked GlcNAc transferase like protein 65 1e-10
At1g22700 unknown protein 48 1e-05
At1g80410 putative N-terminal acetyltransferase (At1g80410) 48 2e-05
At5g56290 peroxisomal targeting signal type 1 receptor 45 1e-04
At4g12400 stress-induced protein sti1 -like protein 45 1e-04
At2g32450 putative O-GlcNAc transferase 43 4e-04
At3g16320 hypothetical protein 43 6e-04
At4g08320 unknown protein 42 0.001
At1g05150 putative O-GlcNAc transferase 40 0.003
At5g48850 unknown protein 40 0.004
At4g37460 unknown protein 39 0.011
At1g62740 TPR-repeat protein 39 0.011
At5g17270 putative protein 37 0.040
At4g32820 putative protein 36 0.053
At1g78770 hypothetical protein 36 0.053
At3g17670 hypothetical protein 35 0.090
At5g37590 kinesin -like protein 35 0.12
>At5g63200 unknown protein
Length = 404
Score = 416 bits (1069), Expect = e-116
Identities = 213/325 (65%), Positives = 254/325 (77%), Gaps = 1/325 (0%)
Query: 3 RLTNDENSQDKSLLS-KDTDSTEGEGKKSHKLGKCRSRPSKTDSLDCGGDADVDQHVQGA 61
RL+N+E+ Q+ +L+ K+ + E E KK K+GKCRSR S DCG DAD D QG
Sbjct: 55 RLSNEESHQEGGILTCKEVEPGEVEAKKISKVGKCRSRSKIESSSDCGVDADGDLANQGV 114
Query: 62 PSSREEKVSSMKTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPE 121
P+SREEK+S++K GLIHVARKMPKNAHAHFILGLM QRL Q QKAI YEKAEEILL E
Sbjct: 115 PASREEKISNLKMGLIHVARKMPKNAHAHFILGLMFQRLGQSQKAIPEYEKAEEILLGCE 174
Query: 122 TEIERPDLLSLVQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVW 181
EI RP+LL LVQIHH QCL+L+ + S KELE EL+EILSKLK+S++ D+RQAAVW
Sbjct: 175 PEIARPELLLLVQIHHGQCLLLDGFGDTDSVKELEGEELEEILSKLKDSIKLDVRQAAVW 234
Query: 182 NTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILK 241
NTLG +LLK G + SAISVLSSLLA+ P+NYDCL NLG+AYLQ G++ELSAKCFQ+L+LK
Sbjct: 235 NTLGLMLLKAGCLMSAISVLSSLLALVPDNYDCLANLGVAYLQSGDMELSAKCFQDLVLK 294
Query: 242 DQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSA 301
D NHP AL+NYAA LLCK++S VAGAGA+ A DQ NVAKECLLAA+++D KSA
Sbjct: 295 DHNHPAALINYAAELLCKHSSTVAGAGANGGADASEDQKAPMNVAKECLLAALRSDPKSA 354
Query: 302 HIWGNLAYAFSISGDHRSSSKCLEK 326
H W NLA ++ + GDHRSSSKCLEK
Sbjct: 355 HAWVNLANSYYMMGDHRSSSKCLEK 379
>At5g63210 putative protein
Length = 262
Score = 346 bits (888), Expect = 2e-95
Identities = 168/250 (67%), Positives = 213/250 (85%)
Query: 335 MSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQ 394
M+TR+AVA R+K+AERSQD S+ LS+ GNEMAS+IR+G+S ++ P AWAGLAM HKAQ
Sbjct: 1 MATRFAVAVQRIKDAERSQDASDQLSWAGNEMASVIREGESVPIDPPIAWAGLAMAHKAQ 60
Query: 395 HEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLK 454
HEI++A+ ++++ LTEMEERAV SLKQAV EDP+D VRWHQLG+HSLC+QQ+K SQKYLK
Sbjct: 61 HEIAAAFVADRNELTEMEERAVYSLKQAVTEDPEDAVRWHQLGLHSLCSQQYKLSQKYLK 120
Query: 455 AAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEK 514
AAV R CSY WSNLG+SLQLS+E S+AE+ YK+AL ++ + QAHAILSNLG YR +K
Sbjct: 121 AAVGRSRECSYAWSNLGISLQLSDEHSEAEEVYKRALTVSKEDQAHAILSNLGNLYRQKK 180
Query: 515 KYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
+Y+ +KAMF+K+LEL+PGYAPA+NNLGLVFVAE EEAK CFEK+L++D LLDAA+SNL
Sbjct: 181 QYEVSKAMFSKALELKPGYAPAYNNLGLVFVAERRWEEAKSCFEKSLEADSLLDAAQSNL 240
Query: 575 VKVVTMSKIC 584
+K TMS++C
Sbjct: 241 LKATTMSRLC 250
>At3g11540 spindly (gibberellin signal transduction protein)
Length = 914
Score = 72.0 bits (175), Expect = 9e-13
Identities = 89/403 (22%), Positives = 167/403 (41%), Gaps = 38/403 (9%)
Query: 185 GFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQN 244
G L + A S + + P N L + GI + + G L +A+ +Q+ ++ D +
Sbjct: 84 GICLQTQNKGNLAFDCFSEAIRLDPHNACALTHCGILHKEEGRLVEAAESYQKALMADAS 143
Query: 245 HPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIW 304
+ A A++L + + AG + EG + A+K D A +
Sbjct: 144 YKPA-AECLAIVLTDLGTSLKLAG-NTQEGI------------QKYYEALKIDPHYAPAY 189
Query: 305 GNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGN 364
NL +S + ++ C EKAA P YA A M +++ E+
Sbjct: 190 YNLGVVYSEMMQYDNALSCYEKAALERP-----MYAEAYCNMGVIYKNRGDLEMAITCYE 244
Query: 365 EMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVA 424
++ + + + + A L K + +++ + V K+A+
Sbjct: 245 RCLAVSPNFEIAKNNMAIALTDLGTKVKLEGDVT---------------QGVAYYKKALY 289
Query: 425 EDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAE 484
+ + LGV +F + + + A + C+ +NLGV + + +A
Sbjct: 290 YNWHYADAMYNLGVAYGEMLKFDMAIVFYELAFHFNPHCAEACNNLGVLYKDRDNLDKAV 349
Query: 485 KAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVF 544
+ Y+ AL + K L+NLG+ Y + K A +M K++ P YA AFNNLG+++
Sbjct: 350 ECYQMALSI--KPNFAQSLNNLGVVYTVQGKMDAAASMIEKAILANPTYAEAFNNLGVLY 407
Query: 545 VAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKICKGL 587
G + A +E+ L+ DP D+ + +++ M+ I +GL
Sbjct: 408 RDAGNITMAIDAYEECLKIDP--DSRNAGQNRLLAMNYINEGL 448
Score = 40.8 bits (94), Expect = 0.002
Identities = 37/173 (21%), Positives = 71/173 (40%), Gaps = 16/173 (9%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHAQCLIL 143
P NA A G++H+ + +A Y+KA L+ + + L++V L L
Sbjct: 108 PHNACALTHCGILHKEEGRLVEAAESYQKA---LMADASYKPAAECLAIVLTDLGTSLKL 164
Query: 144 ESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSS 203
+++ E + K E+++ D A + LG + + + +A+S
Sbjct: 165 AGNTQ-------------EGIQKYYEALKIDPHYAPAYYNLGVVYSEMMQYDNALSCYEK 211
Query: 204 LLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALL 256
P + N+G+ Y G+LE++ C++ + N +A N A L
Sbjct: 212 AALERPMYAEAYCNMGVIYKNRGDLEMAITCYERCLAVSPNFEIAKNNMAIAL 264
Score = 36.2 bits (82), Expect = 0.053
Identities = 15/61 (24%), Positives = 30/61 (48%)
Query: 182 NTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILK 241
N LG + G++ +A S++ + P + NLG+ Y GN+ ++ ++E +
Sbjct: 367 NNLGVVYTVQGKMDAAASMIEKAILANPTYAEAFNNLGVLYRDAGNITMAIDAYEECLKI 426
Query: 242 D 242
D
Sbjct: 427 D 427
Score = 31.2 bits (69), Expect = 1.7
Identities = 46/254 (18%), Positives = 99/254 (38%), Gaps = 37/254 (14%)
Query: 80 ARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHAQ 139
A + P A A+ +G++++ + AI YE+ + + P EI + ++ +
Sbjct: 213 ALERPMYAEAYCNMGVIYKNRGDLEMAITCYERC--LAVSPNFEIAKNNMAIAL------ 264
Query: 140 CLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAIS 199
++ + +LE ++ + ++ K+++ ++ A LG + + AI
Sbjct: 265 -------TDLGTKVKLEG-DVTQGVAYYKKALYYNWHYADAMYNLGVAYGEMLKFDMAIV 316
Query: 200 VLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCK 259
P + NLG+ Y NL+ + +C+Q + N +L N +
Sbjct: 317 FYELAFHFNPHCAEACNNLGVLYKDRDNLDKAVECYQMALSIKPNFAQSLNNLGVVY--- 373
Query: 260 YASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRS 319
+V A+AS + AI A+ A + NL + +G+
Sbjct: 374 --TVQGKMDAAAS----------------MIEKAILANPTYAEAFNNLGVLYRDAGNITM 415
Query: 320 SSKCLEKAAKLEPN 333
+ E+ K++P+
Sbjct: 416 AIDAYEECLKIDPD 429
>At3g04240 O-linked GlcNAc transferase like protein
Length = 977
Score = 64.7 bits (156), Expect = 1e-10
Identities = 83/410 (20%), Positives = 149/410 (36%), Gaps = 84/410 (20%)
Query: 164 LSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYL 223
+++ +E+++ + A + + + G AI + + P D NL AY+
Sbjct: 107 IARNEEALRIQPQFAECYGNMANAWKEKGDTDRAIRYYLIAIELRPNFADAWSNLASAYM 166
Query: 224 QIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAA 283
+ G L + +C Q+ + + LL+ ++++ G L
Sbjct: 167 RKGRLSEATQCCQQAL-----------SLNPLLVDAHSNL----------GNLMKAQGLI 205
Query: 284 NVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVAT 343
+ A C L A++ A W NLA F SGD + + ++A KL+P
Sbjct: 206 HEAYSCYLEAVRIQPTFAIAWSNLAGLFMESGDLNRALQYYKEAVKLKP----------- 254
Query: 344 HRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYES 403
P A+ L V+KA + A
Sbjct: 255 -----------------------------------AFPDAYLNLGNVYKALGRPTEAIMC 279
Query: 404 EQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGC 463
QH L A+ A +++ G Q + ++ K A++ D
Sbjct: 280 YQHALQMRPNSAMAFGNIASI--------YYEQG-------QLDLAIRHYKQALSRDPRF 324
Query: 464 SYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMF 523
++NLG +L+ +A + Y Q L L ++NLG Y A ++F
Sbjct: 325 LEAYNNLGNALKDIGRVDEAVRCYNQCLALQPNHPQ--AMANLGNIYMEWNMMGPASSLF 382
Query: 524 TKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSN 573
+L + G + FNNL +++ +G +A C+ + L+ DPL A N
Sbjct: 383 KATLAVTTGLSAPFNNLAIIYKQQGNYSDAISCYNEVLRIDPLAADALVN 432
Score = 49.3 bits (116), Expect = 6e-06
Identities = 57/264 (21%), Positives = 108/264 (40%), Gaps = 38/264 (14%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHAQCLIL 143
P A+ LG +++ L +P +AI+ Y+ A + +RP + + ++ S+
Sbjct: 254 PAFPDAYLNLGNVYKALGRPTEAIMCYQHA--LQMRPNSAMAFGNIASIYY--------- 302
Query: 144 ESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSS 203
E +L + K+++ D R +N LG L GRV A+ +
Sbjct: 303 ------------EQGQLDLAIRHYKQALSRDPRFLEAYNNLGNALKDIGRVDEAVRCYNQ 350
Query: 204 LLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELI-----LKDQNHPVALV-----NYA 253
LA+ P + + NLG Y++ + ++ F+ + L + +A++ NY+
Sbjct: 351 CLALQPNHPQAMANLGNIYMEWNMMGPASSLFKATLAVTTGLSAPFNNLAIIYKQQGNYS 410
Query: 254 ALLLCKYASVV----AGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAY 309
+ C Y V+ A A + G ++ A + + AI A NLA
Sbjct: 411 DAISC-YNEVLRIDPLAADALVNRGNTYKEIGRVTEAIQDYMHAINFRPTMAEAHANLAS 469
Query: 310 AFSISGDHRSSSKCLEKAAKLEPN 333
A+ SG ++ ++A L P+
Sbjct: 470 AYKDSGHVEAAITSYKQALLLRPD 493
Score = 46.6 bits (109), Expect = 4e-05
Identities = 24/70 (34%), Positives = 41/70 (58%)
Query: 505 NLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSD 564
N+ ++ + RA + ++EL+P +A A++NL ++ +G L EA C ++AL +
Sbjct: 126 NMANAWKEKGDTDRAIRYYLIAIELRPNFADAWSNLASAYMRKGRLSEATQCCQQALSLN 185
Query: 565 PLLDAAKSNL 574
PLL A SNL
Sbjct: 186 PLLVDAHSNL 195
Score = 45.4 bits (106), Expect = 9e-05
Identities = 32/108 (29%), Positives = 54/108 (49%), Gaps = 2/108 (1%)
Query: 467 WSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKS 526
WSNL S + ++A + YK+A+ L K NLG Y+ + A + +
Sbjct: 226 WSNLAGLFMESGDLNRALQYYKEAVKL--KPAFPDAYLNLGNVYKALGRPTEAIMCYQHA 283
Query: 527 LELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
L+++P A AF N+ ++ +G L+ A +++AL DP A +NL
Sbjct: 284 LQMRPNSAMAFGNIASIYYEQGQLDLAIRHYKQALSRDPRFLEAYNNL 331
Score = 43.9 bits (102), Expect = 3e-04
Identities = 33/111 (29%), Positives = 53/111 (47%), Gaps = 4/111 (3%)
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAIL-SNLGIFYRHEKKYQRAKAMFTKS 526
SNLG ++ +A Y +A+ + Q AI SNL + RA + ++
Sbjct: 193 SNLGNLMKAQGLIHEAYSCYLEAVRI---QPTFAIAWSNLAGLFMESGDLNRALQYYKEA 249
Query: 527 LELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKV 577
++L+P + A+ NLG V+ A G EA C++ ALQ P A N+ +
Sbjct: 250 VKLKPAFPDAYLNLGNVYKALGRPTEAIMCYQHALQMRPNSAMAFGNIASI 300
Score = 42.0 bits (97), Expect = 0.001
Identities = 34/142 (23%), Positives = 60/142 (41%), Gaps = 9/142 (6%)
Query: 433 WHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALL 492
++QL + +C + + + + C + W G + +A + Y A+
Sbjct: 97 YYQLQEYDMCIARNEEALRIQPQFAECYGNMANAWKEKG-------DTDRAIRYYLIAIE 149
Query: 493 LATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEE 552
L + SNL Y + + A ++L L P A +NLG + A+GL+ E
Sbjct: 150 L--RPNFADAWSNLASAYMRKGRLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQGLIHE 207
Query: 553 AKYCFEKALQSDPLLDAAKSNL 574
A C+ +A++ P A SNL
Sbjct: 208 AYSCYLEAVRIQPTFAIAWSNL 229
Score = 38.5 bits (88), Expect = 0.011
Identities = 35/174 (20%), Positives = 64/174 (36%), Gaps = 21/174 (12%)
Query: 160 LKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLG 219
+ E S E+V+ A W+ L + +++G + A+ + + P D NLG
Sbjct: 205 IHEAYSCYLEAVRIQPTFAIAWSNLAGLFMESGDLNRALQYYKEAVKLKPAFPDAYLNLG 264
Query: 220 IAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQ 279
Y +G + C+Q + N +A N A++ +G L
Sbjct: 265 NVYKALGRPTEAIMCYQHALQMRPNSAMAFGNIASIYY--------------EQGQL--- 307
Query: 280 VMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN 333
++A A+ D + + NL A G + +C + L+PN
Sbjct: 308 ----DLAIRHYKQALSRDPRFLEAYNNLGNALKDIGRVDEAVRCYNQCLALQPN 357
>At1g22700 unknown protein
Length = 301
Score = 48.1 bits (113), Expect = 1e-05
Identities = 40/150 (26%), Positives = 64/150 (42%), Gaps = 2/150 (1%)
Query: 441 LCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLL--ATKQQ 498
L ++ S K L+ V + LG + + A K +QA+ Q
Sbjct: 145 LVRRELDLSAKELQEQVRSGDASATELFELGAVMLRRKFYPAANKFLQQAIQKWDGDDQD 204
Query: 499 AHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFE 558
+ + LG+ Y E K + A F +++LQPGY A+NNLG + + L A FE
Sbjct: 205 LAQVYNALGVSYVREDKLDKGIAQFEMAVKLQPGYVTAWNNLGDAYEKKKELPLALNAFE 264
Query: 559 KALQSDPLLDAAKSNLVKVVTMSKICKGLL 588
+ L DP A+ + K+ KG++
Sbjct: 265 EVLLFDPNNKVARPRRDALKDRVKLYKGVV 294
Score = 40.0 bits (92), Expect = 0.004
Identities = 31/134 (23%), Positives = 58/134 (43%)
Query: 115 EILLRPETEIERPDLLSLVQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFD 174
++L+R E ++ +L V+ A L ++ P K + +++ D
Sbjct: 143 QVLVRRELDLSAKELQEQVRSGDASATELFELGAVMLRRKFYPAANKFLQQAIQKWDGDD 202
Query: 175 IRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKC 234
A V+N LG ++ ++ I+ + + P NLG AY + L L+
Sbjct: 203 QDLAQVYNALGVSYVREDKLDKGIAQFEMAVKLQPGYVTAWNNLGDAYEKKKELPLALNA 262
Query: 235 FQELILKDQNHPVA 248
F+E++L D N+ VA
Sbjct: 263 FEEVLLFDPNNKVA 276
Score = 28.9 bits (63), Expect = 8.4
Identities = 27/134 (20%), Positives = 54/134 (40%), Gaps = 15/134 (11%)
Query: 359 LSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCS 418
L E+ +R GD+S EL G M+ + + ++ + + +
Sbjct: 150 LDLSAKELQEQVRSGDASATELFEL--GAVMLRRKFYPAANKFLQQ-------------A 194
Query: 419 LKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSE 478
+++ +D D ++ LGV + + + AV G W+NLG + + +
Sbjct: 195 IQKWDGDDQDLAQVYNALGVSYVREDKLDKGIAQFEMAVKLQPGYVTAWNNLGDAYEKKK 254
Query: 479 EPSQAEKAYKQALL 492
E A A+++ LL
Sbjct: 255 ELPLALNAFEEVLL 268
>At1g80410 putative N-terminal acetyltransferase (At1g80410)
Length = 714
Score = 47.8 bits (112), Expect = 2e-05
Identities = 45/196 (22%), Positives = 76/196 (37%), Gaps = 20/196 (10%)
Query: 137 HAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQS 196
H + L ++ + N D++ E +EL + V+ DI+ W+ LG + +
Sbjct: 42 HGETLSMKGLTLNCMDRKTEAYELVRL------GVKNDIKSHVCWHVLGLLYRSDREYRE 95
Query: 197 AISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALL 256
AI + L I P+N + L +L + Q+ +L + Q+L+ NH + + +A
Sbjct: 96 AIKCYRNALRIDPDNLEILRDLSLLQAQMRDLSGFVETRQQLLTLKPNHRMNWIGFAVSQ 155
Query: 257 LCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGD 316
+ A A EG L D N E + Y S+ +
Sbjct: 156 HLNANASKAVEILEAYEGTLEDDYPPENELIEHT--------------EMILYKVSLLEE 201
Query: 317 HRSSSKCLEKAAKLEP 332
S K LE+ K EP
Sbjct: 202 SGSFDKALEELHKKEP 217
Score = 32.0 bits (71), Expect = 0.99
Identities = 26/117 (22%), Positives = 50/117 (42%), Gaps = 2/117 (1%)
Query: 427 PDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKA 486
P +L V S T+Q+K K A + T S G++L + ++A +
Sbjct: 6 PPKEANLFKLIVKSYETKQYKKGLKAADAILKKFPDHGETLSMKGLTLNCMDRKTEAYEL 65
Query: 487 YKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLV 543
+ L + ++H LG+ YR +++Y+ A + +L + P +L L+
Sbjct: 66 VR--LGVKNDIKSHVCWHVLGLLYRSDREYREAIKCYRNALRIDPDNLEILRDLSLL 120
Score = 30.0 bits (66), Expect = 3.8
Identities = 13/48 (27%), Positives = 24/48 (49%)
Query: 286 AKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN 333
A E + +K D+KS W L + ++R + KC A +++P+
Sbjct: 62 AYELVRLGVKNDIKSHVCWHVLGLLYRSDREYREAIKCYRNALRIDPD 109
Score = 29.3 bits (64), Expect = 6.4
Identities = 41/175 (23%), Positives = 67/175 (37%), Gaps = 36/175 (20%)
Query: 112 KAEEILLRPETEIERPDLLSLVQIHHAQCLILESS---SENSSDKELEPHELKEILSKLK 168
KA EIL E +E I H + ++ + S S DK LE KE K+
Sbjct: 163 KAVEILEAYEGTLEDDYPPENELIEHTEMILYKVSLLEESGSFDKALEELHKKE--PKIV 220
Query: 169 ESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENY-------DCLG----- 216
+ + + ++ + +L K GR++ A + LL++ P+NY CLG
Sbjct: 221 DKLSYKEQEVS-------LLSKVGRLEEANKLYRVLLSMNPDNYRYHEGLQKCLGLYSES 273
Query: 217 ---------NLGIAYLQIGNLELSAKCFQEL---ILKDQNHPVALVNYAALLLCK 259
L Y + + + + L+D+N A+ Y LL K
Sbjct: 274 GQYSSDQIEKLNALYQSLSEQYTRSSAVKRIPLDFLQDENFKEAVAKYIKPLLTK 328
>At5g56290 peroxisomal targeting signal type 1 receptor
Length = 728
Score = 44.7 bits (104), Expect = 1e-04
Identities = 28/101 (27%), Positives = 55/101 (53%), Gaps = 2/101 (1%)
Query: 470 LGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLEL 529
LGV LS E +A +++ AL L K +++ + LG + + A + + ++L+L
Sbjct: 596 LGVLYNLSREFDRAITSFQTALQL--KPNDYSLWNKLGATQANSVQSADAISAYQQALDL 653
Query: 530 QPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAA 570
+P Y A+ N+G+ + +G+ +E+ + +AL +P D A
Sbjct: 654 KPNYVRAWANMGISYANQGMYKESIPYYVRALAMNPKADNA 694
Score = 37.4 bits (85), Expect = 0.024
Identities = 48/225 (21%), Positives = 83/225 (36%), Gaps = 51/225 (22%)
Query: 52 ADVDQHVQGAPSSREEKVSSMKTGLIHVAR--------KMPKNAHAHFILGLMHQRLNQP 103
+D++ +V G P +E + GL+ A K P+NA +LG+ H +
Sbjct: 449 SDMNPYV-GHPEPMKEGQELFRKGLLSEAALALEAEVMKNPENAEGWRLLGVTHAENDDD 507
Query: 104 QKAILVYEKAEE-------ILLR----PETEIERPDLLSLV------------------- 133
Q+AI +A+E +LL E+E+ L +
Sbjct: 508 QQAIAAMMRAQEADPTNLEVLLALGVSHTNELEQATALKYLYGWLRNHPKYGAIAPPELA 567
Query: 134 -QIHHAQC--LILESSSENSSDKELE---------PHELKEILSKLKESVQFDIRQAAVW 181
++HA L E+S N D ++ E ++ + ++Q ++W
Sbjct: 568 DSLYHADIARLFNEASQLNPEDADVHIVLGVLYNLSREFDRAITSFQTALQLKPNDYSLW 627
Query: 182 NTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIG 226
N LG + + AIS L + P N+GI+Y G
Sbjct: 628 NKLGATQANSVQSADAISAYQQALDLKPNYVRAWANMGISYANQG 672
Score = 33.5 bits (75), Expect = 0.34
Identities = 37/178 (20%), Positives = 66/178 (36%), Gaps = 23/178 (12%)
Query: 406 HVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSY 465
H + +++A+ ++ +A DP + LGV + T+ KYL Y
Sbjct: 501 HAENDDDQQAIAAMMRAQEADPTNLEVLLALGVSHTNELEQATALKYL-----------Y 549
Query: 466 TWSNLGVSLQLSEEPSQAEKAY---------KQALLLATKQQAHAILSNLGIFYRHEKKY 516
W P A+ Y + + L H +L G+ Y +++
Sbjct: 550 GWLRNHPKYGAIAPPELADSLYHADIARLFNEASQLNPEDADVHIVL---GVLYNLSREF 606
Query: 517 QRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
RA F +L+L+P +N LG +A +++AL P A +N+
Sbjct: 607 DRAITSFQTALQLKPNDYSLWNKLGATQANSVQSADAISAYQQALDLKPNYVRAWANM 664
>At4g12400 stress-induced protein sti1 -like protein
Length = 558
Score = 44.7 bits (104), Expect = 1e-04
Identities = 22/75 (29%), Positives = 39/75 (51%)
Query: 500 HAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEK 559
H + SN Y +Y+ A + K++EL+P ++ ++ LG F+ +EA ++K
Sbjct: 36 HILYSNRSASYASLHRYEEALSDAKKTIELKPDWSKGYSRLGAAFIGLSKFDEAVDSYKK 95
Query: 560 ALQSDPLLDAAKSNL 574
L+ DP + KS L
Sbjct: 96 GLEIDPSNEMLKSGL 110
>At2g32450 putative O-GlcNAc transferase
Length = 802
Score = 43.1 bits (100), Expect = 4e-04
Identities = 38/145 (26%), Positives = 70/145 (48%), Gaps = 6/145 (4%)
Query: 445 QFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHA-IL 503
+++ + K L+ A+ + +L SL E +A + +++A+ L + H L
Sbjct: 354 EYRAAVKALEEAIYLKPDYADAHCDLASSLHAMGEDERAIEVFQRAIDL---KPGHVDAL 410
Query: 504 SNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQS 563
NLG Y ++QRA M+T+ L + P + A N + + G EEAK ++AL+
Sbjct: 411 YNLGGLYMDLGRFQRASEMYTRVLAVWPNHWRAQLNKAVSLLGAGETEEAKRALKEALKM 470
Query: 564 DPLLDA--AKSNLVKVVTMSKICKG 586
++ A S+L ++ K+ KG
Sbjct: 471 TNRVELHDAVSHLKQLQKKKKVKKG 495
Score = 36.6 bits (83), Expect = 0.040
Identities = 25/99 (25%), Positives = 44/99 (44%)
Query: 159 ELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNL 218
E + + L+E++ A L L G + AI V + + P + D L NL
Sbjct: 354 EYRAAVKALEEAIYLKPDYADAHCDLASSLHAMGEDERAIEVFQRAIDLKPGHVDALYNL 413
Query: 219 GIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLL 257
G Y+ +G + +++ + ++ NH A +N A LL
Sbjct: 414 GGLYMDLGRFQRASEMYTRVLAVWPNHWRAQLNKAVSLL 452
>At3g16320 hypothetical protein
Length = 727
Score = 42.7 bits (99), Expect = 6e-04
Identities = 32/145 (22%), Positives = 64/145 (44%), Gaps = 2/145 (1%)
Query: 420 KQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEE 479
++ ++ D P W +G + T+ K + A+ + +Y + G EE
Sbjct: 489 QELISVDRLSPESWCAVGNCYSLRKDHDTALKMFQRAIQLNERFTYAHTLCGHEFAALEE 548
Query: 480 PSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNN 539
AE+ Y++AL + T+ + LG+ Y ++K++ A+ F +L++ P +
Sbjct: 549 FEDAERCYRKALGIDTRH--YNAWYGLGMTYLRQEKFEFAQHQFQLALQINPRSSVIMCY 606
Query: 540 LGLVFVAEGLLEEAKYCFEKALQSD 564
G+ +EA EKA+ +D
Sbjct: 607 YGIALHESKRNDEALMMMEKAVLTD 631
Score = 35.8 bits (81), Expect = 0.069
Identities = 25/110 (22%), Positives = 56/110 (50%), Gaps = 4/110 (3%)
Query: 457 VACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATK-QQAHAILSNLGIFYRHEKK 515
++ DR +W +G L ++ A K +++A+ L + AH + G + ++
Sbjct: 492 ISVDRLSPESWCAVGNCYSLRKDHDTALKMFQRAIQLNERFTYAHTLC---GHEFAALEE 548
Query: 516 YQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
++ A+ + K+L + + A+ LG+ ++ + E A++ F+ ALQ +P
Sbjct: 549 FEDAERCYRKALGIDTRHYNAWYGLGMTYLRQEKFEFAQHQFQLALQINP 598
Score = 35.0 bits (79), Expect = 0.12
Identities = 31/139 (22%), Positives = 62/139 (44%), Gaps = 8/139 (5%)
Query: 408 LTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTW 467
L E E+ C ++A+ D W+ LG+ L ++F+ +Q + A+ + S
Sbjct: 546 LEEFEDAERC-YRKALGIDTRHYNAWYGLGMTYLRQEKFEFAQHQFQLALQINPRSSVIM 604
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQ------QAHAILSNLGIFYRHEKKYQRAKA 521
G++L S+ +A ++A+L K +AH IL++LG +++ +K + K
Sbjct: 605 CYYGIALHESKRNDEALMMMEKAVLTDAKNPLPKYYKAH-ILTSLGDYHKAQKVLEELKE 663
Query: 522 MFTKSLELQPGYAPAFNNL 540
+ + +N L
Sbjct: 664 CAPQESSVHASLGKIYNQL 682
>At4g08320 unknown protein
Length = 266
Score = 41.6 bits (96), Expect = 0.001
Identities = 21/51 (41%), Positives = 33/51 (64%), Gaps = 1/51 (1%)
Query: 525 KSLELQPGYAPAFNNLGLVFVAEGLLEEA-KYCFEKALQSDPLLDAAKSNL 574
KS+E+ P Y+ A++ LGL + A+G EA + F+KAL DP ++ K N+
Sbjct: 74 KSIEIDPNYSKAYSRLGLAYYAQGKYAEAIEKGFKKALLLDPHNESVKENI 124
>At1g05150 putative O-GlcNAc transferase
Length = 808
Score = 40.4 bits (93), Expect = 0.003
Identities = 32/119 (26%), Positives = 58/119 (47%), Gaps = 4/119 (3%)
Query: 445 QFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHA-IL 503
+++ + K L+ A+ + +L SL E +A + +++A+ L + H L
Sbjct: 359 EYRAAVKALEEAIYLKPDYADAHCDLASSLHSMGEDERAIEVFQRAIDL---KPGHVDAL 415
Query: 504 SNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQ 562
NLG Y ++QRA M+T+ L + P + A N + + G EEAK ++AL+
Sbjct: 416 YNLGGLYMDLGRFQRASEMYTRVLTVWPNHWRAQLNKAVSLLGAGETEEAKRALKEALK 474
Score = 37.0 bits (84), Expect = 0.031
Identities = 25/99 (25%), Positives = 44/99 (44%)
Query: 159 ELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNL 218
E + + L+E++ A L L G + AI V + + P + D L NL
Sbjct: 359 EYRAAVKALEEAIYLKPDYADAHCDLASSLHSMGEDERAIEVFQRAIDLKPGHVDALYNL 418
Query: 219 GIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLL 257
G Y+ +G + +++ + ++ NH A +N A LL
Sbjct: 419 GGLYMDLGRFQRASEMYTRVLTVWPNHWRAQLNKAVSLL 457
>At5g48850 unknown protein
Length = 306
Score = 40.0 bits (92), Expect = 0.004
Identities = 20/84 (23%), Positives = 44/84 (51%), Gaps = 1/84 (1%)
Query: 479 EPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFN 538
+P++ +++ + + +Q+ +L NLG Y + KY A+A++ K+ ++P A
Sbjct: 146 KPTKTARSHGKKFQVTVQQEISRLLGNLGWAYMQQAKYLSAEAVYRKAQMVEPD-ANKSC 204
Query: 539 NLGLVFVAEGLLEEAKYCFEKALQ 562
NL + + +G EE + + L+
Sbjct: 205 NLAMCLIKQGRFEEGRLVLDDVLE 228
>At4g37460 unknown protein
Length = 869
Score = 38.5 bits (88), Expect = 0.011
Identities = 27/92 (29%), Positives = 42/92 (45%)
Query: 479 EPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFN 538
+ S+ K Y A + T + + GI +E Y +A ++F K L+ +P Y A
Sbjct: 276 DESRKNKKYTIARISGTHSISVDFRLSRGIAQVNEGNYTKAISIFDKVLKEEPTYPEALI 335
Query: 539 NLGLVFVAEGLLEEAKYCFEKALQSDPLLDAA 570
G + + LE A F KA+QS+P A
Sbjct: 336 GRGTAYAFQRELESAIADFTKAIQSNPAASEA 367
Score = 34.7 bits (78), Expect = 0.15
Identities = 35/173 (20%), Positives = 64/173 (36%), Gaps = 21/173 (12%)
Query: 159 ELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNL 218
EL+ ++ +++Q + + W G G A+ L+ L P + D L
Sbjct: 346 ELESAIADFTKAIQSNPAASEAWKRRGQARAALGEYVEAVEDLTKALVFEPNSPDVLHER 405
Query: 219 GIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALAD 278
GI + + + K + LK + + Y L A AS G
Sbjct: 406 GIVNFKSKDFTAAVKDLS-ICLKQEKDNKSAYTYLGL-------------AFASLGEYKK 451
Query: 279 QVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLE 331
A+E L +I+ D W +LA + DH + +C+E+ +++
Sbjct: 452 -------AEEAHLKSIQLDSNYLEAWLHLAQFYQELADHCKALECIEQVLQVD 497
Score = 31.2 bits (69), Expect = 1.7
Identities = 29/152 (19%), Positives = 63/152 (41%), Gaps = 2/152 (1%)
Query: 413 ERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGV 472
E A+ +A+ +P W + G ++ + + L A+ + G+
Sbjct: 348 ESAIADFTKAIQSNPAASEAWKRRGQARAALGEYVEAVEDLTKALVFEPNSPDVLHERGI 407
Query: 473 SLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPG 532
S++ + A K ++ L ++ + + LG+ + +Y++A+ KS++L
Sbjct: 408 VNFKSKDFTAAVKDL--SICLKQEKDNKSAYTYLGLAFASLGEYKKAEEAHLKSIQLDSN 465
Query: 533 YAPAFNNLGLVFVAEGLLEEAKYCFEKALQSD 564
Y A+ +L + +A C E+ LQ D
Sbjct: 466 YLEAWLHLAQFYQELADHCKALECIEQVLQVD 497
Score = 28.9 bits (63), Expect = 8.4
Identities = 37/176 (21%), Positives = 65/176 (36%), Gaps = 26/176 (14%)
Query: 219 GIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALAD 278
GIA + GN + F +++ ++ +P AL+ A A E A+AD
Sbjct: 304 GIAQVNEGNYTKAISIFDKVLKEEPTYPEALIGRG----------TAYAFQRELESAIAD 353
Query: 279 QVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTR 338
AI+++ ++ W A + G++ + + L KA EPN
Sbjct: 354 -----------FTKAIQSNPAASEAWKRRGQARAALGEYVEAVEDLTKALVFEPNSPDVL 402
Query: 339 YAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQ 394
+ K + + +L E +D S+ L A+A L KA+
Sbjct: 403 HERGIVNFKSKDFTAAVKDLSICLKQE-----KDNKSAYTYLGLAFASLGEYKKAE 453
Score = 28.9 bits (63), Expect = 8.4
Identities = 18/59 (30%), Positives = 28/59 (46%)
Query: 507 GIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
G Y +++ + A A FTK+++ P + A+ G A G EA KAL +P
Sbjct: 338 GTAYAFQRELESAIADFTKAIQSNPAASEAWKRRGQARAALGEYVEAVEDLTKALVFEP 396
>At1g62740 TPR-repeat protein
Length = 571
Score = 38.5 bits (88), Expect = 0.011
Identities = 20/75 (26%), Positives = 34/75 (44%)
Query: 500 HAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEK 559
H + SN + Y A + K++EL+P + ++ LG + +EA + K
Sbjct: 36 HVLFSNRSAAHASLNHYDEALSDAKKTVELKPDWGKGYSRLGAAHLGLNQFDEAVEAYSK 95
Query: 560 ALQSDPLLDAAKSNL 574
L+ DP + KS L
Sbjct: 96 GLEIDPSNEGLKSGL 110
>At5g17270 putative protein
Length = 899
Score = 36.6 bits (83), Expect = 0.040
Identities = 38/190 (20%), Positives = 72/190 (37%), Gaps = 15/190 (7%)
Query: 155 LEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDC 214
L+ ++++ L +VQ D WN + + + + + + L +++
Sbjct: 619 LKARDVQKALDAFTFAVQLDPDNGEAWNNIACLHMIKKKSKESFIAFKEALKFKRDSWQM 678
Query: 215 LGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALL--------LCKYASVVAG 266
N + +GN++ + + Q+++ +N V +V ++ CK +S
Sbjct: 679 WENFSHVAMDVGNIDQAFEAIQQILKMSKNKRVDVVLLDRIMTELEKRNSACKSSSSSTE 738
Query: 267 AGASASEGALADQVMAANVAK----ECLLAAIKADVK---SAHIWGNLAYAFSISGDHRS 319
AS+ E A E L I+ VK +A IWG A I GD
Sbjct: 739 TEASSDESTETKPCTATPAETQRQLELLGKVIQQIVKTESTAEIWGLYARWSRIKGDLTV 798
Query: 320 SSKCLEKAAK 329
S+ L K +
Sbjct: 799 CSEALLKQVR 808
Score = 30.4 bits (67), Expect = 2.9
Identities = 40/222 (18%), Positives = 77/222 (34%), Gaps = 33/222 (14%)
Query: 299 KSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN----------------CMSTRYAVA 342
+S +W NL Y + + G ++ + PN C V+
Sbjct: 510 ESLELWDNLIYCYCLLGKKSAAVDLINARLLERPNDPRLWCSLGDVTINDSCYEKALEVS 569
Query: 343 THRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVEL-PTAWAGLAMVHKAQHEISSAY 401
+ A+R+ S + G E + ++ + +L L P W L ++
Sbjct: 570 NDKSVRAKRALARSAY-NRGDFEKSKMLWEAAMALNSLYPDGWFALGAAALKARDV---- 624
Query: 402 ESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDR 461
++A+ + AV DPD+ W+ + + ++ K S K A+ R
Sbjct: 625 -----------QKALDAFTFAVQLDPDNGEAWNNIACLHMIKKKSKESFIAFKEALKFKR 673
Query: 462 GCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAIL 503
W N QA +A +Q L ++ ++ +L
Sbjct: 674 DSWQMWENFSHVAMDVGNIDQAFEAIQQILKMSKNKRVDVVL 715
Score = 30.4 bits (67), Expect = 2.9
Identities = 35/162 (21%), Positives = 65/162 (39%), Gaps = 18/162 (11%)
Query: 406 HVLTEMEERAVCSLKQAVAEDPDDPVRWHQLG---VHSLCTQQFK--TSQKYLKAAVACD 460
+ L + AV + + E P+DP W LG ++ C ++ ++ K ++A A
Sbjct: 522 YCLLGKKSAAVDLINARLLERPNDPRLWCSLGDVTINDSCYEKALEVSNDKSVRAKRALA 581
Query: 461 RGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAK 520
R +Y + S L E Y LG + Q+A
Sbjct: 582 RS-AYNRGDFEKSKMLWEAAMALNSLYPDGWFA------------LGAAALKARDVQKAL 628
Query: 521 AMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQ 562
FT +++L P A+NN+ + + + +E+ F++AL+
Sbjct: 629 DAFTFAVQLDPDNGEAWNNIACLHMIKKKSKESFIAFKEALK 670
>At4g32820 putative protein
Length = 1817
Score = 36.2 bits (82), Expect = 0.053
Identities = 31/140 (22%), Positives = 64/140 (45%), Gaps = 14/140 (10%)
Query: 110 YEKAEEILLRPETEIERPDLLSLV--------QIHHAQCLILESSSENSSDKELEPHELK 161
Y+KA E+L E+ ++ P + + +HH + L L++ + + EL +
Sbjct: 46 YDKARELL---ESILKDPIITNSKVETIANDNHLHHLRFLALKNLA--TVFLELGSSHYE 100
Query: 162 EILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIA 221
L+ +++ D + + +WN LG + G + + L +P N++C+ L
Sbjct: 101 NALNCYLQAIDLDAKDSVLWNHLGTLSCSMGLLSISRWAFEQGLLCSPNNWNCMEKLLEV 160
Query: 222 YLQIGNLELSAKCFQELILK 241
+ +G+ E+S LIL+
Sbjct: 161 LIAVGD-EVSCLSVANLILR 179
>At1g78770 hypothetical protein
Length = 542
Score = 36.2 bits (82), Expect = 0.053
Identities = 32/157 (20%), Positives = 73/157 (46%), Gaps = 24/157 (15%)
Query: 380 LPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVH 439
LPT + G+ + ++++ + M+ +A+C P DP+ +++LGV
Sbjct: 377 LPTLYIGMEYMRTHSYKLADQFF--------MQAKAIC---------PSDPLVYNELGVV 419
Query: 440 SLCTQQFKTSQKYLKAAVA-CDRGCSYTWS----NLGVSLQLSEEPSQAEKAYKQALLLA 494
+ +++ + ++ + +A + +W NL + + + +A Y++AL L+
Sbjct: 420 AYHMKEYGKAVRWFEKTLAHIPSALTESWEPTVVNLAHAYRKLRKDREAISYYERALTLS 479
Query: 495 TKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQP 531
TK + + S L Y + + A + + K+L L+P
Sbjct: 480 TK--SLSTYSGLAYTYHLQGNFSAAISYYHKALWLKP 514
Score = 29.3 bits (64), Expect = 6.4
Identities = 27/116 (23%), Positives = 49/116 (41%), Gaps = 10/116 (8%)
Query: 436 LGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALL--- 492
+G+ + T +K + ++ A A ++ LGV +E +A + +++ L
Sbjct: 382 IGMEYMRTHSYKLADQFFMQAKAICPSDPLVYNELGVVAYHMKEYGKAVRWFEKTLAHIP 441
Query: 493 LATKQQAHAILSNLGIFYRHEKK-------YQRAKAMFTKSLELQPGYAPAFNNLG 541
A + + NL YR +K Y+RA + TKSL G A ++ G
Sbjct: 442 SALTESWEPTVVNLAHAYRKLRKDREAISYYERALTLSTKSLSTYSGLAYTYHLQG 497
Score = 28.9 bits (63), Expect = 8.4
Identities = 16/60 (26%), Positives = 29/60 (47%)
Query: 506 LGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
+G +Y KKY A+ F+K+ + ++PA G F A+ ++A + A + P
Sbjct: 314 VGCYYYCIKKYAEARRYFSKATGIDGSFSPARIGYGNSFAAQEEGDQAMSAYRTAARLFP 373
>At3g17670 hypothetical protein
Length = 366
Score = 35.4 bits (80), Expect = 0.090
Identities = 26/102 (25%), Positives = 45/102 (43%)
Query: 481 SQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNL 540
S+AE QAL L H I + + Y A +++L L P Y+ +
Sbjct: 257 SEAEALLTQALELKPYGGLHRIFKHRSVAKLGMLDYSGALEDISQALALAPNYSEPYICQ 316
Query: 541 GLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSK 582
G V+VA+G + A+ + L+ DP L +K ++ + +
Sbjct: 317 GDVYVAKGQYDLAEKSYLTCLEIDPSLRRSKPFKARIAKLQQ 358
>At5g37590 kinesin -like protein
Length = 225
Score = 35.0 bits (79), Expect = 0.12
Identities = 18/63 (28%), Positives = 37/63 (58%), Gaps = 8/63 (12%)
Query: 504 SNLGIFYRHEKKYQRAKAMFTKSLE-LQPGYAP-------AFNNLGLVFVAEGLLEEAKY 555
+NL YR +K++ +A+ ++ +++ L+ Y P +NLG +++ + LEEA+
Sbjct: 143 NNLAELYRVKKEFDKAEPLYLEAVSILEEFYGPDDVRVGATLHNLGQLYLVQRKLEEARA 202
Query: 556 CFE 558
C+E
Sbjct: 203 CYE 205
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,307,634
Number of Sequences: 26719
Number of extensions: 496566
Number of successful extensions: 1988
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 1788
Number of HSP's gapped (non-prelim): 189
length of query: 590
length of database: 11,318,596
effective HSP length: 105
effective length of query: 485
effective length of database: 8,513,101
effective search space: 4128853985
effective search space used: 4128853985
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0174.1


