

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0172.11
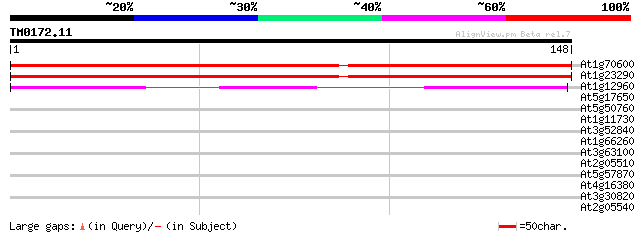
(148 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g70600 60S ribosomal protein L27A 260 2e-70
At1g23290 60s ribosomal protein l27a. 254 1e-68
At1g12960 60S ribosomal protein L27a, putative 127 2e-30
At5g17650 glycine/proline-rich protein 35 0.011
At5g50760 putative protein 28 2.3
At1g11730 Avr9 elicitor response-like protein 27 3.1
At3g52840 beta-galactosidase precursor - like protein 27 4.0
At1g66260 RNA and export factor binding protein, putative 27 4.0
At3g63100 putative protein 27 5.2
At2g05510 putative glycine-rich protein 27 5.2
At5g57870 eukaryotic initiation factor 4, eIF4-like protein 26 6.8
At4g16380 unknown protein 26 6.8
At3g30820 hypothetical protein 26 8.9
At2g05540 putative glycine-rich protein 26 8.9
>At1g70600 60S ribosomal protein L27A
Length = 146
Score = 260 bits (665), Expect = 2e-70
Identities = 123/148 (83%), Positives = 134/148 (90%), Gaps = 2/148 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKLRN+FFCP VN+DKLWSL+P++VK A + K P+IDVT +G+FKVLGKG LP N+
Sbjct: 61 FHKLRNKFFCPIVNLDKLWSLVPEDVK--AKSTKDNVPLIDVTQHGFFKVLGKGHLPENK 118
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P VVKAKLISK AEKKIKE GGAV+LTA
Sbjct: 119 PFVVKAKLISKTAEKKIKEAGGAVVLTA 146
>At1g23290 60s ribosomal protein l27a.
Length = 146
Score = 254 bits (649), Expect = 1e-68
Identities = 120/148 (81%), Positives = 131/148 (88%), Gaps = 2/148 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M T KKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MATALKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKLRN+FFCP VN+DKLWSL+P++VK A + K P+IDVT +G+FKVLGKG LP N+
Sbjct: 61 FHKLRNKFFCPIVNLDKLWSLVPEDVK--AKSSKDNVPLIDVTQHGFFKVLGKGHLPENK 118
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P VVKAKLISK AEKKIKE GGAV+LTA
Sbjct: 119 PFVVKAKLISKTAEKKIKEAGGAVVLTA 146
>At1g12960 60S ribosomal protein L27a, putative
Length = 104
Score = 127 bits (320), Expect = 2e-30
Identities = 74/147 (50%), Positives = 81/147 (54%), Gaps = 47/147 (31%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTT KK R R HVS GHGR GKHRK PG RGNAG VGMRY
Sbjct: 1 MTTSRKKTRNLREHVSVGHGRFGKHRKLPGSRGNAG-------------------VGMRY 41
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKLRN+F+C VN+DKLWS+ VLGKG LP N+
Sbjct: 42 FHKLRNKFYCQIVNLDKLWSM----------------------------VLGKGFLPENK 73
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLT 147
PVVVKAKL+S EKKIKE G AV+LT
Sbjct: 74 PVVVKAKLVSNTDEKKIKEAGSAVVLT 100
>At5g17650 glycine/proline-rich protein
Length = 173
Score = 35.4 bits (80), Expect = 0.011
Identities = 22/56 (39%), Positives = 27/56 (47%), Gaps = 5/56 (8%)
Query: 14 HVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRYFHKLRNQFF 69
H+S HG G H H G G G H + K+ G FGK GM F K + +FF
Sbjct: 119 HMSHHHGHYGHHHGHGYGYGYHG---HGKFKHGKFKHGKFGKHGM--FGKHKGKFF 169
>At5g50760 putative protein
Length = 183
Score = 27.7 bits (60), Expect = 2.3
Identities = 16/55 (29%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query: 86 VKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQPVVVKAKLISKIAEKKIKEN 140
++D +N+ P + ++G+F V V P Q +VVK KL++ K + E+
Sbjct: 38 LEDAKSNESKGKPKKESPSHGFFTVY---VGPTKQRIVVKTKLLNHPLFKNLLED 89
>At1g11730 Avr9 elicitor response-like protein
Length = 384
Score = 27.3 bits (59), Expect = 3.1
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Query: 47 KYH-PGY--FGKVGMRYFHKLRNQFFCPTVNIDKLWSLLPQEVKDQAAND 93
KYH P Y FG+VG +YF QF+ + ++ + L+ Q++ + AN+
Sbjct: 260 KYHEPEYWKFGEVGNKYFRHATGQFYAISKDL-ATYILINQDLLHKYANE 308
>At3g52840 beta-galactosidase precursor - like protein
Length = 727
Score = 26.9 bits (58), Expect = 4.0
Identities = 16/59 (27%), Positives = 26/59 (43%)
Query: 8 NRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRYFHKLRN 66
N +G V IG+H RGN G ++ I +K + G+ R++H R+
Sbjct: 639 NTMGKGQVWVNGHNIGRHWPAYTARGNCGRCNYAGIYNEKKCLSHCGEPSQRWYHVPRS 697
>At1g66260 RNA and export factor binding protein, putative
Length = 295
Score = 26.9 bits (58), Expect = 4.0
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 1/35 (2%)
Query: 4 RFKKNRKKRGHVSAGHGRI-GKHRKHPGGRGNAGG 37
R + ++G V+AG G G+ R + GGRGN G
Sbjct: 231 RLPLQQNQQGGVTAGRGGFRGRGRGNGGGRGNKSG 265
>At3g63100 putative protein
Length = 199
Score = 26.6 bits (57), Expect = 5.2
Identities = 15/36 (41%), Positives = 18/36 (49%), Gaps = 5/36 (13%)
Query: 12 RGHVSA---GHGRIGKHRKHPGGRGN--AGGMHHHR 42
RGH GH R G+ +H RG+ G HHHR
Sbjct: 96 RGHGHGHGHGHRRHGRDHRHGRDRGHHRGHGHHHHR 131
>At2g05510 putative glycine-rich protein
Length = 127
Score = 26.6 bits (57), Expect = 5.2
Identities = 12/28 (42%), Positives = 12/28 (42%)
Query: 13 GHVSAGHGRIGKHRKHPGGRGNAGGMHH 40
GH G G G H GG GG HH
Sbjct: 83 GHYGGGGGHYGGGGGHGGGGHYGGGGHH 110
>At5g57870 eukaryotic initiation factor 4, eIF4-like protein
Length = 780
Score = 26.2 bits (56), Expect = 6.8
Identities = 13/28 (46%), Positives = 18/28 (63%), Gaps = 1/28 (3%)
Query: 11 KRGHVSAGHGRIGKHRKHPGGRGNAGGM 38
+RG VS+G G + +PGGR AGG+
Sbjct: 476 RRGMVSSG-GPVSPGPVYPGGRPGAGGL 502
>At4g16380 unknown protein
Length = 192
Score = 26.2 bits (56), Expect = 6.8
Identities = 17/51 (33%), Positives = 25/51 (48%), Gaps = 5/51 (9%)
Query: 54 GKVGMRYFHKLRNQFFCPTV--NIDKLWSLLPQEVKDQAANDKSKAPVIDV 102
GKV M KL+ CP + K+ P E++DQ ++KS +I V
Sbjct: 34 GKVTMM---KLKVDLDCPKCYKKVKKVLCKFPSEIRDQLFDEKSNIVIIKV 81
>At3g30820 hypothetical protein
Length = 405
Score = 25.8 bits (55), Expect = 8.9
Identities = 11/27 (40%), Positives = 17/27 (62%), Gaps = 1/27 (3%)
Query: 16 SAGHGRIGKHRKHPGGRGNAGGMHHHR 42
S GH R+G+ R+ PG + ++ HHR
Sbjct: 380 SLGH-RLGRSRRQPGEQSSSESPTHHR 405
>At2g05540 putative glycine-rich protein
Length = 135
Score = 25.8 bits (55), Expect = 8.9
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 5/47 (10%)
Query: 13 GHVSAGHG-RIGKHRKHPGGRGNAGGMHHHR----ILFDKYHPGYFG 54
G+ G+G R G +R GG GN GG + +R + + GY+G
Sbjct: 65 GNPGGGYGNRGGGYRNRDGGYGNRGGGYGNRGGGYCRYGCCYRGYYG 111
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,793,927
Number of Sequences: 26719
Number of extensions: 167603
Number of successful extensions: 445
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 424
Number of HSP's gapped (non-prelim): 21
length of query: 148
length of database: 11,318,596
effective HSP length: 90
effective length of query: 58
effective length of database: 8,913,886
effective search space: 517005388
effective search space used: 517005388
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0172.11


