

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0173b.8
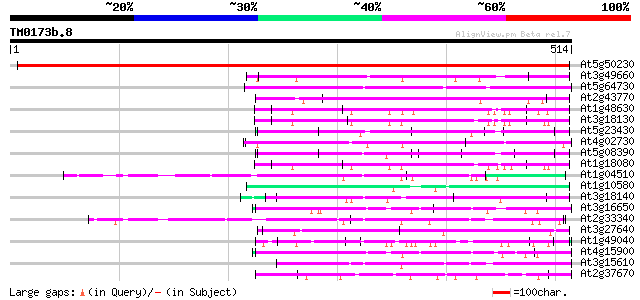
(514 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g50230 putative protein 766 0.0
At3g49660 putative WD-40 repeat - protein 119 3e-27
At5g64730 unknown protein 117 2e-26
At2g43770 putative splicing factor 115 4e-26
At1g48630 unknown protein 102 4e-22
At3g18130 protein kinase C-receptor/G-protein, putative 102 7e-22
At5g23430 unknown protein 100 1e-21
At4g02730 putative WD-repeat protein 100 3e-21
At5g08390 katanin p80 subunit - like protein 100 3e-21
At1g18080 putative guanine nucleotide-binding protein beta sub... 93 4e-19
At1g04510 putative pre-mRNA splicing factor PRP19 92 7e-19
At1g10580 putative pre-mRNA splicing factor 88 1e-17
At3g18140 unknown protein 87 2e-17
At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2 87 3e-17
At2g33340 putative PRP19-like spliceosomal protein 87 3e-17
At3g27640 unknown protein 84 2e-16
At1g49040 hypothetical protein; similar to EST emb|Z26090.1 84 2e-16
At4g15900 PRL1 protein 83 4e-16
At3g15610 unknown protein 82 7e-16
At2g37670 putative WD-40 repeat protein 80 2e-15
>At5g50230 putative protein
Length = 515
Score = 766 bits (1977), Expect = 0.0
Identities = 358/506 (70%), Positives = 445/506 (87%)
Query: 8 QEEIAQKAINHALKSLRKRHLLEEAAHAPAFLALSRPIVSQGSEWKEKAENLQVELQQCY 67
QEE A +AIN AL++LRKRHLLEE AHAPA ALS+P++SQGSEWKEK E L+ ELQQCY
Sbjct: 9 QEEKAMEAINDALRALRKRHLLEEGAHAPAISALSKPLISQGSEWKEKTEKLETELQQCY 68
Query: 68 KAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEMRDEFSQLKVDLEQKIKELE 127
KAQSRLSEQLV+EVA+SR SKA++QEK+ I DLQKELT+ R++ ++L+ +LE+K K ++
Sbjct: 69 KAQSRLSEQLVIEVAESRTSKAILQEKELLINDLQKELTQRREDCTRLQEELEEKTKTVD 128
Query: 128 VIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANALYEEMVERLR 187
V+++EN E+++QLE+MT++ +KAE ENKMLIDRWML+KM+DAERLNEAN LYEEM+ +L+
Sbjct: 129 VLIAENLEIRSQLEEMTSRVQKAETENKMLIDRWMLQKMQDAERLNEANDLYEEMLAKLK 188
Query: 188 ASGLEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASILFEFNSSRLIT 247
A+GLE LARQQVDG+VRR E+G + F++S IPSTC R++AHEGGC SI+FE+NS L T
Sbjct: 189 ANGLETLARQQVDGIVRRNEDGTDHFVESTIPSTCANRIHAHEGGCGSIVFEYNSGTLFT 248
Query: 248 GGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVR 307
GGQD++VK+WDTN+G++ +L+G +G++LD+ +THDN+SVIAA+SSNNL+VWD++SGRVR
Sbjct: 249 GGQDRAVKMWDTNSGTLIKSLYGSLGNILDMAVTHDNKSVIAATSSNNLFVWDVSSGRVR 308
Query: 308 HTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMD 367
HTLTGH DKVCAVDVSK SSRHVVSAAYDRTIK+WDL KGYCTNT++F SNCNA+C S+D
Sbjct: 309 HTLTGHTDKVCAVDVSKFSSRHVVSAAYDRTIKLWDLHKGYCTNTVLFTSNCNAICLSID 368
Query: 368 GQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDV 427
G T++SGH+DGNLRLWDIQ+GKLLSEVA HS AVTS+SLSRNGN +LTSGRDNVHN+FD
Sbjct: 369 GLTVFSGHMDGNLRLWDIQTGKLLSEVAGHSSAVTSVSLSRNGNRILTSGRDNVHNVFDT 428
Query: 428 RSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTS 487
R+LE+C + R +GNR+ASNWSRSCISPD+ +VAAGSADGSVH+WS+ K IVS LKE TS
Sbjct: 429 RTLEICGTLRASGNRLASNWSRSCISPDDDYVAAGSADGSVHVWSLSKGNIVSILKEQTS 488
Query: 488 SVLCCRWSGIGKPLASADKNGIVCIW 513
+LCC WSGIGKPLASADKNG VC W
Sbjct: 489 PILCCSWSGIGKPLASADKNGYVCTW 514
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 119 bits (299), Expect = 3e-27
Identities = 72/255 (28%), Positives = 126/255 (49%), Gaps = 16/255 (6%)
Query: 229 HEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVI 288
HE G + + F ++ +++ D+++K+WD TGS+ L G + + ++
Sbjct: 70 HENGISDVAFSSDARFIVSASDDKTLKLWDVETGSLIKTLIGHTNYAFCVNFNPQSNMIV 129
Query: 289 AASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGY 348
+ S + +WD+ +G+ L H D V AVD ++ S +VS++YD ++WD G+
Sbjct: 130 SGSFDETVRIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSL-IVSSSYDGLCRIWDSGTGH 188
Query: 349 CTNTIIFASN--CNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISL 406
C T+I N + + FS +G+ I G +D LRLW+I S K L H A IS
Sbjct: 189 CVKTLIDDENPPVSFVRFSPNGKFILVGTLDNTLRLWNISSAKFLKTYTGHVNAQYCISS 248
Query: 407 S---RNGNVVLTSGRDNVHNLFDVRS---LEVCSSFRDTGNRVASNWSRSCISPDNSHVA 460
+ NG +++ DN +++++ S L+ +T VA + P + +A
Sbjct: 249 AFSVTNGKRIVSGSEDNCVHMWELNSKKLLQKLEGHTETVMNVACH-------PTENLIA 301
Query: 461 AGSADGSVHIWSVHK 475
+GS D +V IW+ K
Sbjct: 302 SGSLDKTVRIWTQKK 316
Score = 117 bits (293), Expect = 2e-26
Identities = 79/311 (25%), Positives = 143/311 (45%), Gaps = 18/311 (5%)
Query: 218 IPSTCKFR-------LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNT-----GSVC 265
IP+T F L +H +S+ F + L + D++++ + NT
Sbjct: 5 IPATASFTPYVHSQTLTSHNRAVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPV 64
Query: 266 SNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKV 325
G + D+ + D R +++AS L +WD+ +G + TL GH + V+ +
Sbjct: 65 QEFTGHENGISDVAFSSDARFIVSASDDKTLKLWDVETGSLIKTLIGHTNYAFCVNFNPQ 124
Query: 326 SSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASN-CNALCFSMDGQTIYSGHVDGNLRLWD 384
S+ +VS ++D T+++WD+ G C + S+ A+ F+ DG I S DG R+WD
Sbjct: 125 SNM-IVSGSFDETVRIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWD 183
Query: 385 IQSGKLLSE-VAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRV 443
+G + + + V+ + S NG +L DN L+++ S + ++ TG+
Sbjct: 184 SGTGHCVKTLIDDENPPVSFVRFSPNGKFILVGTLDNTLRLWNISSAKFLKTY--TGHVN 241
Query: 444 ASNWSRSCISPDNS-HVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLA 502
A S S N + +GS D VH+W ++ +++ L+ HT +V+ +A
Sbjct: 242 AQYCISSAFSVTNGKRIVSGSEDNCVHMWELNSKKLLQKLEGHTETVMNVACHPTENLIA 301
Query: 503 SADKNGIVCIW 513
S + V IW
Sbjct: 302 SGSLDKTVRIW 312
>At5g64730 unknown protein
Length = 299
Score = 117 bits (293), Expect = 2e-26
Identities = 78/300 (26%), Positives = 144/300 (48%), Gaps = 6/300 (2%)
Query: 216 SNIPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSV 275
+ +P+ L HEG + F + + +T G+D+++++W+ + G + V
Sbjct: 4 TELPTKEAHILKGHEGAVLAARFNGDGNYALTCGKDRTIRLWNPHRGILIKTYKSHGREV 63
Query: 276 LDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAY 335
D+ +T DN + +Y WD+++GRV GH +V AV + SS VVSA +
Sbjct: 64 RDVHVTSDNAKFCSCGGDRQVYYWDVSTGRVIRKFRGHDGEVNAVKFND-SSSVVVSAGF 122
Query: 336 DRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQT-IYSGHVDGNLRLWDIQSGKLLSEV 394
DR+++VWD I + + + + +T I G VDG +R +D++ G+ +S+
Sbjct: 123 DRSLRVWDCRSHSVEPVQIIDTFLDTVMSVVLTKTEIIGGSVDGTVRTFDMRIGREMSDN 182
Query: 395 AAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISP 454
V IS+S +GN VL D+ L D + E+ ++ G+ S + C++
Sbjct: 183 LGQ--PVNCISISNDGNCVLAGCLDSTLRLLDRTTGELLQVYK--GHISKSFKTDCCLTN 238
Query: 455 DNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIWK 514
++HV GS DG V W + +++S + H V + + ++ +G + +WK
Sbjct: 239 SDAHVIGGSEDGLVFFWDLVDAKVLSKFRAHDLVVTSVSYHPKEDCMLTSSVDGTIRVWK 298
>At2g43770 putative splicing factor
Length = 343
Score = 115 bits (289), Expect = 4e-26
Identities = 72/274 (26%), Positives = 126/274 (45%), Gaps = 9/274 (3%)
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSN---LHGCIGSVLDLTITH 282
L+ H ++ F + + +G D+ + +W + C N L G ++LDL T
Sbjct: 49 LSGHPSAVYTMKFNPAGTLIASGSHDREIFLWRVHGD--CKNFMVLKGHKNAILDLHWTS 106
Query: 283 DNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVW 342
D +++AS + WD+ +G+ + H V + ++ ++S + D T K+W
Sbjct: 107 DGSQIVSASPDKTVRAWDVETGKQIKKMAEHSSFVNSCCPTRRGPPLIISGSDDGTAKLW 166
Query: 343 DLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVT 402
D+ + T A+ FS I++G VD ++++WD++ G+ + H +T
Sbjct: 167 DMRQRGAIQTFPDKYQITAVSFSDAADKIFTGGVDNDVKVWDLRKGEATMTLEGHQDTIT 226
Query: 403 SISLSRNGNVVLTSGRDNVHNLFDVRSL----EVCSSFRDTGNRVASNWSRSCISPDNSH 458
+SLS +G+ +LT+G DN ++D+R F + N + SPD +
Sbjct: 227 GMSLSPDGSYLLTNGMDNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSWSPDGTK 286
Query: 459 VAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCC 492
V AGS+D VHIW + L HT SV C
Sbjct: 287 VTAGSSDRMVHIWDTTSRRTIYKLPGHTGSVNEC 320
Score = 70.9 bits (172), Expect = 2e-12
Identities = 54/238 (22%), Positives = 106/238 (43%), Gaps = 17/238 (7%)
Query: 287 VIAASSSNNLYVWDLNSGRVRH-TLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
+ + S +++W ++ L GHK+ + + + S+ +VSA+ D+T++ WD+
Sbjct: 68 IASGSHDREIFLWRVHGDCKNFMVLKGHKNAILDLHWTSDGSQ-IVSASPDKTVRAWDVE 126
Query: 346 KGYCTNTII-FASNCNALCFSMDGQT-IYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTS 403
G + +S N+ C + G I SG DG +LWD++ + + +T+
Sbjct: 127 TGKQIKKMAEHSSFVNSCCPTRRGPPLIISGSDDGTAKLWDMRQRGAI-QTFPDKYQITA 185
Query: 404 ISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGS 463
+S S + + T G DN ++D+R E + + + +SPD S++
Sbjct: 186 VSFSDAADKIFTGGVDNDVKVWDLRKGEATMTLEGHQDTITG----MSLSPDGSYLLTNG 241
Query: 464 ADGSVHIWSVH----KNEIVSTLKEHT----SSVLCCRWSGIGKPLASADKNGIVCIW 513
D + +W + +N V + H ++L C WS G + + + +V IW
Sbjct: 242 MDNKLCVWDMRPYAPQNRCVKIFEGHQHNFEKNLLKCSWSPDGTKVTAGSSDRMVHIW 299
Score = 34.3 bits (77), Expect = 0.17
Identities = 28/126 (22%), Positives = 51/126 (40%), Gaps = 14/126 (11%)
Query: 394 VAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCI- 452
++ H AV ++ + G ++ + D L+ V D N + ++ I
Sbjct: 49 LSGHPSAVYTMKFNPAGTLIASGSHDREIFLWRVHG--------DCKNFMVLKGHKNAIL 100
Query: 453 ----SPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPL-ASADKN 507
+ D S + + S D +V W V + + + EH+S V C + G PL S +
Sbjct: 101 DLHWTSDGSQIVSASPDKTVRAWDVETGKQIKKMAEHSSFVNSCCPTRRGPPLIISGSDD 160
Query: 508 GIVCIW 513
G +W
Sbjct: 161 GTAKLW 166
Score = 32.3 bits (72), Expect = 0.64
Identities = 15/61 (24%), Positives = 29/61 (46%)
Query: 241 NSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWD 300
+ +++ G D+ V +WDT + L G GSV + + + SS N+Y+ +
Sbjct: 283 DGTKVTAGSSDRMVHIWDTTSRRTIYKLPGHTGSVNECVFHPTEPIIGSCSSDKNIYLGE 342
Query: 301 L 301
+
Sbjct: 343 I 343
>At1g48630 unknown protein
Length = 326
Score = 102 bits (255), Expect = 4e-22
Identities = 80/303 (26%), Positives = 141/303 (46%), Gaps = 38/303 (12%)
Query: 241 NSSRLITGGQDQSVKVW-----DTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNN 295
NS ++T +D+S+ +W D + G + G V D+ ++ D + ++ S
Sbjct: 27 NSDVIVTSSRDKSIILWKLTKEDKSYGVAQRRMTGHSHFVQDVVLSSDGQFALSGSWDGE 86
Query: 296 LYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIF 355
L +WDL +G GH V +V S +R +VSA+ DRTIK+W+ + G C TI
Sbjct: 87 LRLWDLATGESTRRFVGHTKDVLSVAFS-TDNRQIVSASRDRTIKLWNTL-GECKYTISE 144
Query: 356 ASN----CNALCFSMDG--QTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRN 409
A + + FS + TI S D +++W++Q+ KL + +A HS + ++++S +
Sbjct: 145 ADGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNTLAGHSGYLNTVAVSPD 204
Query: 410 GNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVH 469
G++ + G+D V L+D+ + S + G+ + S C SP N + + + S+
Sbjct: 205 GSLCASGGKDGVILLWDLAEGKKLYSL-EAGSIIHS----LCFSP-NRYWLCAATENSIR 258
Query: 470 IWSVHKNEIVSTLK-----------------EHTSSVLC--CRWSGIGKPLASADKNGIV 510
IW + +V LK T + C WS G L S +G++
Sbjct: 259 IWDLESKSVVEDLKVDLKAEAEKTDGSTGIGNKTKVIYCTSLNWSADGNTLFSGYTDGVI 318
Query: 511 CIW 513
+W
Sbjct: 319 RVW 321
Score = 93.2 bits (230), Expect = 3e-19
Identities = 71/274 (25%), Positives = 121/274 (43%), Gaps = 33/274 (12%)
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDN 284
R+ H ++ + ++G D +++WD TG G VL + + DN
Sbjct: 58 RMTGHSHFVQDVVLSSDGQFALSGSWDGELRLWDLATGESTRRFVGHTKDVLSVAFSTDN 117
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTLT---GHKDKVCAVDVS-KVSSRHVVSAAYDRTIK 340
R +++AS + +W+ G ++T++ GHK+ V V S +VSA++D+T+K
Sbjct: 118 RQIVSASRDRTIKLWN-TLGECKYTISEADGHKEWVSCVRFSPNTLVPTIVSASWDKTVK 176
Query: 341 VWDLMKGYCTNTIIFASN-CNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSL 399
VW+L NT+ S N + S DG SG DG + LWD+ GK L + A S+
Sbjct: 177 VWNLQNCKLRNTLAGHSGYLNTVAVSPDGSLCASGGKDGVILLWDLAEGKKLYSLEAGSI 236
Query: 400 AVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR--------------DTGNR--- 442
+ S+ S N L + +N ++D+ S V + GN+
Sbjct: 237 -IHSLCFSPN-RYWLCAATENSIRIWDLESKSVVEDLKVDLKAEAEKTDGSTGIGNKTKV 294
Query: 443 ---VASNWSRSCISPDNSHVAAGSADGSVHIWSV 473
+ NW S D + + +G DG + +W +
Sbjct: 295 IYCTSLNW-----SADGNTLFSGYTDGVIRVWGI 323
Score = 73.2 bits (178), Expect = 3e-13
Identities = 55/218 (25%), Positives = 103/218 (47%), Gaps = 14/218 (6%)
Query: 306 VRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK-----GYCTNTIIFASN-C 359
++ T+ H D V A+ +S +V+++ D++I +W L K G + S+
Sbjct: 7 LKGTMCAHTDMVTAIATPVDNSDVIVTSSRDKSIILWKLTKEDKSYGVAQRRMTGHSHFV 66
Query: 360 NALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRD 419
+ S DGQ SG DG LRLWD+ +G+ H+ V S++ S + ++++ RD
Sbjct: 67 QDVVLSSDGQFALSGSWDGELRLWDLATGESTRRFVGHTKDVLSVAFSTDNRQIVSASRD 126
Query: 420 NVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCI--SPDN--SHVAAGSADGSVHIWSVHK 475
L++ + G++ W SC+ SP+ + + S D +V +W++
Sbjct: 127 RTIKLWNTLGECKYTISEADGHK---EWV-SCVRFSPNTLVPTIVSASWDKTVKVWNLQN 182
Query: 476 NEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ +TL H+ + S G AS K+G++ +W
Sbjct: 183 CKLRNTLAGHSGYLNTVAVSPDGSLCASGGKDGVILLW 220
>At3g18130 protein kinase C-receptor/G-protein, putative
Length = 326
Score = 102 bits (253), Expect = 7e-22
Identities = 80/303 (26%), Positives = 138/303 (45%), Gaps = 38/303 (12%)
Query: 241 NSSRLITGGQDQSVKVW-----DTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNN 295
NS ++T +D+S+ +W D + G L G V D+ ++ D + ++ S
Sbjct: 27 NSDIIVTASRDKSIILWKLTKDDKSYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDGE 86
Query: 296 LYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIF 355
L +WDL +G GH V +V S +R +VSA+ DRTIK+W+ + G C TI
Sbjct: 87 LRLWDLATGETTRRFVGHTKDVLSVAFS-TDNRQIVSASRDRTIKLWNTL-GECKYTISE 144
Query: 356 AS------NCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRN 409
+C + TI S D +++W++Q+ KL + + HS + ++++S +
Sbjct: 145 GDGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNSLVGHSGYLNTVAVSPD 204
Query: 410 GNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVH 469
G++ + G+D V L+D+ + S + G+ + S C SP N + + + S+
Sbjct: 205 GSLCASGGKDGVILLWDLAEGKKLYSL-EAGSIIHS----LCFSP-NRYWLCAATENSIR 258
Query: 470 IWSVHKNEIVSTLKEHTSS----------------VLCC---RWSGIGKPLASADKNGIV 510
IW + +V LK S V+ C WS G L S +G+V
Sbjct: 259 IWDLESKSVVEDLKVDLKSEAEKNEGGVGTGNQKKVIYCTSLNWSADGSTLFSGYTDGVV 318
Query: 511 CIW 513
+W
Sbjct: 319 RVW 321
Score = 98.2 bits (243), Expect = 1e-20
Identities = 74/274 (27%), Positives = 123/274 (44%), Gaps = 33/274 (12%)
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDN 284
RL H ++ + ++G D +++WD TG G VL + + DN
Sbjct: 58 RLTGHSHFVEDVVLSSDGQFALSGSWDGELRLWDLATGETTRRFVGHTKDVLSVAFSTDN 117
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTLT---GHKDKVCAVDVS-KVSSRHVVSAAYDRTIK 340
R +++AS + +W+ G ++T++ GHK+ V V S +VSA++D+T+K
Sbjct: 118 RQIVSASRDRTIKLWN-TLGECKYTISEGDGHKEWVSCVRFSPNTLVPTIVSASWDKTVK 176
Query: 341 VWDLMKGYCTNTIIFASN-CNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSL 399
VW+L N+++ S N + S DG SG DG + LWD+ GK L + A S+
Sbjct: 177 VWNLQNCKLRNSLVGHSGYLNTVAVSPDGSLCASGGKDGVILLWDLAEGKKLYSLEAGSI 236
Query: 400 AVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR--------------DTGNR--- 442
+ S+ S N L + +N ++D+ S V + TGN+
Sbjct: 237 -IHSLCFSPN-RYWLCAATENSIRIWDLESKSVVEDLKVDLKSEAEKNEGGVGTGNQKKV 294
Query: 443 ---VASNWSRSCISPDNSHVAAGSADGSVHIWSV 473
+ NW S D S + +G DG V +W +
Sbjct: 295 IYCTSLNW-----SADGSTLFSGYTDGVVRVWGI 323
Score = 71.2 bits (173), Expect = 1e-12
Identities = 56/214 (26%), Positives = 101/214 (47%), Gaps = 14/214 (6%)
Query: 310 LTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK-----GYCTNTIIFASN-CNALC 363
+ H D V A+ +S +V+A+ D++I +W L K G + S+ +
Sbjct: 11 MRAHTDIVTAIATPIDNSDIIVTASRDKSIILWKLTKDDKSYGVAQRRLTGHSHFVEDVV 70
Query: 364 FSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHN 423
S DGQ SG DG LRLWD+ +G+ H+ V S++ S + ++++ RD
Sbjct: 71 LSSDGQFALSGSWDGELRLWDLATGETTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIK 130
Query: 424 LFDVRSLEVCSSFRDTGNRVASNWSRSCI--SPDN--SHVAAGSADGSVHIWSVHKNEIV 479
L++ +L C G+ W SC+ SP+ + + S D +V +W++ ++
Sbjct: 131 LWN--TLGECKYTISEGDG-HKEWV-SCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLR 186
Query: 480 STLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++L H+ + S G AS K+G++ +W
Sbjct: 187 NSLVGHSGYLNTVAVSPDGSLCASGGKDGVILLW 220
>At5g23430 unknown protein
Length = 837
Score = 100 bits (250), Expect = 1e-21
Identities = 58/210 (27%), Positives = 99/210 (46%), Gaps = 3/210 (1%)
Query: 228 AHEGGCASILFEFNSSR-LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRS 286
AH + SSR L+TGG+D V +W + +L+G + +T
Sbjct: 14 AHSAAVNCLKIGRKSSRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEVL 73
Query: 287 VIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK 346
V A ++S + +WDL ++ TLTGH+ +VD S + D +K+WD+ K
Sbjct: 74 VAAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFHPF-GEFFASGSLDTNLKIWDIRK 132
Query: 347 GYCTNTII-FASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSIS 405
C +T N L F+ DG+ + SG D +++WD+ +GKLL+E +H + S+
Sbjct: 133 KGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVKVWDLTAGKLLTEFKSHEGQIQSLD 192
Query: 406 LSRNGNVVLTSGRDNVHNLFDVRSLEVCSS 435
+ ++ T D +D+ + E+ S
Sbjct: 193 FHPHEFLLATGSADRTVKFWDLETFELIGS 222
Score = 84.0 bits (206), Expect = 2e-16
Identities = 55/219 (25%), Positives = 104/219 (47%), Gaps = 10/219 (4%)
Query: 284 NRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAV--DVSKVSSRHVVSAAYDRTIKV 341
+R ++ + + +W + +L GH + +V D S+V V + A TIK+
Sbjct: 29 SRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEVL---VAAGAASGTIKL 85
Query: 342 WDLMKGYCTNTII-FASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLA 400
WDL + T+ SNC ++ F G+ SG +D NL++WDI+ + H+
Sbjct: 86 WDLEEAKIVRTLTGHRSNCISVDFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRG 145
Query: 401 VTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVA 460
V + + +G V++ G DN+ ++D+ + ++ + F+ ++ S P +A
Sbjct: 146 VNVLRFTPDGRWVVSGGEDNIVKVWDLTAGKLLTEFKSHEGQIQS----LDFHPHEFLLA 201
Query: 461 AGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGK 499
GSAD +V W + E++ + T+ V C ++ GK
Sbjct: 202 TGSADRTVKFWDLETFELIGSGGPETAGVRCLSFNPDGK 240
Score = 69.7 bits (169), Expect = 4e-12
Identities = 51/238 (21%), Positives = 94/238 (39%), Gaps = 12/238 (5%)
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
L H G S+ F+ + + G ++K+WD + L G + + +
Sbjct: 55 LYGHSSGIDSVTFDASEVLVAAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFHPFGE 114
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
+ S NL +WD+ HT GH V + + R VVS D +KVWDL
Sbjct: 115 FFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTP-DGRWVVSGGEDNIVKVWDLT 173
Query: 346 KG-YCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSI 404
G T +L F + +G D ++ WD+++ +L+ + V +
Sbjct: 174 AGKLLTEFKSHEGQIQSLDFHPHEFLLATGSADRTVKFWDLETFELIGSGGPETAGVRCL 233
Query: 405 SLSRNGNVVLTSGRDNV----------HNLFDVRSLEVCSSFRDTGNRVASNWSRSCI 452
S + +G VL ++++ H+ DV + G + ++++SC+
Sbjct: 234 SFNPDGKTVLCGLQESLKIFSWEPIRCHDGVDVGWSRLSDMNVHEGKLLGCSYNQSCV 291
Score = 55.5 bits (132), Expect = 7e-08
Identities = 35/145 (24%), Positives = 64/145 (44%), Gaps = 4/145 (2%)
Query: 369 QTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVR 428
+ + +G D + LW I + + HS + S++ + +V L+D+
Sbjct: 30 RVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEVLVAAGAASGTIKLWDLE 89
Query: 429 SLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSS 488
++ + TG+R SN P A+GS D ++ IW + K + T K HT
Sbjct: 90 EAKIVRTL--TGHR--SNCISVDFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRG 145
Query: 489 VLCCRWSGIGKPLASADKNGIVCIW 513
V R++ G+ + S ++ IV +W
Sbjct: 146 VNVLRFTPDGRWVVSGGEDNIVKVW 170
Score = 37.4 bits (85), Expect = 0.020
Identities = 25/103 (24%), Positives = 40/103 (38%), Gaps = 10/103 (9%)
Query: 211 EFF----LDSNIP------STCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTN 260
EFF LD+N+ C H G + F + +++GG+D VKVWD
Sbjct: 114 EFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVKVWDLT 173
Query: 261 TGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNS 303
G + + G + L + S+ + WDL +
Sbjct: 174 AGKLLTEFKSHEGQIQSLDFHPHEFLLATGSADRTVKFWDLET 216
>At4g02730 putative WD-repeat protein
Length = 333
Score = 100 bits (248), Expect = 3e-21
Identities = 72/307 (23%), Positives = 138/307 (44%), Gaps = 11/307 (3%)
Query: 217 NIPSTCKFR----LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCI 272
N+P +R L H + + F + + L + D+++ +W S+ G
Sbjct: 26 NVPIYKPYRHLKTLEGHTAAISCVKFSNDGNLLASASVDKTMILWSATNYSLIHRYEGHS 85
Query: 273 GSVLDLTITHDNRSVIAASSSNNLYVWDLNSG-RVRHTLTGHKDKVCAVDVSKVSSRHVV 331
+ DL + D+ +AS L +WD S L GH + V V+ + S+ +V
Sbjct: 86 SGISDLAWSSDSHYTCSASDDCTLRIWDARSPYECLKVLRGHTNFVFCVNFNPPSNL-IV 144
Query: 332 SAAYDRTIKVWDLMKGYCTNTI-IFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKL 390
S ++D TI++W++ G C I + +++ F+ DG I S DG+ ++WD + G
Sbjct: 145 SGSFDETIRIWEVKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSCKIWDAKEGTC 204
Query: 391 LSE-VAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSR 449
L + S AV+ S NG +L + D+ L + + + + N+V S
Sbjct: 205 LKTLIDDKSPAVSFAKFSPNGKFILVATLDSTLKLSNYATGKFLKVYTGHTNKVFCITSA 264
Query: 450 SCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADK--N 507
++ + ++ +GS D V++W + I+ L+ HT +V+ + ++S+ +
Sbjct: 265 FSVT-NGKYIVSGSEDNCVYLWDLQARNILQRLEGHTDAVISVSCHPVQNEISSSGNHLD 323
Query: 508 GIVCIWK 514
+ IWK
Sbjct: 324 KTIRIWK 330
Score = 74.7 bits (182), Expect = 1e-13
Identities = 53/209 (25%), Positives = 93/209 (44%), Gaps = 8/209 (3%)
Query: 215 DSNIPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGS 274
D+ P C L H + F S+ +++G D+++++W+ TG +
Sbjct: 113 DARSPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRIWEVKTGKCVRMIKAHSMP 172
Query: 275 VLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVS--SRHVVS 332
+ + D +++AS + +WD G TL DK AV +K S + ++
Sbjct: 173 ISSVHFNRDGSLIVSASHDGSCKIWDAKEGTCLKTLI--DDKSPAVSFAKFSPNGKFILV 230
Query: 333 AAYDRTIKVWDLMKGYCTNTIIFASN---CNALCFSM-DGQTIYSGHVDGNLRLWDIQSG 388
A D T+K+ + G +N C FS+ +G+ I SG D + LWD+Q+
Sbjct: 231 ATLDSTLKLSNYATGKFLKVYTGHTNKVFCITSAFSVTNGKYIVSGSEDNCVYLWDLQAR 290
Query: 389 KLLSEVAAHSLAVTSISLSRNGNVVLTSG 417
+L + H+ AV S+S N + +SG
Sbjct: 291 NILQRLEGHTDAVISVSCHPVQNEISSSG 319
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 99.8 bits (247), Expect = 3e-21
Identities = 64/237 (27%), Positives = 105/237 (44%), Gaps = 7/237 (2%)
Query: 228 AHEGGCASILFEFNSSR-LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRS 286
AH + SSR L+TGG+D V +W + +L+G + +T
Sbjct: 107 AHSAAVNCLKIGRKSSRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEGL 166
Query: 287 VIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK 346
V A ++S + +WDL +V TLTGH+ +V+ S + D +K+WD+ K
Sbjct: 167 VAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFHPF-GEFFASGSLDTNLKIWDIRK 225
Query: 347 GYCTNTII-FASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSIS 405
C +T N L F+ DG+ I SG D +++WD+ +GKLL E +H + S+
Sbjct: 226 KGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQSLD 285
Query: 406 LSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAG 462
+ ++ T D +D+ + E+ S G + +PD V G
Sbjct: 286 FHPHEFLLATGSADKTVKFWDLETFELIGS----GGTETTGVRCLTFNPDGKSVLCG 338
Score = 83.2 bits (204), Expect = 3e-16
Identities = 53/217 (24%), Positives = 100/217 (45%), Gaps = 6/217 (2%)
Query: 284 NRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWD 343
+R ++ + + +W + +L GH + +V S V + A TIK+WD
Sbjct: 122 SRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFD-ASEGLVAAGAASGTIKLWD 180
Query: 344 LMKGYCTNTII-FASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVT 402
L + T+ SNC ++ F G+ SG +D NL++WDI+ + H+ V
Sbjct: 181 LEEAKVVRTLTGHRSNCVSVNFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVN 240
Query: 403 SISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAG 462
+ + +G +++ G DNV ++D+ + ++ F+ ++ S P +A G
Sbjct: 241 VLRFTPDGRWIVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQS----LDFHPHEFLLATG 296
Query: 463 SADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGK 499
SAD +V W + E++ + T+ V C ++ GK
Sbjct: 297 SADKTVKFWDLETFELIGSGGTETTGVRCLTFNPDGK 333
Score = 67.8 bits (164), Expect = 1e-11
Identities = 43/190 (22%), Positives = 76/190 (39%), Gaps = 2/190 (1%)
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
L H G S+ F+ + + G ++K+WD V L G + + +
Sbjct: 148 LYGHSSGIDSVTFDASEGLVAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFHPFGE 207
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
+ S NL +WD+ HT GH V + + R +VS D +KVWDL
Sbjct: 208 FFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTP-DGRWIVSGGEDNVVKVWDLT 266
Query: 346 KGYCTNTI-IFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSI 404
G + +L F + +G D ++ WD+++ +L+ + V +
Sbjct: 267 AGKLLHEFKSHEGKIQSLDFHPHEFLLATGSADKTVKFWDLETFELIGSGGTETTGVRCL 326
Query: 405 SLSRNGNVVL 414
+ + +G VL
Sbjct: 327 TFNPDGKSVL 336
Score = 62.8 bits (151), Expect = 4e-10
Identities = 40/154 (25%), Positives = 65/154 (41%), Gaps = 10/154 (6%)
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
L H C S+ F +G D ++K+WD G V L T D R
Sbjct: 190 LTGHRSNCVSVNFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGR 249
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWD-- 343
+++ N + VWDL +G++ H H+ K+ ++D +A D+T+K WD
Sbjct: 250 WIVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQSLDFHPHEFLLATGSA-DKTVKFWDLE 308
Query: 344 ---LMKGYCTNTIIFASNCNALCFSMDGQTIYSG 374
L+ T T + L F+ DG+++ G
Sbjct: 309 TFELIGSGGTET----TGVRCLTFNPDGKSVLCG 338
Score = 57.0 bits (136), Expect = 2e-08
Identities = 35/145 (24%), Positives = 64/145 (44%), Gaps = 4/145 (2%)
Query: 369 QTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVR 428
+ + +G D + LW I + + HS + S++ + +V L+D+
Sbjct: 123 RVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEGLVAAGAASGTIKLWDLE 182
Query: 429 SLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSS 488
+V + TG+R SN P A+GS D ++ IW + K + T K HT
Sbjct: 183 EAKVVRTL--TGHR--SNCVSVNFHPFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRG 238
Query: 489 VLCCRWSGIGKPLASADKNGIVCIW 513
V R++ G+ + S ++ +V +W
Sbjct: 239 VNVLRFTPDGRWIVSGGEDNVVKVW 263
Score = 37.0 bits (84), Expect = 0.026
Identities = 25/103 (24%), Positives = 39/103 (37%), Gaps = 10/103 (9%)
Query: 211 EFF----LDSNIP------STCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTN 260
EFF LD+N+ C H G + F + +++GG+D VKVWD
Sbjct: 207 EFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNVVKVWDLT 266
Query: 261 TGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNS 303
G + G + L + S+ + WDL +
Sbjct: 267 AGKLLHEFKSHEGKIQSLDFHPHEFLLATGSADKTVKFWDLET 309
>At1g18080 putative guanine nucleotide-binding protein beta
subunit
Length = 327
Score = 92.8 bits (229), Expect = 4e-19
Identities = 78/304 (25%), Positives = 136/304 (44%), Gaps = 39/304 (12%)
Query: 241 NSSRLITGGQDQSVKVW-----DTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNN 295
N+ +++ +D+S+ +W D G L G V D+ ++ D + ++ S
Sbjct: 27 NADIIVSASRDKSIILWKLTKDDKAYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSWDGE 86
Query: 296 LYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIF 355
L +WDL +G GH V +V S + +R +VSA+ DRTIK+W+ + G C TI
Sbjct: 87 LRLWDLAAGVSTRRFVGHTKDVLSVAFS-LDNRQIVSASRDRTIKLWNTL-GECKYTISE 144
Query: 356 AS-------NCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSR 408
+C + TI S D +++W++ + KL S +A H+ V+++++S
Sbjct: 145 GGEGHRDWVSCVRFSPNTLQPTIVSASWDKTVKVWNLSNCKLRSTLAGHTGYVSTVAVSP 204
Query: 409 NGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSV 468
+G++ + G+D V L+D+ + S N V C SP+ + A + G +
Sbjct: 205 DGSLCASGGKDGVVLLWDLAEGKKLYSLE--ANSVI---HALCFSPNRYWLCAATEHG-I 258
Query: 469 HIWSVHKNEIVSTLKEHTSS----------------VLCC---RWSGIGKPLASADKNGI 509
IW + IV LK + V+ C WS G L S +G+
Sbjct: 259 KIWDLESKSIVEDLKVDLKAEAEKADNSGPAATKRKVIYCTSLNWSADGSTLFSGYTDGV 318
Query: 510 VCIW 513
+ +W
Sbjct: 319 IRVW 322
Score = 83.6 bits (205), Expect = 2e-16
Identities = 66/275 (24%), Positives = 119/275 (43%), Gaps = 34/275 (12%)
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDN 284
RL H ++ + ++G D +++WD G G VL + + DN
Sbjct: 58 RLTGHSHFVEDVVLSSDGQFALSGSWDGELRLWDLAAGVSTRRFVGHTKDVLSVAFSLDN 117
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTLT----GHKDKVCAVDVS-KVSSRHVVSAAYDRTI 339
R +++AS + +W+ G ++T++ GH+D V V S +VSA++D+T+
Sbjct: 118 RQIVSASRDRTIKLWN-TLGECKYTISEGGEGHRDWVSCVRFSPNTLQPTIVSASWDKTV 176
Query: 340 KVWDLMKGYCTNTII-FASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHS 398
KVW+L +T+ + + S DG SG DG + LWD+ GK L + A+S
Sbjct: 177 KVWNLSNCKLRSTLAGHTGYVSTVAVSPDGSLCASGGKDGVVLLWDLAEGKKLYSLEANS 236
Query: 399 LAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR----------DTGNRVAS--- 445
+ + ++ S N L + ++ ++D+ S + + D A+
Sbjct: 237 V-IHALCFSPN-RYWLCAATEHGIKIWDLESKSIVEDLKVDLKAEAEKADNSGPAATKRK 294
Query: 446 -------NWSRSCISPDNSHVAAGSADGSVHIWSV 473
NW S D S + +G DG + +W +
Sbjct: 295 VIYCTSLNW-----SADGSTLFSGYTDGVIRVWGI 324
Score = 82.8 bits (203), Expect = 4e-16
Identities = 62/218 (28%), Positives = 103/218 (46%), Gaps = 13/218 (5%)
Query: 306 VRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK-----GYCTNTIIFASN-C 359
++ T+ H D V A+ ++ +VSA+ D++I +W L K G + S+
Sbjct: 7 LKGTMRAHTDMVTAIATPIDNADIIVSASRDKSIILWKLTKDDKAYGVAQRRLTGHSHFV 66
Query: 360 NALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRD 419
+ S DGQ SG DG LRLWD+ +G H+ V S++ S + ++++ RD
Sbjct: 67 EDVVLSSDGQFALSGSWDGELRLWDLAAGVSTRRFVGHTKDVLSVAFSLDNRQIVSASRD 126
Query: 420 NVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCI--SPDNSH--VAAGSADGSVHIWSVHK 475
L++ +L C G +W SC+ SP+ + + S D +V +W++
Sbjct: 127 RTIKLWN--TLGECKYTISEGGEGHRDWV-SCVRFSPNTLQPTIVSASWDKTVKVWNLSN 183
Query: 476 NEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ STL HT V S G AS K+G+V +W
Sbjct: 184 CKLRSTLAGHTGYVSTVAVSPDGSLCASGGKDGVVLLW 221
>At1g04510 putative pre-mRNA splicing factor PRP19
Length = 523
Score = 92.0 bits (227), Expect = 7e-19
Identities = 100/395 (25%), Positives = 167/395 (41%), Gaps = 47/395 (11%)
Query: 50 SEWKE-KAENLQVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEM 108
+EW N +E QQ + A+ LS L A R IA L+KE
Sbjct: 78 TEWDSLMLSNFALE-QQLHTARQELSHALYQHDAACRV-----------IARLKKE---- 121
Query: 109 RDEFSQLKVDLEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKD 168
RDE QL + E+++ + + N+ L + K ID
Sbjct: 122 RDESRQLLAEAERQLPAAPEVATSNAAL---------------SNGKRGIDDGEQGPNAK 166
Query: 169 AERLNEANALYEEMVERLRASGLEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNA 228
RL + + E+ + A+ +Q ++Q+ + + +F S+ P
Sbjct: 167 KMRLGISAEVITELTD-CNAALSQQRKKRQIPKTLASVDALEKFTQLSSHPLH-----KT 220
Query: 229 HEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVI 288
++ G S+ + + TGG D + ++D +G + S L G V + D V+
Sbjct: 221 NKPGIFSMDILHSKDVIATGGIDTTAVLFDRPSGQILSTLTGHSKKVTSIKFVGDTDLVL 280
Query: 289 AASSSNNLYVWDLNSG---RVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
ASS + +W + RHTL H +V AV V ++++ VSA+ D T +DL
Sbjct: 281 TASSDKTVRIWGCSEDGNYTSRHTLKDHSAEVRAVTVH-ATNKYFVSASLDSTWCFYDLS 339
Query: 346 KGYCTNTIIFAS----NCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAV 401
G C + AS N A F DG + +G +++WD++S +++ H+ +
Sbjct: 340 SGLCLAQVTDASENDVNYTAAAFHPDGLILGTGTAQSIVKIWDVKSQANVAKFGGHNGEI 399
Query: 402 TSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSF 436
TSIS S NG + T+ D V L+D+R L+ +F
Sbjct: 400 TSISFSENGYFLATAALDGV-RLWDLRKLKNFRTF 433
Score = 43.1 bits (100), Expect = 4e-04
Identities = 48/194 (24%), Positives = 75/194 (37%), Gaps = 48/194 (24%)
Query: 364 FSMD----GQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRD 419
FSMD I +G +D L+D SG++LS + HS VTSI + ++VLT+ D
Sbjct: 226 FSMDILHSKDVIATGGIDTTAVLFDRPSGQILSTLTGHSKKVTSIKFVGDTDLVLTASSD 285
Query: 420 NV---------------HNLFD----VRSLEVCSS--------------FRDTGNRVAS- 445
H L D VR++ V ++ F D + +
Sbjct: 286 KTVRIWGCSEDGNYTSRHTLKDHSAEVRAVTVHATNKYFVSASLDSTWCFYDLSSGLCLA 345
Query: 446 ----------NWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWS 495
N++ + PD + G+A V IW V V+ H + +S
Sbjct: 346 QVTDASENDVNYTAAAFHPDGLILGTGTAQSIVKIWDVKSQANVAKFGGHNGEITSISFS 405
Query: 496 GIGKPLASADKNGI 509
G LA+A +G+
Sbjct: 406 ENGYFLATAALDGV 419
>At1g10580 putative pre-mRNA splicing factor
Length = 616
Score = 88.2 bits (217), Expect = 1e-17
Identities = 79/314 (25%), Positives = 126/314 (39%), Gaps = 29/314 (9%)
Query: 218 IPSTCKFRLNAHEGGCASI-LFEFNSSRLITGGQDQSVKVWDT-NTGSVCSNLHGCIGSV 275
IP + H G ++I F L++ G D VK+WD N+G G +V
Sbjct: 270 IPKRLVHTWSGHTKGVSAIRFFPKQGHLLLSAGMDCKVKIWDVYNSGKCMRTYMGHAKAV 329
Query: 276 LDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTL-TGHKDKVCAVDVSKVSSRHVVSAA 334
D+ ++D + A N+ WD +G+V T TG V ++ +++
Sbjct: 330 RDICFSNDGSKFLTAGYDKNIKYWDTETGQVISTFSTGKIPYVVKLNPDDDKQNILLAGM 389
Query: 335 YDRTIKVWDLMKGYCT----------NTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWD 384
D+ I WD+ G T NTI F N S D D +LR+W+
Sbjct: 390 SDKKIVQWDINTGEVTQEYDQHLGAVNTITFVDNNRRFVTSSD---------DKSLRVWE 440
Query: 385 IQSG---KLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRS-LEVCSSFRDTG 440
K +SE HS+ SIS+ NGN + DN ++ R ++ R G
Sbjct: 441 FGIPVVIKYISEPHMHSMP--SISVHPNGNWLAAQSLDNQILIYSTRERFQLNKKKRFAG 498
Query: 441 NRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGK- 499
+ VA + SPD V +G +G W ++ TLK H + W + +
Sbjct: 499 HIVAGYACQVNFSPDGRFVMSGDGEGKCWFWDWKSCKVFRTLKCHNGVCIGAEWHPLEQS 558
Query: 500 PLASADKNGIVCIW 513
+A+ +G++ W
Sbjct: 559 KVATCGWDGLIKYW 572
>At3g18140 unknown protein
Length = 305
Score = 87.0 bits (214), Expect = 2e-17
Identities = 68/275 (24%), Positives = 126/275 (45%), Gaps = 8/275 (2%)
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG 304
L T D +++ W+ TG + V L IT D + +AA+ + ++ ++D+NS
Sbjct: 8 LATASYDHTIRFWEAETGRCYRTIQYPDSHVNRLEITPD-KHYLAAACNPHIRLFDVNSN 66
Query: 305 RVRHTLT--GHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNAL 362
+ +T H + V AV + ++ + S + D T+K+WDL C + N +
Sbjct: 67 SPQPVMTYDSHTNNVMAVGF-QCDAKWMYSGSEDGTVKIWDLRAPGCQKEYESVAAVNTV 125
Query: 363 CFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAH-SLAVTSISLSRNGN-VVLTSGRDN 420
+ + SG +GN+R+WD+++ E+ AV S+++ +G VV + R
Sbjct: 126 VLHPNQTELISGDQNGNIRVWDLRANSCSCELVPEVDTAVRSLTVMWDGTMVVAANNRGT 185
Query: 421 VHNLFDVRSLEVCSSFRDTGNRVASNWS--RSCISPDNSHVAAGSADGSVHIWSVHKNEI 478
+ +R + + F A N + +SP N ++A S+D +V IW+V ++
Sbjct: 186 CYVWRLLRGKQTMTEFEPLHKLQAHNGHILKCLLSPANKYLATASSDKTVKIWNVDGFKL 245
Query: 479 VSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
L H V C +S G+ L +A + +W
Sbjct: 246 EKVLTGHQRWVWDCVFSVDGEFLVTASSDMTARLW 280
Score = 68.6 bits (166), Expect = 8e-12
Identities = 46/184 (25%), Positives = 88/184 (47%), Gaps = 16/184 (8%)
Query: 235 SILFEFNSSRLITGGQDQSVKVWDTNTGSV-CSNLHGCIGSVLDLTITHDNRSVIAASSS 293
+++ N + LI+G Q+ +++VWD S C + +V LT+ D V+AA++
Sbjct: 124 TVVLHPNQTELISGDQNGNIRVWDLRANSCSCELVPEVDTAVRSLTVMWDGTMVVAANNR 183
Query: 294 NNLYVWDLNSGRVR-------HTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDL-- 344
YVW L G+ H L H + +S +++++ +A+ D+T+K+W++
Sbjct: 184 GTCYVWRLLRGKQTMTEFEPLHKLQAHNGHILKCLLSP-ANKYLATASSDKTVKIWNVDG 242
Query: 345 --MKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVT 402
++ T + +C FS+DG+ + + D RLW + +GK + H A
Sbjct: 243 FKLEKVLTGHQRWVWDC---VFSVDGEFLVTASSDMTARLWSMPAGKEVKVYQGHHKATV 299
Query: 403 SISL 406
+L
Sbjct: 300 CCAL 303
Score = 55.8 bits (133), Expect = 5e-08
Identities = 54/282 (19%), Positives = 106/282 (37%), Gaps = 42/282 (14%)
Query: 212 FFLDSNIPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGC 271
F ++SN P ++H ++ F+ ++ + +G +D +VK+WD C +
Sbjct: 61 FDVNSNSPQPV-MTYDSHTNNVMAVGFQCDAKWMYSGSEDGTVKIWDLRAPG-CQKEYES 118
Query: 272 IGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVV 331
+ +V + + + +I+ + N+ VWDL + L D VV
Sbjct: 119 VAAVNTVVLHPNQTELISGDQNGNIRVWDLRANSCSCELVPEVDTAVRSLTVMWDGTMVV 178
Query: 332 SAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLL 391
+A T VW L++G T T + L
Sbjct: 179 AANNRGTCYVWRLLRGKQTMT----------------------------------EFEPL 204
Query: 392 SEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSC 451
++ AH+ + LS + T+ D +++V ++ TG++ W C
Sbjct: 205 HKLQAHNGHILKCLLSPANKYLATASSDKTVKIWNVDGFKLEKVL--TGHQ---RWVWDC 259
Query: 452 I-SPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCC 492
+ S D + S+D + +WS+ + V + H + +CC
Sbjct: 260 VFSVDGEFLVTASSDMTARLWSMPAGKEVKVYQGHHKATVCC 301
>At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2
Length = 479
Score = 86.7 bits (213), Expect = 3e-17
Identities = 72/306 (23%), Positives = 129/306 (41%), Gaps = 32/306 (10%)
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
L H G S+ F+ ++ TG D+++K+WD TG + L G IG V L +++ +
Sbjct: 166 LQGHLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGVLKLTLTGHIGQVRGLAVSNRHT 225
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
+ +A + WDL +V + GH V + + V++ D +VWD+
Sbjct: 226 YMFSAGDDKQVKCWDLEQNKVIRSYHGHLHGVYCLALHPTLD-VVLTGGRDSVCRVWDIR 284
Query: 346 KGYCTNTIIFASNCNALCFSM-----DGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLA 400
T IF ++ FS+ D Q I H D ++ WD++ GK ++ + H
Sbjct: 285 ----TKMQIFVLPHDSDVFSVLARPTDPQVITGSH-DSTIKFWDLRYGKSMATITNHKKT 339
Query: 401 VTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSF----RDTGNRVASNWSRSCISPDN 456
V +++L N +++ DN+ F + E C + RD N VA N ++
Sbjct: 340 VRAMALHPKENDFVSASADNIKK-FSLPKGEFCHNMLSLQRDIINAVAVN--------ED 390
Query: 457 SHVAAGSADGSVHIW---SVHKNEIVSTL-----KEHTSSVLCCRWSGIGKPLASADKNG 508
+ G G + W S H + T+ E + + + G L + + +
Sbjct: 391 GVMVTGGDKGGLWFWDWKSGHNFQRAETIVQPGSLESEAGIYAACYDQTGSRLVTCEGDK 450
Query: 509 IVCIWK 514
+ +WK
Sbjct: 451 TIKMWK 456
Score = 53.1 bits (126), Expect = 4e-07
Identities = 54/209 (25%), Positives = 79/209 (36%), Gaps = 46/209 (22%)
Query: 223 KFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSV------- 275
K L H G + + + + G D+ VK WD V + HG + V
Sbjct: 205 KLTLTGHIGQVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNKVIRSYHGHLHGVYCLALHP 264
Query: 276 -LDLTIT----------------------HD-----------NRSVIAASSSNNLYVWDL 301
LD+ +T HD + VI S + + WDL
Sbjct: 265 TLDVVLTGGRDSVCRVWDIRTKMQIFVLPHDSDVFSVLARPTDPQVITGSHDSTIKFWDL 324
Query: 302 NSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKG-YCTNTIIFASN-C 359
G+ T+T HK V A+ + VSA+ D IK + L KG +C N + +
Sbjct: 325 RYGKSMATITNHKKTVRAMALHP-KENDFVSASAD-NIKKFSLPKGEFCHNMLSLQRDII 382
Query: 360 NALCFSMDGQTIYSGHVDGNLRLWDIQSG 388
NA+ + DG + G G L WD +SG
Sbjct: 383 NAVAVNEDGVMVTGGD-KGGLWFWDWKSG 410
>At2g33340 putative PRP19-like spliceosomal protein
Length = 565
Score = 86.7 bits (213), Expect = 3e-17
Identities = 110/450 (24%), Positives = 192/450 (42%), Gaps = 51/450 (11%)
Query: 73 LSEQLVVEVADSRASKALVQEKD--NAIADLQKELTEMRDEFSQLKVDLEQKIKELEVIV 130
L +QL A S AL Q IA L+KE RDE QL ++E+ I V
Sbjct: 90 LEQQL--HTARQELSHALYQHDSACRVIARLKKE----RDEARQLLAEVERHIPAAPEAV 143
Query: 131 SENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANALYEEMVERLRASG 190
+ N+ L + K+A + ++ D L AE + E + ++ +
Sbjct: 144 TANAAL--------SNGKRAAVDEELGPDAKKLCPGISAEIITELTDCNAALSQKRKKRQ 195
Query: 191 LEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASILFEFNSSRLITGGQ 250
+ Q +D + R + + +N P C S+ + + TGG
Sbjct: 196 IPQTLAS-IDTLERFTQLSSHPLHKTNKPGIC------------SMDILHSKDVIATGGV 242
Query: 251 DQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG----RV 306
D + ++D +G + S L G V + D+ V+ AS+ + +W N G
Sbjct: 243 DATAVLFDRPSGQILSTLTGHSKKVTSVKFVGDSDLVLTASADKTVRIWR-NPGDGNYAC 301
Query: 307 RHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFAS---NCNALC 363
+TL H +V AV V ++++ VSA+ D T +DL G C + S + A
Sbjct: 302 GYTLNDHSAEVRAVTVHP-TNKYFVSASLDGTWCFYDLSSGSCLAQVSDDSKNVDYTAAA 360
Query: 364 FSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHN 423
F DG + +G +++WD++S +++ H+ VT+IS S NG + T+ D V
Sbjct: 361 FHPDGLILGTGTSQSVVKIWDVKSQANVAKFDGHTGEVTAISFSENGYFLATAAEDGV-R 419
Query: 424 LFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNE--IVST 481
L+D+R L SF ++++ + P S++ ++D V+ + K E ++ T
Sbjct: 420 LWDLRKLRNFKSF------LSADANSVEFDPSGSYLGIAASDIKVYQTASVKAEWNLIKT 473
Query: 482 LKE--HTSSVLCCRWSGIGKPLA--SADKN 507
L + T C ++ + +A S D+N
Sbjct: 474 LPDLSGTGKATCVKFGSDAQYVAVGSMDRN 503
Score = 50.1 bits (118), Expect = 3e-06
Identities = 41/204 (20%), Positives = 85/204 (41%), Gaps = 13/204 (6%)
Query: 313 HKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTII-FASNCNALCFSMDGQTI 371
+K +C++D+ S + + D T ++D G +T+ + ++ F D +
Sbjct: 221 NKPGICSMDILH-SKDVIATGGVDATAVLFDRPSGQILSTLTGHSKKVTSVKFVGDSDLV 279
Query: 372 YSGHVDGNLRLW------DIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLF 425
+ D +R+W + G L++ HS V ++++ +++ D +
Sbjct: 280 LTASADKTVRIWRNPGDGNYACGYTLND---HSAEVRAVTVHPTNKYFVSASLDGTWCFY 336
Query: 426 DVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEH 485
D+ S + D V +++ + PD + G++ V IW V V+ H
Sbjct: 337 DLSSGSCLAQVSDDSKNV--DYTAAAFHPDGLILGTGTSQSVVKIWDVKSQANVAKFDGH 394
Query: 486 TSSVLCCRWSGIGKPLASADKNGI 509
T V +S G LA+A ++G+
Sbjct: 395 TGEVTAISFSENGYFLATAAEDGV 418
Score = 28.5 bits (62), Expect = 9.3
Identities = 14/56 (25%), Positives = 27/56 (48%)
Query: 459 VAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIWK 514
+A G D + ++ +I+STL H+ V ++ G + +A + V IW+
Sbjct: 237 IATGGVDATAVLFDRPSGQILSTLTGHSKKVTSVKFVGDSDLVLTASADKTVRIWR 292
>At3g27640 unknown protein
Length = 535
Score = 84.0 bits (206), Expect = 2e-16
Identities = 79/302 (26%), Positives = 131/302 (43%), Gaps = 34/302 (11%)
Query: 228 AHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSV 287
AH I + S L+T DQ++KVWD L G G+V + N +
Sbjct: 126 AHYNAIFDISWIKGDSCLLTASGDQTIKVWDVEENKCTGVLIGHTGTVKSMCSHPTNSDL 185
Query: 288 IAASSSNNLY-VWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK 346
+ + S + + +WDL R + HK++ C V H+ + + I+
Sbjct: 186 LVSGSRDGCFALWDL-----RCKSSSHKEEFCINSTGMVKGAHL--SPLSKRIR------ 232
Query: 347 GYCTNTIIFASNCNALCFSMDGQTI-YSGHVDGNLRLWDIQSGKLLSEVAA--------- 396
+S+ ++ + D TI +G D L+ WDI+ K A+
Sbjct: 233 ----RRKAASSSITSVLYVKDEITIATAGAPDSALKFWDIRKLKAPFAQASPQSDPTNTK 288
Query: 397 --HSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISP 454
S + S+S +G + S +DN L++ L+ +G R+ S + R+ ISP
Sbjct: 289 EKRSHGIVSLSQDSSGTYLTASCKDNRIYLYNTLRLDKGPVQSFSGCRIDSFFVRTMISP 348
Query: 455 DNSHVAAGSADGSVHIWSVHKNEI-VSTLKEHTSSVLCCRW--SGIGKPLASADKNGIVC 511
D +V +GS+DG+ +IW V+K ++ LK H V W S IGK +A+A + V
Sbjct: 349 DGEYVLSGSSDGNAYIWQVNKPQVDPIILKGHDFEVTAVDWSPSEIGK-VATASDDFTVR 407
Query: 512 IW 513
+W
Sbjct: 408 LW 409
Score = 51.2 bits (121), Expect = 1e-06
Identities = 41/132 (31%), Positives = 65/132 (49%), Gaps = 7/132 (5%)
Query: 232 GCASILFEFNSSRLITGGQDQSVKVWDT---NTGSVCSNLHGC-IGSVLDLT-ITHDNRS 286
G S+ + + + L +D + +++T + G V S GC I S T I+ D
Sbjct: 294 GIVSLSQDSSGTYLTASCKDNRIYLYNTLRLDKGPVQS-FSGCRIDSFFVRTMISPDGEY 352
Query: 287 VIAASSSNNLYVWDLNSGRVRHT-LTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
V++ SS N Y+W +N +V L GH +V AVD S V +A+ D T+++W++
Sbjct: 353 VLSGSSDGNAYIWQVNKPQVDPIILKGHDFEVTAVDWSPSEIGKVATASDDFTVRLWNIE 412
Query: 346 KGYCTNTIIFAS 357
CTN AS
Sbjct: 413 NNICTNANATAS 424
Score = 38.1 bits (87), Expect = 0.012
Identities = 34/127 (26%), Positives = 52/127 (40%), Gaps = 13/127 (10%)
Query: 399 LAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSS-----------FRDTGNRVASNW 447
LAV+ SRN + S D +LF+ S + SS FRD + +
Sbjct: 73 LAVSFCKTSRNSQLFAVSDEDGHVSLFNSSSKKFASSATHQENTEKARFRDWIAHYNAIF 132
Query: 448 SRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSV-LCCRWSGIGKPLASADK 506
S I D+ + A S D ++ +W V +N+ L HT +V C L S +
Sbjct: 133 DISWIKGDSCLLTA-SGDQTIKVWDVEENKCTGVLIGHTGTVKSMCSHPTNSDLLVSGSR 191
Query: 507 NGIVCIW 513
+G +W
Sbjct: 192 DGCFALW 198
>At1g49040 hypothetical protein; similar to EST emb|Z26090.1
Length = 1264
Score = 83.6 bits (205), Expect = 2e-16
Identities = 72/266 (27%), Positives = 123/266 (46%), Gaps = 27/266 (10%)
Query: 246 ITGGQDQSVKVWDTNT--GSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNS 303
I+G D VK+WD + + + L G G+V I+ D +++ S ++ VWD +
Sbjct: 948 ISGSTDCLVKIWDPSLRGSELRATLKGHTGTVR--AISSDRGKIVSGSDDLSVIVWDKQT 1005
Query: 304 GRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALC 363
++ L GH +V V + +S V++AA+D T+K+WD+ C T+ C++
Sbjct: 1006 TQLLEELKGHDSQVSCVKM--LSGERVLTAAHDGTVKMWDVRTDMCVATV---GRCSSAI 1060
Query: 364 FSM--DGQT--IYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRD 419
S+ D T + + D +WDI+SGK + ++ H+ + SI + + L +G D
Sbjct: 1061 LSLEYDDSTGILAAAGRDTVANIWDIRSGKQMHKLKGHTKWIRSIRMVED---TLITGSD 1117
Query: 420 NVHNLFDVRSLEVCSSFRDTGNRVASNWSRSC-ISPDNSHVAAGSADGSVHIWSVHKNEI 478
+ + R V D + +S SP + + GSADG + W + I
Sbjct: 1118 D----WTARVWSVSRGSCDAVLACHAGPVQSVEYSPFDKGIITGSADGLLRFWENDEGGI 1173
Query: 479 --VSTLKEHTSSVLCC----RWSGIG 498
V + H+SS+L W GIG
Sbjct: 1174 KCVKNITLHSSSILSINAGENWLGIG 1199
Score = 77.4 bits (189), Expect = 2e-14
Identities = 62/269 (23%), Positives = 112/269 (41%), Gaps = 35/269 (13%)
Query: 226 LNAHEG--GCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHD 283
L H+ C +L + R++T D +VK+WD T + + C ++L L
Sbjct: 1012 LKGHDSQVSCVKML---SGERVLTAAHDGTVKMWDVRTDMCVATVGRCSSAILSLEYDDS 1068
Query: 284 NRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWD 343
+ AA +WD+ SG+ H L GH + ++ ++ +++ + D T +VW
Sbjct: 1069 TGILAAAGRDTVANIWDIRSGKQMHKLKGHTKWIRSI---RMVEDTLITGSDDWTARVWS 1125
Query: 344 LMKGYCTNTI-IFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSG--KLLSEVAAHSLA 400
+ +G C + A ++ +S + I +G DG LR W+ G K + + HS +
Sbjct: 1126 VSRGSCDAVLACHAGPVQSVEYSPFDKGIITGSADGLLRFWENDEGGIKCVKNITLHSSS 1185
Query: 401 VTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSR----------- 449
+ SI+ N + DN +LF S + G +V S W
Sbjct: 1186 ILSINAGENWLGI--GAADNSMSLFHRPS--------NAGTKV-SGWQLYRVPQRTAAVV 1234
Query: 450 SCISPD--NSHVAAGSADGSVHIWSVHKN 476
C++ D + +G +G + +W N
Sbjct: 1235 RCVASDLERKRICSGGRNGVLRLWDATIN 1263
Score = 57.8 bits (138), Expect = 1e-08
Identities = 47/249 (18%), Positives = 101/249 (39%), Gaps = 50/249 (20%)
Query: 308 HTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWD----------LMKGYCTNTIIFAS 357
H++T + +VC + + + +S + D +K+WD +KG+ +S
Sbjct: 927 HSVT--RREVCDLVGDREDAGFFISGSTDCLVKIWDPSLRGSELRATLKGHTGTVRAISS 984
Query: 358 NCNAL-------------------------------CFSM-DGQTIYSGHVDGNLRLWDI 385
+ + C M G+ + + DG +++WD+
Sbjct: 985 DRGKIVSGSDDLSVIVWDKQTTQLLEELKGHDSQVSCVKMLSGERVLTAAHDGTVKMWDV 1044
Query: 386 QSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVAS 445
++ ++ V S A+ S+ + ++ +GRD V N++D+RS + + +
Sbjct: 1045 RTDMCVATVGRCSSAILSLEYDDSTGILAAAGRDTVANIWDIRSGKQMHKLKG-----HT 1099
Query: 446 NWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASAD 505
W RS +++ + GS D + +WSV + + L H V +S K + +
Sbjct: 1100 KWIRSIRMVEDT-LITGSDDWTARVWSVSRGSCDAVLACHAGPVQSVEYSPFDKGIITGS 1158
Query: 506 KNGIVCIWK 514
+G++ W+
Sbjct: 1159 ADGLLRFWE 1167
Score = 44.7 bits (104), Expect = 1e-04
Identities = 44/201 (21%), Positives = 93/201 (45%), Gaps = 18/201 (8%)
Query: 322 VSKVSSRHVVSAAYDRTIKVWDLMKGY-----CTNTIIFASNCNALCFSMDGQTIYSGHV 376
VS +S + +AA + ++KG+ +++ C+ + D SG
Sbjct: 893 VSDNASSDITAAAQKKIQTNVRVLKGHGGAVTALHSVTRREVCDLVGDREDAGFFISGST 952
Query: 377 DGNLRLWD--IQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNL-FDVRSLEVC 433
D +++WD ++ +L + + H+ V +IS R G +V SG D++ + +D ++ ++
Sbjct: 953 DCLVKIWDPSLRGSELRATLKGHTGTVRAISSDR-GKIV--SGSDDLSVIVWDKQTTQLL 1009
Query: 434 SSFRDTGNRVASNWSRSCISP-DNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCC 492
+ ++V SC+ V + DG+V +W V + V+T+ +S++L
Sbjct: 1010 EELKGHDSQV------SCVKMLSGERVLTAAHDGTVKMWDVRTDMCVATVGRCSSAILSL 1063
Query: 493 RWSGIGKPLASADKNGIVCIW 513
+ LA+A ++ + IW
Sbjct: 1064 EYDDSTGILAAAGRDTVANIW 1084
Score = 30.8 bits (68), Expect = 1.9
Identities = 26/130 (20%), Positives = 52/130 (40%), Gaps = 7/130 (5%)
Query: 221 TCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSV--CSNLHGCIGSVLDL 278
+C L H G S+ + +ITG D ++ W+ + G + N+ S+L +
Sbjct: 1130 SCDAVLACHAGPVQSVEYSPFDKGIITGSADGLLRFWENDEGGIKCVKNITLHSSSILSI 1189
Query: 279 TITHDNRSVIAASSSNNLYVWDLN-----SGRVRHTLTGHKDKVCAVDVSKVSSRHVVSA 333
+ + AA +S +L+ N SG + + V S + + + S
Sbjct: 1190 NAGENWLGIGAADNSMSLFHRPSNAGTKVSGWQLYRVPQRTAAVVRCVASDLERKRICSG 1249
Query: 334 AYDRTIKVWD 343
+ +++WD
Sbjct: 1250 GRNGVLRLWD 1259
>At4g15900 PRL1 protein
Length = 486
Score = 82.8 bits (203), Expect = 4e-16
Identities = 66/303 (21%), Positives = 125/303 (40%), Gaps = 25/303 (8%)
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
+ H G S+ F+ ++ TG D+++K+WD TG + L G I V L +++ +
Sbjct: 172 IQGHLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGVLKLTLTGHIEQVRGLAVSNRHT 231
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
+ +A + WDL +V + GH V + + +++ D +VWD+
Sbjct: 232 YMFSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLALHPTLD-VLLTGGRDSVCRVWDIR 290
Query: 346 KGYCTNTIIFASN------CNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSL 399
T IFA + C+ D Q + H D ++ WD++ GK +S + H
Sbjct: 291 ----TKMQIFALSGHDNTVCSVFTRPTDPQVVTGSH-DTTIKFWDLRYGKTMSTLTHHKK 345
Query: 400 AVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHV 459
+V +++L N ++ DN F + E C + + + ++ D V
Sbjct: 346 SVRAMTLHPKENAFASASADNTKK-FSLPKGEFCHNMLSQQKTII---NAMAVNEDGVMV 401
Query: 460 AAGSADGSVHIW---SVHKNEIVSTL-----KEHTSSVLCCRWSGIGKPLASADKNGIVC 511
G +GS+ W S H + T+ E + + + G L + + + +
Sbjct: 402 TGGD-NGSIWFWDWKSGHSFQQSETIVQPGSLESEAGIYAACYDNTGSRLVTCEADKTIK 460
Query: 512 IWK 514
+WK
Sbjct: 461 MWK 463
Score = 64.3 bits (155), Expect = 2e-10
Identities = 57/268 (21%), Positives = 104/268 (38%), Gaps = 15/268 (5%)
Query: 223 KFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITH 282
K L H + + + + G D+ VK WD V + HG + V L + H
Sbjct: 211 KLTLTGHIEQVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNKVIRSYHGHLSGVYCLAL-H 269
Query: 283 DNRSVIAASSSNNL-YVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKV 341
V+ +++ VWD+ + L+GH + VC+V ++ + VV+ ++D TIK
Sbjct: 270 PTLDVLLTGGRDSVCRVWDIRTKMQIFALSGHDNTVCSV-FTRPTDPQVVTGSHDTTIKF 328
Query: 342 WDLMKGYCTNTII-FASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLA 400
WDL G +T+ + A+ S D N + + + G+ + +
Sbjct: 329 WDLRYGKTMSTLTHHKKSVRAMTLHPKENAFASASAD-NTKKFSLPKGEFCHNMLSQQKT 387
Query: 401 VTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRS-------CIS 453
+ + V++T G + +D +S SF+ + V S C
Sbjct: 388 IINAMAVNEDGVMVTGGDNGSIWFWDWKS---GHSFQQSETIVQPGSLESEAGIYAACYD 444
Query: 454 PDNSHVAAGSADGSVHIWSVHKNEIVST 481
S + AD ++ +W +N T
Sbjct: 445 NTGSRLVTCEADKTIKMWKEDENATPET 472
>At3g15610 unknown protein
Length = 341
Score = 82.0 bits (201), Expect = 7e-16
Identities = 68/274 (24%), Positives = 128/274 (45%), Gaps = 13/274 (4%)
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG 304
LI+ +D + + TG G G+V + ++ +AS+ + +WD +G
Sbjct: 34 LISASKDSQPMLRNGETGDWIGTFEGHKGAVWSSCLDNNALRAASASADFSAKLWDALTG 93
Query: 305 RVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFAS--NCNAL 362
V H+ HK V A S+ ++++++ +++ ++V+DL + T I S + L
Sbjct: 94 DVLHSFE-HKHIVRACAFSQ-DTKYLITGGFEKILRVFDLNRLDAPPTEIDKSPGSIRTL 151
Query: 363 CFSMDGQTIYSGHVD-GNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNV 421
+ QTI S D G +RLWD++SGK++ + S VTS +S++G + T+ V
Sbjct: 152 TWLHGDQTILSSCTDIGGVRLWDVRSGKIVQTLETKS-PVTSAEVSQDGRYITTADGSTV 210
Query: 422 HNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSH-VAAGSADGSVHIWSVHKNEIVS 480
+D + S+ + N + + P + + AG D V ++ H + +
Sbjct: 211 -KFWDANHFGLVKSYD-----MPCNIESASLEPKSGNKFVAGGEDMWVRLFDFHTGKEIG 264
Query: 481 TLKEHTSSVLCCRWSGIGKPLASADKNGIVCIWK 514
K H V C R++ G+ AS ++G + IW+
Sbjct: 265 CNKGHHGPVHCVRFAPTGESYASGSEDGTIRIWQ 298
>At2g37670 putative WD-40 repeat protein
Length = 903
Score = 80.5 bits (197), Expect = 2e-15
Identities = 74/320 (23%), Positives = 139/320 (43%), Gaps = 40/320 (12%)
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHG---------CIGSVL 276
+ AHEG +I F ++ L +GG D+ + VW+ + S G C S
Sbjct: 400 IQAHEGAVWTIKFSQDAHYLASGGADRVIHVWEVQECELMSMNEGSLTPIHPSLCDSSGN 459
Query: 277 DLTITHDNRSVIAASSSNNLYVWDL---------NSGRVRHTLTGHKDKVCAVDVSKVSS 327
++T+ + +S + ++ D S + +L GH D + +D+S S
Sbjct: 460 EITVVEKKKKGKGSSGRRHNHIPDYVHVPETVFSFSDKPVCSLKGHLDAI--LDLSWSKS 517
Query: 328 RHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFS---MDGQTIYSGHVDGNLRLWD 384
+ ++S++ D+T+++WD+ C +FA N C +D SG +D +R+W
Sbjct: 518 QLLLSSSMDKTVRLWDIETKTCLK--LFAHNDYVTCIQFSPVDENYFLSGSLDAKIRIWS 575
Query: 385 IQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFD--------VRSLEVCSSF 436
IQ ++ H + VT+ + +G L + +D ++V S+
Sbjct: 576 IQDRHVVEWSDLHEM-VTAACYTPDGQGALIGSHKGICRAYDTEDCKLSQTNQIDVQSNK 634
Query: 437 RDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLK--EHTSSVLCCRW 494
+ R +++ S ++P S V SAD + I + +E++ K +T S L +
Sbjct: 635 KSQAKRKITSFQFSPVNP--SEVLVTSADSRIRI--LDGSEVIHKFKGFRNTCSQLSASY 690
Query: 495 SGIGKPLASADKNGIVCIWK 514
S GK + A ++ V +WK
Sbjct: 691 SQDGKYIICASEDSQVYLWK 710
Score = 63.9 bits (154), Expect = 2e-10
Identities = 52/241 (21%), Positives = 104/241 (42%), Gaps = 19/241 (7%)
Query: 264 VCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVS 323
VCS L G + ++LDL+ + ++ ++++S + +WD+ + + L H D V + S
Sbjct: 499 VCS-LKGHLDAILDLSWSK-SQLLLSSSMDKTVRLWDIET-KTCLKLFAHNDYVTCIQFS 555
Query: 324 KVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLW 383
V + +S + D I++W + + A C++ DGQ G G R +
Sbjct: 556 PVDENYFLSGSLDAKIRIWSIQDRHVVEWSDLHEMVTAACYTPDGQGALIGSHKGICRAY 615
Query: 384 DIQSGKL---------LSEVAAHSLAVTSISLS--RNGNVVLTSGRDNVHNLFDVRSLEV 432
D + KL ++ + +TS S V++TS + L +
Sbjct: 616 DTEDCKLSQTNQIDVQSNKKSQAKRKITSFQFSPVNPSEVLVTSADSRIRILDGSEVIHK 675
Query: 433 CSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCC 492
FR+T +++++++S+ D ++ S D V++W + STL + C
Sbjct: 676 FKGFRNTCSQLSASYSQ-----DGKYIICASEDSQVYLWKNDFHRTRSTLTTQSHEHFHC 730
Query: 493 R 493
+
Sbjct: 731 K 731
Score = 29.3 bits (64), Expect = 5.4
Identities = 13/45 (28%), Positives = 27/45 (59%)
Query: 390 LLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCS 434
+ E+ AH AV +I S++ + + + G D V ++++V+ E+ S
Sbjct: 396 MCQEIQAHEGAVWTIKFSQDAHYLASGGADRVIHVWEVQECELMS 440
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,948,869
Number of Sequences: 26719
Number of extensions: 445941
Number of successful extensions: 4567
Number of sequences better than 10.0: 507
Number of HSP's better than 10.0 without gapping: 264
Number of HSP's successfully gapped in prelim test: 246
Number of HSP's that attempted gapping in prelim test: 2025
Number of HSP's gapped (non-prelim): 1599
length of query: 514
length of database: 11,318,596
effective HSP length: 104
effective length of query: 410
effective length of database: 8,539,820
effective search space: 3501326200
effective search space used: 3501326200
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0173b.8


