

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0173b.7
(1526 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
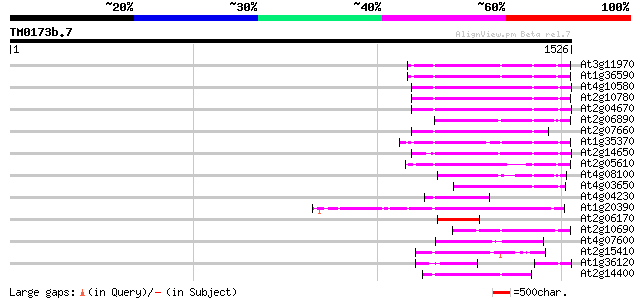
Score E
Sequences producing significant alignments: (bits) Value
At3g11970 hypothetical protein 211 2e-54
At1g36590 hypothetical protein 211 2e-54
At4g10580 putative reverse-transcriptase -like protein 210 6e-54
At2g10780 pseudogene 208 2e-53
At2g04670 putative retroelement pol polyprotein 206 6e-53
At2g06890 putative retroelement integrase 195 1e-49
At2g07660 putative retroelement pol polyprotein 188 2e-47
At1g35370 hypothetical protein 182 1e-45
At2g14650 putative retroelement pol polyprotein 182 1e-45
At2g05610 putative retroelement pol polyprotein 171 4e-42
At4g08100 putative polyprotein 157 6e-38
At4g03650 putative reverse transcriptase 144 5e-34
At4g04230 putative transposon protein 121 3e-27
At1g20390 hypothetical protein 118 3e-26
At2g06170 putative Ty3-gypsy-like retroelement pol polyprotein 110 8e-24
At2g10690 putative retroelement pol polyprotein 106 1e-22
At4g07600 104 4e-22
At2g15410 putative retroelement pol polyprotein 99 1e-20
At1g36120 putative reverse transcriptase gb|AAD22339.1 98 4e-20
At2g14400 putative retroelement pol polyprotein 92 3e-18
>At3g11970 hypothetical protein
Length = 1499
Score = 211 bits (538), Expect = 2e-54
Identities = 136/445 (30%), Positives = 224/445 (49%), Gaps = 20/445 (4%)
Query: 1082 FWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWS 1141
F E+ +H ++L + P RP + + + + DLL ++ S SP++
Sbjct: 585 FREKHNHKIKL------LEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYA 638
Query: 1142 CAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKS 1201
V K+ GT RL ++Y+ LN +PIP +DL+ L A IFSK D+++
Sbjct: 639 SPVVLVKKKD----GTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRA 694
Query: 1202 GFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYI 1260
G+ Q+++ D KTAF G +E+ VMPFGL NAP+ FQ +MN IF P KF +V+
Sbjct: 695 GYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFF 754
Query: 1261 DDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINR 1320
DD+L++S S+++H +HL +++ N + +K + K+ +LGH I I
Sbjct: 755 DDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPA 814
Query: 1321 AIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPPPWSDIHTNV 1380
I+ ++P K QL+ FLG Y F I PLH K D W+ +
Sbjct: 815 KIKAVKEWPQPTTLK-QLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTAVAQQA 873
Query: 1381 VKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQ 1440
+ +K + P L LP +VETDA G G +L Q + +A+ S+ Q
Sbjct: 874 FEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQ----EGHPLAYISRQLKGKQ 929
Query: 1441 QNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQA 1500
+ S +KE+LA++ ++ ++ L+ F+++ D +S K +L++ + + I +W
Sbjct: 930 LHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRL----NTPIQQQWLP 985
Query: 1501 ILSVFDFEIEYIKGSTNSLPDFLTR 1525
L FD+EI+Y +G N + D L+R
Sbjct: 986 KLLEFDYEIQYRQGKENVVADALSR 1010
>At1g36590 hypothetical protein
Length = 1499
Score = 211 bits (538), Expect = 2e-54
Identities = 136/445 (30%), Positives = 224/445 (49%), Gaps = 20/445 (4%)
Query: 1082 FWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWS 1141
F E+ +H ++L + P RP + + + + DLL ++ S SP++
Sbjct: 585 FREKHNHKIKL------LEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYA 638
Query: 1142 CAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKS 1201
V K+ GT RL ++Y+ LN +PIP +DL+ L A IFSK D+++
Sbjct: 639 SPVVLVKKKD----GTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRA 694
Query: 1202 GFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYI 1260
G+ Q+++ D KTAF G +E+ VMPFGL NAP+ FQ +MN IF P KF +V+
Sbjct: 695 GYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFF 754
Query: 1261 DDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINR 1320
DD+L++S S+++H +HL +++ N + +K + K+ +LGH I I
Sbjct: 755 DDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPA 814
Query: 1321 AIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPPPWSDIHTNV 1380
I+ ++P K QL+ FLG Y F I PLH K D W+ +
Sbjct: 815 KIKAVKEWPQPTTLK-QLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTAVAQQA 873
Query: 1381 VKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQ 1440
+ +K + P L LP +VETDA G G +L Q + +A+ S+ Q
Sbjct: 874 FEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQ----EGHPLAYISRQLKGKQ 929
Query: 1441 QNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQA 1500
+ S +KE+LA++ ++ ++ L+ F+++ D +S K +L++ + + I +W
Sbjct: 930 LHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRL----NTPIQQQWLP 985
Query: 1501 ILSVFDFEIEYIKGSTNSLPDFLTR 1525
L FD+EI+Y +G N + D L+R
Sbjct: 986 KLLEFDYEIQYRQGKENVVADALSR 1010
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 210 bits (534), Expect = 6e-54
Identities = 137/436 (31%), Positives = 221/436 (50%), Gaps = 15/436 (3%)
Query: 1093 PYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAE 1152
P+ + P P +M + ++++ DLL K IR S SPW +V K+
Sbjct: 480 PFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKD- 538
Query: 1153 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1212
G+ RL I+Y+ LN+ RYP+P +LL +L A FSK D+ SG+ QI + E D
Sbjct: 539 ---GSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEAD 595
Query: 1213 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSID 1271
KTAF +G +E+ VMPFGL NAP+ F R+MN +F +F I++IDD+L++S+S +
Sbjct: 596 VRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPE 655
Query: 1272 QHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQ 1331
+ HL + +++ + +K S +Q ++ FLGH + + IE +P +
Sbjct: 656 EQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWP-R 714
Query: 1332 IIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKN 1390
+ T+++ FLG Y F +++ +P+ KD P WS +K + +
Sbjct: 715 PTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTS 774
Query: 1391 LPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEV 1450
P L LP +V TDAS +G G +L Q ++IA+ S+ + NY T E+
Sbjct: 775 TPVLALPEHGQPYMVYTDASRVGLGCVLMQ----HGKVIAYASRQLMKHEGNYPTHDLEM 830
Query: 1451 LAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIE 1510
A++ ++ ++S L K V D KS K I + NL + RW +++ +D EI
Sbjct: 831 AAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQR----RWMELVADYDLEIA 886
Query: 1511 YIKGSTNSLPDFLTRE 1526
Y G N + D L+R+
Sbjct: 887 YHPGKANVVVDALSRK 902
>At2g10780 pseudogene
Length = 1611
Score = 208 bits (529), Expect = 2e-53
Identities = 132/435 (30%), Positives = 221/435 (50%), Gaps = 15/435 (3%)
Query: 1093 PYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAE 1152
P+ + P P +M + ++++ +LL K IR S SPW +V K+
Sbjct: 648 PFTIELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKD- 706
Query: 1153 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1212
G+ RL I+Y+ LN+ +YP+P +L+ +L A+ FSK D+ SG+ QI ++ D
Sbjct: 707 ---GSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTD 763
Query: 1213 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSID 1271
KTAF + +E+ VMPFGL NAP+ F ++MN +F +F I++I+D+L++S+S +
Sbjct: 764 VRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWE 823
Query: 1272 QHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQ 1331
H +HL + ++++ + +K S +Q + FLGH I + I ++P +
Sbjct: 824 AHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWP-R 882
Query: 1332 IIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKN 1390
+ T+++ FLG Y F +++ +PL KD WSD ++K + N
Sbjct: 883 PRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTN 942
Query: 1391 LPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEV 1450
P L LP V TDAS +G G +L Q K +IA+ S+ ++NY T E+
Sbjct: 943 APVLVLPEEGEPYTVYTDASIVGLGCVLMQ----KGSVIAYASRQLRKHEKNYPTHDLEM 998
Query: 1451 LAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIE 1510
A+V + ++S L K + D KS K I + NL + RW +++ ++ +I
Sbjct: 999 AAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQR----RWMELVADYNLDIA 1054
Query: 1511 YIKGSTNSLPDFLTR 1525
Y G N + D L+R
Sbjct: 1055 YHPGKANQVADALSR 1069
Score = 32.3 bits (72), Expect = 2.2
Identities = 15/40 (37%), Positives = 24/40 (59%), Gaps = 3/40 (7%)
Query: 668 SKPHSTQAAKT--PPENRPQGKDV-TCYNCGKPGHISRYC 704
SK H+ Q+ P +++ G++ TC+ CGK GH+ R C
Sbjct: 404 SKDHAIQSCPKMEPGQSKVLGEETRTCFYCGKTGHLKREC 443
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 206 bits (525), Expect = 6e-53
Identities = 135/436 (30%), Positives = 218/436 (49%), Gaps = 15/436 (3%)
Query: 1093 PYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAE 1152
P+ + P P +M + ++++ DLL K IR S SPW +V K+
Sbjct: 506 PFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKD- 564
Query: 1153 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1212
G+ RL I+Y+ LN +YP+P +LL +L A FSK D+ SG+ QI + E D
Sbjct: 565 ---GSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEAD 621
Query: 1213 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSID 1271
KTAF +G +E+ VMPF L NAP+ F R+MN +F +F I++IDD+L++S+S +
Sbjct: 622 VRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPE 681
Query: 1272 QHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQ 1331
+H HL + +++ + +K S +Q +I FLGH + + IE +P +
Sbjct: 682 EHEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWP-R 740
Query: 1332 IIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKN 1390
+ T+++ FL Y F +++ +P+ KD P WS +K + +
Sbjct: 741 PTNATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTS 800
Query: 1391 LPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEV 1450
P L LP +V TDAS +G G +L Q + ++IA+ S+ + NY T E+
Sbjct: 801 TPVLALPEHGQPYMVYTDASRVGLGCVLMQ----RGKVIAYASRQLRKHEGNYPTHDLEM 856
Query: 1451 LAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIE 1510
++ ++ ++S L K V D KS K I + NL RW +++ +D EI
Sbjct: 857 AVVIFALKIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQ----MRWMELVADYDLEIA 912
Query: 1511 YIKGSTNSLPDFLTRE 1526
Y G N + D L+R+
Sbjct: 913 YHPGKANVVADALSRK 928
>At2g06890 putative retroelement integrase
Length = 1215
Score = 195 bits (496), Expect = 1e-49
Identities = 121/374 (32%), Positives = 195/374 (51%), Gaps = 15/374 (4%)
Query: 1156 GTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYK 1215
G+ R+ + + +N +PIP D+L LH + IFSK D+KSG+ QI++ E D +K
Sbjct: 467 GSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKIDLKSGYHQIRMNEGDEWK 526
Query: 1216 TAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSIDQHF 1274
TAF G YEW VMPFGL +APS F R+MN + F IVY DD+L++S+S+ +H
Sbjct: 527 TAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFVIVYFDDILVYSESLREHI 586
Query: 1275 KHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIID 1334
+HL++ +++++K + + K + + FLG + + ++ +P
Sbjct: 587 EHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVKVDEEKVKAIRDWPS---P 643
Query: 1335 KT--QLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNL 1391
KT +++ F G + F STI+ PL + +KKD W + +K ++ N
Sbjct: 644 KTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKAQEEAFQSLKDKLTNA 703
Query: 1392 PCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVL 1451
P L L +E DAS IG G +L Q +++IAF S+ A NY T KE+
Sbjct: 704 PVLILSEFLKTFEIECDASGIGIGAVLMQ----DQKLIAFFSEKLGGATLNYPTYDKELY 759
Query: 1452 AIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEY 1511
A+V ++ +Q L + F++ D +S K + K + L +H ARW + F + I+Y
Sbjct: 760 ALVRALQRWQHYLWPKVFVIHTDHESLKHL--KGQQKLNKRH--ARWVEFIETFAYVIKY 815
Query: 1512 IKGSTNSLPDFLTR 1525
KG N + D L++
Sbjct: 816 KKGKDNVVADALSQ 829
Score = 40.8 bits (94), Expect = 0.006
Identities = 43/164 (26%), Positives = 67/164 (40%), Gaps = 29/164 (17%)
Query: 609 QQLTKEKSQNRRDLGTFCEQFGIQGCPKKPKPRKQDPPPKQQWRRKSSQNHDHRKPKPRS 668
+Q K KS +R G+ G KP ++++ R S N P+ S
Sbjct: 90 EQQVKRKSSSRSSYGS--------GTIAKPTYQREE-------RTSSYHNKPIVSPRAES 134
Query: 669 KPHST-QAAKTPPE-NRPQGKDVTCYNCGKPGHISRYCRLKRRI-----SELHLEPEIED 721
KP++ Q K E + + +DV CY C GH + C KR + E+ E EI D
Sbjct: 135 KPYAAVQDHKGKAEISTSRVRDVRCYKCQGKGHYANECPNKRVMILLDNGEIEPEEEIPD 194
Query: 722 KINNLLIQTSDEEESVPSDSE--VSEDLNQIQNDDDQSSSSINV 763
++L E E +P+ E V+ +Q D+ N+
Sbjct: 195 SPSSL-----KENEELPAQGELLVARRTLSVQTKTDEQEQRKNL 233
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 188 bits (478), Expect = 2e-47
Identities = 118/375 (31%), Positives = 195/375 (51%), Gaps = 11/375 (2%)
Query: 1093 PYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAE 1152
P+ + P P +M + ++++ +LL K IR S SPW +V K+
Sbjct: 143 PFTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKD- 201
Query: 1153 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1212
G+ RL I+Y+ LN+ +YP+P +L+ +L A+ FSK D+ SG+ QI ++ D
Sbjct: 202 ---GSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTD 258
Query: 1213 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSID 1271
KTAF +G +E+ VMPFGL NAP+ F ++MN +F +F I++IDD+L+ S+S +
Sbjct: 259 VRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWE 318
Query: 1272 QHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQ 1331
H +HL + ++++ + +K S +Q + FLGH I + I ++P +
Sbjct: 319 AHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWP-R 377
Query: 1332 IIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKN 1390
+ T+++ FLG Y F +++ +PL KD WSD ++K + N
Sbjct: 378 PRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLIN 437
Query: 1391 LPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEV 1450
P L LP V TDAS +G G +L Q K +IA+ S+ ++NY T E+
Sbjct: 438 APVLVLPEEGEPYTVYTDASIVGLGCVLMQ----KGSVIAYASRQLRKHEKNYPTHDLEM 493
Query: 1451 LAIVLSISNFQSDLI 1465
A+V ++ ++S LI
Sbjct: 494 AAVVFALKIWRSYLI 508
>At1g35370 hypothetical protein
Length = 1447
Score = 182 bits (463), Expect = 1e-45
Identities = 137/469 (29%), Positives = 224/469 (47%), Gaps = 31/469 (6%)
Query: 1060 KSRIENILENIQSSIC--FDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELL 1117
+S ++NI+E DLP F E+ H ++L + P RP +
Sbjct: 541 ESVVQNIVEEFPDVFAEPTDLP-PFREKHDHKIKL------LEGANPVNQRPYRYVVHQK 593
Query: 1118 QFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYP 1177
+ + D+++ I+ S SP++ V K+ GT RL ++Y LN R+
Sbjct: 594 DEIDKIVQDMIKSGTIQVSSSPFASPVVLVKKKD----GTWRLCVDYTELNGMTVKDRFL 649
Query: 1178 IPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNA 1237
IP +DL+ L + +FSK D+++G+ Q+++ D KTAF G +E+ VM FGL NA
Sbjct: 650 IPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNA 709
Query: 1238 PSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKV 1296
P+ FQ +MN +F KF +V+ DD+LI+S SI++H +HL +++ + + +K
Sbjct: 710 PATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLHKLFAKGSK- 768
Query: 1297 SLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLS 1356
LGH I I I+ ++P K Q++ FLG Y F
Sbjct: 769 -------EHLGHFISAREIETDPAKIQAVKEWPTPTTVK-QVRGFLGFAGYYRRFVRNFG 820
Query: 1357 TIIKPLHDRLKKDPPPWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGG 1416
I PLH K D WS + +K + N P L LP +VETDA G
Sbjct: 821 VIAGPLHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRA 880
Query: 1417 ILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCK 1476
+L Q K +A+ S+ Q + S +KE+LA + ++ ++ L+ F+++ D +
Sbjct: 881 VLMQ----KGHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQR 936
Query: 1477 SAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTR 1525
S K +L++ + + +W L FD+EI+Y +G N + D L+R
Sbjct: 937 SLKYLLEQRLNTPVQQ----QWLPKLLEFDYEIQYRQGKENLVADALSR 981
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 182 bits (462), Expect = 1e-45
Identities = 125/437 (28%), Positives = 214/437 (48%), Gaps = 29/437 (6%)
Query: 1093 PYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAE 1152
P+ + P P +M + ++++ DLL ++ +V K+
Sbjct: 467 PFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAPVL------------FVKKKD- 513
Query: 1153 IERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKD 1212
G+ RL I+Y+ LN +YP+P +LL +L A FSK D+ SG+ I + E D
Sbjct: 514 ---GSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEAD 570
Query: 1213 RYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSID 1271
KTAF +G +E+ VMPFGL NAP+ F R+MN +F +F I++IDD+L++S+S++
Sbjct: 571 VRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLE 630
Query: 1272 QHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIH-QGTIIPINRAIEFTDKFPD 1330
+H HL + +++ + +K S +Q ++ FLGH + +G + + D
Sbjct: 631 EHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTP 690
Query: 1331 QIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIK 1389
+ T+++ FLG Y F +++ +P+ KD P WS +K +
Sbjct: 691 --TNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLT 748
Query: 1390 NLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKE 1449
+ P L LP +V TDAS +G G +L Q + ++IA+ S+ + NY T E
Sbjct: 749 STPVLALPEHGEPYMVYTDASGVGLGCVLMQ----RGKVIAYASRQLRKHEGNYPTHDLE 804
Query: 1450 VLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEI 1509
+ A++ ++ ++S L V D KS K I + NL + +W +++ +D EI
Sbjct: 805 MAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQR----QWMELVADYDLEI 860
Query: 1510 EYIKGSTNSLPDFLTRE 1526
Y G N + D L+ +
Sbjct: 861 AYHPGKANVVADALSHK 877
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 171 bits (432), Expect = 4e-42
Identities = 125/450 (27%), Positives = 207/450 (45%), Gaps = 72/450 (16%)
Query: 1077 DLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRS 1136
DLP F H +EL + + P RP + + ++D+L I+ S
Sbjct: 23 DLP-PFRAPHDHKIELLEDSN------PVNQRPYRYVVHQKNEIDKIVDDMLASGTIQAS 75
Query: 1137 KSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSK 1196
SP++ V K+ GT RL ++Y+ LN R+PIP +DL+ L + ++SK
Sbjct: 76 SSPYASPVVLVKKKD----GTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNVYSK 131
Query: 1197 FDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKF 1255
D+++G+ Q+++ D +KTAF G YE+ VMPFGL NAP+ FQ +MN F P KF
Sbjct: 132 IDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFLRKF 191
Query: 1256 AIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTI 1315
+V+ DD+LI+S S+++H KHL +++ + + +K + ++ +LGH I I
Sbjct: 192 VLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISGEGI 251
Query: 1316 IPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPPPWSD 1375
I+ +P ++ QL FLG Y F
Sbjct: 252 ATDPAKIKAVQDWPVP-VNLKQLCGFLGLTGYYRRFF----------------------- 287
Query: 1376 IHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKH 1435
+VE DA G G +L Q + +AF S+
Sbjct: 288 ----------------------------VVEMDACGHGIGAVLMQ----EGHPLAFISRQ 315
Query: 1436 WNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIF 1495
Q + S +KE+LA++ + ++ LI+ F+++ D +S K +L++ + + I
Sbjct: 316 LKGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRL----NTPIQ 371
Query: 1496 ARWQAILSVFDFEIEYIKGSTNSLPDFLTR 1525
+W L FD+EI+Y +G N + D L+R
Sbjct: 372 QQWLPKLLEFDYEIQYKQGKENLVADALSR 401
>At4g08100 putative polyprotein
Length = 1054
Score = 157 bits (396), Expect = 6e-38
Identities = 103/350 (29%), Positives = 166/350 (47%), Gaps = 39/350 (11%)
Query: 1165 KPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQ 1224
+ +N R+PIP D L +LH + IFSK D+KSG+ Q +++E D +KTA
Sbjct: 529 RAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQRL 588
Query: 1225 YEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISI 1283
YEW VMPFGL NAP+ F R+MN + F IVY DD+L++S++++ H HL + +
Sbjct: 589 YEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVLDL 648
Query: 1284 IKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLG 1343
++K + + K + + FLG + I ++ ++P+ K ++ +G
Sbjct: 649 LRKEKLYANLKKCTFCTDNLVFLGFVVSADGIKVDEEKVKAIREWPN---PKNVSEKDIG 705
Query: 1344 CLNYVADFCPQLSTIIKPLHDRLKKDPPPWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFK 1403
W D N + +K ++ N LPN
Sbjct: 706 F---------------------------KWEDAQENAFQALKEKLTNSSVPILPNFMKSF 738
Query: 1404 IVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSD 1463
+E DAS +G G +L Q + IA+ S+ A NY T KE+ A+V ++ +Q
Sbjct: 739 EIECDASGLGIGAVLMQ----DHKPIAYFSEKLGGATLNYPTYDKELYALVRALQTWQHY 794
Query: 1464 LINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIK 1513
L +KF++ D +S K + K + L +H ARW + F + I+Y K
Sbjct: 795 LWPKKFVIHTDHESLKHL--KGQQKLNKRH--ARWVEFIETFPYVIKYKK 840
>At4g03650 putative reverse transcriptase
Length = 839
Score = 144 bits (362), Expect = 5e-34
Identities = 95/306 (31%), Positives = 156/306 (50%), Gaps = 11/306 (3%)
Query: 1208 LQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIF 1266
L E D KTAF +G +E+ VMPFGL NAP+ F R+MN +F +F I++IDD+L++
Sbjct: 516 LAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVY 575
Query: 1267 SQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTD 1326
S+S ++H HL + +++ + +K S +Q ++ FLGH + + IE
Sbjct: 576 SKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIR 635
Query: 1327 KFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIK 1385
+P + + T+++ FLG Y F +++ +P+ KD P WS +K
Sbjct: 636 DWP-RPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLK 694
Query: 1386 LRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYST 1445
+ + P L LP +V TDAS +G G L Q + ++IA+ S+ + NY T
Sbjct: 695 EMLTSTPVLALPEHGEPYMVYTDASGVGLGCALMQ----RGKVIAYASRQLRKHEGNYPT 750
Query: 1446 VKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVF 1505
E+ A++ ++ ++S L K V D KS K I + NL + RW +++ +
Sbjct: 751 HDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQR----RWMELVADY 806
Query: 1506 DFEIEY 1511
D EI Y
Sbjct: 807 DLEIAY 812
>At4g04230 putative transposon protein
Length = 315
Score = 121 bits (304), Expect = 3e-27
Identities = 67/179 (37%), Positives = 100/179 (55%), Gaps = 5/179 (2%)
Query: 1128 LQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLAR 1187
+ K IR S SP + V K+ GT R+ ++ + +N R+PIP D+L
Sbjct: 125 IHKGYIRESLSPCAVPVLLVPKKD----GTWRMCLDCRAINNITIKYRHPIPRLYDMLDE 180
Query: 1188 LHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNE 1247
L A IFSK D++SG+ Q++++E D +KTAF G YE VMPFGL NAPS F R+MN+
Sbjct: 181 LSGAIIFSKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQ 240
Query: 1248 IFNP-*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRF 1305
+ KF +VY DD+LI+++S H + L + ++K G+ + K + K F
Sbjct: 241 VLRSFIGKFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTFCSDKFFF 299
>At1g20390 hypothetical protein
Length = 1791
Score = 118 bits (295), Expect = 3e-26
Identities = 152/708 (21%), Positives = 297/708 (41%), Gaps = 56/708 (7%)
Query: 824 KPVTIQDLQSEVHILKA-------EVKSLKQIQTSQQLILEKLTEENSNGGSSSSSSSTS 876
+PV ++ +VH++ V+S+KQ + ++++ SS+S++ +
Sbjct: 500 EPVVVR----QVHVIMGGLQNCSDSVRSIKQYRKKAEMVV---------AWPSSTSTTRN 546
Query: 877 TSNRAPNNNVGDFLEIINYVTIQKFYINITIIIGDFILETPALFDTGADSSCISEGL--- 933
+ AP + LE ++ T + + +II D + T L DTG+ I + +
Sbjct: 547 PNQSAPISFTDVDLEGLD--TPHNDPLVVELIISDSRV-TRVLIDTGSSVDLIFKDVLTA 603
Query: 934 --IPTRYFEKTTEKLSGAEGSKLIIKYKIPSAIIKNDSLEIETSFLLVRNLTHKVIIETP 991
I R + ++ L+G +G ++ I I + ++ + + VI+ TP
Sbjct: 604 MNITDRQIKPVSKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTP 663
Query: 992 FIKKL--FPYNTDEKGITVQHHGQPIVFKFSKPPFVKTLNIISYKEKQINF-----LKEE 1044
+I ++ P + H+G +F P KT + SY+E ++ + E
Sbjct: 664 WIHQMQAIPSTYHQCVKFPTHNG---IFTLRAPKEAKTPSR-SYEESELCRTEMVNIDES 719
Query: 1045 ISYKNIEVQLQ-QPSVKSRIENILENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQI 1103
+ + V + PS++ + +L+ + + + + + +E +
Sbjct: 720 DPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDM---KGIDPAITAHELNVDPTFK 776
Query: 1104 PTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSP-WSCAAFYVNKQAEIERGTPRLVI 1162
P K + ++ E + E+ LL+ I K P W V K+ G R+ +
Sbjct: 777 PVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKK----NGKWRVCV 832
Query: 1163 NYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPF 1222
+Y LN+A YP+P+ L+ + S D SG+ QI + + D+ KT+F
Sbjct: 833 DYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDR 892
Query: 1223 GQYEWNVMPFGLKNAPSEFQRIMNEIF-NP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFI 1281
G Y + VM FGLKNA + +QR +N++ + + VYIDD+L+ S + H +HL+
Sbjct: 893 GTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCF 952
Query: 1282 SIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRF 1341
++ GM ++ TK + T FLG+ + + I + I + P + ++QR
Sbjct: 953 DVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSP-RNAREVQRL 1011
Query: 1342 LGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQ 1400
G + + F + + P ++ LK+ W +++K + P L P
Sbjct: 1012 TGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVG 1071
Query: 1401 AFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNF 1460
+ SD +L ++ +++ I +TSK A+ Y ++K LA+V S
Sbjct: 1072 ETLYLYIAVSDHAVSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKL 1131
Query: 1461 QSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFE 1508
+ + V D L+ + + + +W LS +D +
Sbjct: 1132 RPYFQSHTIAVLTD-----QPLRVALHSPSQSGRMTKWAVELSEYDID 1174
>At2g06170 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 587
Score = 110 bits (274), Expect = 8e-24
Identities = 50/113 (44%), Positives = 76/113 (67%), Gaps = 1/113 (0%)
Query: 1165 KPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQ 1224
K +N+ R+PIP D+L L +K+FSK D++SG+ QI+++ D +KT F G
Sbjct: 401 KAINKITIKYRFPIPQLDDMLDELSGSKVFSKIDLRSGYHQIRIRPGDEWKTDFKSKDGL 460
Query: 1225 YEWNVMPFGLKNAPSEFQRIMNEIFNP*S-KFAIVYIDDVLIFSQSIDQHFKH 1276
YEW VMPFG+ NAPS F R+MN+I P + F +VY DD+LI+S++ +++ +H
Sbjct: 461 YEWQVMPFGMSNAPSTFMRLMNQILRPFTGSFVVVYFDDILIYSKTKEEYLEH 513
>At2g10690 putative retroelement pol polyprotein
Length = 622
Score = 106 bits (264), Expect = 1e-22
Identities = 95/328 (28%), Positives = 151/328 (45%), Gaps = 19/328 (5%)
Query: 1204 WQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP*-SKFAIVYIDD 1262
+++Q + KT FT P G + + M FGL NAP FQR M IF+ + V++DD
Sbjct: 154 FKLQFTLMIKKKTTFTCPNGTFAYKRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMDD 213
Query: 1263 VLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRA- 1321
FS + +L + ++ + ++ K + LGH I G I +++A
Sbjct: 214 FSGFSSCL----LNLCRVLERCEETNLVLNWEKCHFMVHEGIVLGHKI-SGKGIEVDKAK 268
Query: 1322 --IEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHT 1378
+ + P + D ++ FLG + F S I +PL L K+ + +
Sbjct: 269 IDVMIQLQPPKTVKD---IRSFLGHAEFYRRFIKDFSKIARPLTRLLCKETEFNFDEDCL 325
Query: 1379 NVVKQIKLRIKNLPCLYLPN-PQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWN 1437
IK + + P PN F+I+ DA D G +L QKI DK +I + S+ +
Sbjct: 326 KAFHLIKETLVSAPIFQAPNWDHPFEIM-CDAFDYAVGAVLDQKIDDKLHVIYYASRTMD 384
Query: 1438 PAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFAR 1497
AQ Y+T++KE+LA+V + +S L+ K V +D + + I K +K R
Sbjct: 385 EAQTRYATIEKELLAVVFAFEKIRSYLVGFKVKVYIDHAALRHIYAKK----ETKSRLLR 440
Query: 1498 WQAILSVFDFEIEYIKGSTNSLPDFLTR 1525
W +L FD EI KG N + D L+R
Sbjct: 441 WILLLQEFDMEIIDKKGVENGVADHLSR 468
>At4g07600
Length = 630
Score = 104 bits (259), Expect = 4e-22
Identities = 83/294 (28%), Positives = 132/294 (44%), Gaps = 21/294 (7%)
Query: 1159 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1218
R+ I+Y+ LN A +P+P +L RL + + D SGF+QI + D KT F
Sbjct: 356 RMCIDYRNLNAASRNDHFPLPFTDQMLERLANHPYYCFLDGYSGFFQIPIHPNDHEKTTF 415
Query: 1219 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSIDQHFKHL 1277
T P+G + + MPFGL NAP+ FQR M IF+ + V++DD ++ S +L
Sbjct: 416 TCPYGTFAYERMPFGLCNAPATFQRCMTSIFSDIIEEMVEVFMDDFSVYGPSFSSCLLNL 475
Query: 1278 NTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQ 1337
++ ++ + ++ K + LGH I + + IE +DK
Sbjct: 476 GRVLTRCEETNLVLNWEKCHFMVKEGIMLGHKISE-------KGIE---------VDK-- 517
Query: 1338 LQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYL 1396
FLG + F S I +PL L K+ + + IK + + P +
Sbjct: 518 -GCFLGHAGFYRRFIKDFSKIARPLTRLLCKETEFEFDEDCPKSFHTIKEALVSAPVVRA 576
Query: 1397 PNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEV 1450
N + DA D G +L QKI K +I + S+ + AQ Y+T +KE+
Sbjct: 577 TNWDYPFEIMCDALDYAVGAVLGQKIDKKLHVIYYASRTLDEAQGRYATTEKEL 630
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 99.4 bits (246), Expect = 1e-20
Identities = 92/362 (25%), Positives = 161/362 (44%), Gaps = 40/362 (11%)
Query: 1104 PTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSP-WSCAAFYVNKQAEIERGTPRLVI 1162
P K + ++ + + E+ LL I + P W V K+ G R+ I
Sbjct: 782 PLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKK----NGKWRVCI 837
Query: 1163 NYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPF 1222
++ LN+A +P+P+ L+ ++ S D SG+ QI + + DR KT F
Sbjct: 838 DFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQ 897
Query: 1223 GQYEWNVMPFGLKNAPSEFQRIMNEIFNP*SKFAI-VYIDDVLIFSQSIDQHFKHLNTFI 1281
G Y + VMPFGLKNA + + R++N++F ++ VYIDD+L+ S ++H HL
Sbjct: 898 GTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCF 957
Query: 1282 SIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIID------K 1335
++ + M ++ +K + T FLG+ + R IE K IID
Sbjct: 958 QVLNRYNMKLNPSKCTFGVTSGEFLGY-------LVTRRGIEANPKQISAIIDLPSPRNT 1010
Query: 1336 TQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCL 1394
++QR +G + + F + + P + L+ + W + + + L
Sbjct: 1011 REVQRLIGRIAALNRFISRSTDKCLPFYQLLRANKRFEWDE-----------KCEEGETL 1059
Query: 1395 YLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIV 1454
YL + V T A G+L ++ ++ I + SK + A+ Y T++K A+V
Sbjct: 1060 YL-----YIAVSTSA----VSGVLVREDRGEQHPIFYVSKTLDGAELRYPTLEKLAFAVV 1110
Query: 1455 LS 1456
+S
Sbjct: 1111 IS 1112
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 97.8 bits (242), Expect = 4e-20
Identities = 58/171 (33%), Positives = 87/171 (49%), Gaps = 23/171 (13%)
Query: 1104 PTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVIN 1163
P P +M + ++++ DLL K IR S S W G P L
Sbjct: 535 PLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRW---------------GAPGL--- 576
Query: 1164 YKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFG 1223
N+ +YP+P +LL +L A FSK D+ G+ Q + E D KTAF +G
Sbjct: 577 ----NRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPIAEADVRKTAFRTRYG 632
Query: 1224 QYEWNVMPFGLKNAPSEFQRIMNEIFNP-*SKFAIVYIDDVLIFSQSIDQH 1273
+E+ VMPFGL NAP+ R+MN +F +F I++IDD+L++ +S ++H
Sbjct: 633 HFEFVVMPFGLTNAPTALMRLMNSVFQEFLDEFVIIFIDDILVYFKSPEEH 683
Score = 53.1 bits (126), Expect = 1e-06
Identities = 32/100 (32%), Positives = 51/100 (51%), Gaps = 4/100 (4%)
Query: 1427 QIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDV 1486
++I + S+ + NY T E+ AI+ ++ + S L K V D KS KDI +
Sbjct: 723 KVIVYASRQLRKHEGNYPTHDLEMAAIIFALKIWGSYLYGGKVQVFTDHKSLKDIFTQPE 782
Query: 1487 KNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
NL + RW +++ +D EI Y G TN + D L+R+
Sbjct: 783 LNLRQR----RWMELVADYDLEIAYHPGKTNVVADALSRK 818
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 91.7 bits (226), Expect = 3e-18
Identities = 80/299 (26%), Positives = 135/299 (44%), Gaps = 8/299 (2%)
Query: 1123 EINDLLQKKLIRRSKSP-WSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 1181
E++ LL+ IR + P W V K+ G R+ I++ LN+A +P+P+
Sbjct: 483 EVDKLLKIGSIREVQYPDWLANTVVVKKK----NGKDRVCIDFTDLNKACPKDSFPLPHI 538
Query: 1182 KDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEF 1241
L+ ++ S D SG+ QI + +D+ KT F G Y + VMPFGL+NA + +
Sbjct: 539 DRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNAGATY 598
Query: 1242 QRIMNEIFNP-*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQ 1300
R++N++F+ K VYIDD+LI S + H KHL +I+ + M ++ K +
Sbjct: 599 PRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAKCTFGV 658
Query: 1301 TKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIK 1360
FLG+ + + I I P K ++QR G + + F + +
Sbjct: 659 PSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFK-EVQRLTGRIAALNRFISRSTDKSL 717
Query: 1361 PLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGIL 1418
P + LK + W + Q+K + + P L+ P + S+ G+L
Sbjct: 718 PFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSNHAVSGVL 776
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,787,780
Number of Sequences: 26719
Number of extensions: 1675502
Number of successful extensions: 8297
Number of sequences better than 10.0: 159
Number of HSP's better than 10.0 without gapping: 82
Number of HSP's successfully gapped in prelim test: 78
Number of HSP's that attempted gapping in prelim test: 7812
Number of HSP's gapped (non-prelim): 433
length of query: 1526
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1414
effective length of database: 8,326,068
effective search space: 11773060152
effective search space used: 11773060152
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0173b.7


