

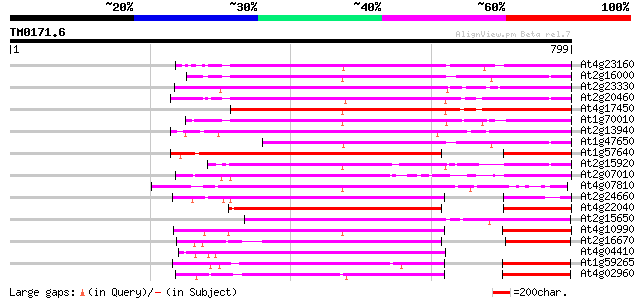
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0171.6
(799 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g23160 putative protein 421 e-118
At2g16000 putative retroelement pol polyprotein 396 e-110
At2g23330 putative retroelement pol polyprotein 392 e-109
At2g20460 putative retroelement pol polyprotein 390 e-108
At4g17450 retrotransposon like protein 387 e-107
At1g70010 hypothetical protein 385 e-107
At2g13940 putative retroelement pol polyprotein 375 e-104
At1g47650 hypothetical protein 342 6e-94
At1g57640 335 6e-92
At2g15920 putative retroelement pol polyprotein 334 1e-91
At2g07010 putative retroelement pol polyprotein 310 3e-84
At4g07810 putative polyprotein 304 1e-82
At2g24660 putative retroelement pol polyprotein 303 3e-82
At4g22040 LTR retrotransposon like protein 301 9e-82
At2g15650 putative retroelement pol polyprotein 290 2e-78
At4g10990 putative retrotransposon polyprotein 281 8e-76
At2g16670 putative retroelement pol polyprotein 260 2e-69
At4g04410 putative polyprotein 254 1e-67
At1g59265 polyprotein, putative 253 2e-67
At4g02960 putative polyprotein of LTR transposon 252 7e-67
>At4g23160 putative protein
Length = 1240
Score = 421 bits (1083), Expect = e-118
Identities = 246/584 (42%), Positives = 343/584 (58%), Gaps = 63/584 (10%)
Query: 237 ASSSNSLDDLSVSIPSADATSVQVPLPSPESPSTTSVSDHTRRPTRPRHQPSHLRNYVLH 296
AS+S+S D+ +PSA+ +Q +P P S HT R +P++L++Y H
Sbjct: 7 ASTSSSSIDI---MPSAN---IQNDVPEP--------SVHTSH--RRTRKPAYLQDYYCH 50
Query: 297 TVSSSCKASQTSSGIKYPISNYMSYSNLSIPHHAYAMSLSLDSEPHTYAEASKHKCWVDA 356
+V+S + IS ++SY +S +H++ + ++ EP TY EA + W A
Sbjct: 51 SVASLTI---------HDISQFLSYEKVSPLYHSFLVCIAKAKEPSTYNEAKEFLVWCGA 101
Query: 357 MNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEGI 416
M+ EI A+E TW + LPPN PI KWVYKIK ++GT+ERYKARLVA GY Q EGI
Sbjct: 102 MDDEIGAMETTHTWEICTLPPNKKPIGCKWVYKIKYNSDGTIERYKARLVAKGYTQQEGI 161
Query: 417 YYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVSC--- 473
+ +TFSP KLT V+++LA++++ N+ LHQLD++NAFL GDL E++YMK+P G +
Sbjct: 162 DFIETFSPVCKLTSVKLILAISAIYNFTLHQLDISNAFLNGDLDEEIYMKLPPGYAARQG 221
Query: 474 --VDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILL 531
+ VC L KS YGLKQASRQW+ KF+ L+ G+ Q+HSDH+ F K F +L
Sbjct: 222 DSLPPNAVCYLKKSIYGLKQASRQWFLKFSVTLIGFGFVQSHSDHTYFLKITATLFLCVL 281
Query: 532 IYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQRKYCLD 591
+YVDDII+ N +K+ L + FK++DLG LKYFLGLE++ SA GI++CQRKY LD
Sbjct: 282 VYVDDIIICSNNDAAVDELKSQLKSCFKLRDLGPLKYFLGLEIARSAAGINICQRKYALD 341
Query: 592 LVHDSGVLGSKPVSTPLDPSSRLSQDGGGATL*GCFFIQKTHRKTV-LSYYNQV*YNLCS 650
L+ ++G+LG KP S P+DPS S GG F K +R+ + Y Q+ S
Sbjct: 342 LLDETGLLGCKPSSVPMDPSVTFSAHSGGD-----FVDAKAYRRLIGRLMYLQITRLDIS 396
Query: 651 SAAKSVPLQSYCDSLCCSS*NSKV---------------SERESKKRVIFPKEFCSTIVG 695
A + S L K+ S+ E + +V F S
Sbjct: 397 FAVNKLSQFSEAPRLAHQQAVMKILHYIKGTVGQGLFYSSQAEMQLQVFSDASFQS---- 452
Query: 696 V**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RL 755
C DTRRS YC F+G SLI W+SKKQQ +SKSS+EA YRAL+ AT E+ L
Sbjct: 453 --------CKDTRRSTNGYCMFLGTSLISWKSKKQQVVSKSSAEAEYRALSFATDEMMWL 504
Query: 756 TYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
++LQ+ KP++++CDN +A+HIA N VFHERTKH+E +C
Sbjct: 505 AQFFRELQLPLSKPTLLFCDNTAAIHIATNAVFHERTKHIESDC 548
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 396 bits (1018), Expect = e-110
Identities = 226/564 (40%), Positives = 323/564 (57%), Gaps = 45/564 (7%)
Query: 253 ADATSVQVPLPSPESPSTTSVSDHTRRPTRPRHQPSHLRNYVLHTVSSSCKASQTSSGIK 312
+D T LPS S +S R R P+HL +Y +T+ S K
Sbjct: 866 SDTTHSPSSLPSQISDLPPQISSQ-----RVRKPPAHLNDYHCNTMQSDHK--------- 911
Query: 313 YPISNYMSYSNLSIPHHAYAMSLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSL 372
YPIS+ +SYS +S H Y +++ P YAEA K W +A++ EI A+E TW +
Sbjct: 912 YPISSTISYSKISPSHMCYINNITKIPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEI 971
Query: 373 VPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVR 432
LP + KWV+ +K A+G +ERYKARLVA GY Q EG+ Y DTFSP AK+T ++
Sbjct: 972 TTLPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIK 1031
Query: 433 MVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVS-----CVDSGKVCKLHKSSY 487
++L +++ W L QLDV+NAFL G+L E+++MK+PEG + + S V +L +S Y
Sbjct: 1032 LLLKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIY 1091
Query: 488 GLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEF 547
GLKQASRQW+ KF+S L++ G+K+ H DH+LF K F I+L+YVDDI++A
Sbjct: 1092 GLKQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDGEFVIVLVYVDDIVIASTSEAAA 1151
Query: 548 TRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTP 607
++ LD FK++DLG LKYFLGLEV+ + GIS+CQRKY L+L+ +G+L KPVS P
Sbjct: 1152 AQLTEELDQRFKLRDLGDLKYFLGLEVARTTAGISICQRKYALELLQSTGMLACKPVSVP 1211
Query: 608 LDPSSRLSQDGGGATL*GCFFIQKTHRKTVLSYYNQV*YNLCSSAAKSVPLQSYCDSLCC 667
+ P+ ++ +D G + Y ++ L + + LC
Sbjct: 1212 MIPNLKMRKDDGDLI-------------EDIEQYRRIVGKLMYLTITRPDITFAVNKLCQ 1258
Query: 668 SS*NSKVSERESKKRVI------------FPKEFCSTIVGV**CRLGGCVDTRRSVTSYC 715
S + + + RV+ + T+ G C D+RRS TS+
Sbjct: 1259 FSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQDSRRSTTSFT 1318
Query: 716 FFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEPVKPSVIYCD 775
F+G+SLI WRSKKQ T+S+SS+EA YRALA ATCE+ L LL LQ P P ++Y D
Sbjct: 1319 MFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQASPPVP-ILYSD 1377
Query: 776 NQSALHIAANPVFHERTKHLEIEC 799
+ +A++IA NPVFHERTKH++++C
Sbjct: 1378 STAAIYIATNPVFHERTKHIKLDC 1401
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 392 bits (1008), Expect = e-109
Identities = 236/596 (39%), Positives = 341/596 (56%), Gaps = 42/596 (7%)
Query: 235 VPASSSNSLDDLSVSIPSADATSVQVPLPSPESPSTTSVSDHTRRPTRPRHQPSHLRNYV 294
VP+SS + D S S SA T+ + +PS+ + + + R + + L+++V
Sbjct: 860 VPSSSPSRSIDRSTSDLSASDTTELLSTGESSTPSSPGLPELLGKGCREKKKSVLLKDFV 919
Query: 295 LHTVS---------------------SSCKASQTSSGIKYPISNYMSYSNLSIPHHAYAM 333
+T S +S A S YP+S++++ S S H A+
Sbjct: 920 TNTTSKKKTASHNIHSPSQVLPSGLPTSLSADSVSGKTLYPLSDFLTNSGYSANHIAFMA 979
Query: 334 SLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRR 393
++ +EP + +A K W +AM+ EI ALEAN TW + LP I +KWVYK+K
Sbjct: 980 AILDSNEPKHFKDAILIKEWCEAMSKEIDALEANHTWDITDLPHGKKAISSKWVYKLKYN 1039
Query: 394 ANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNA 453
++GT+ER+KARLV G +Q EG+ + +TF+P AKLT VR +LA+A+ +W +HQ+DV+NA
Sbjct: 1040 SDGTLERHKARLVVMGNHQKEGVDFKETFAPVAKLTTVRTILAVAAAKDWEVHQMDVHNA 1099
Query: 454 FLLGDLYEDVYMKVPEGVSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAH 513
FL GDL E+VYM++P G C D KVC+L KS YGLKQA R W++K ++ L G+ Q++
Sbjct: 1100 FLHGDLEEEVYMRLPPGFKCSDPSKVCRLRKSLYGLKQAPRCWFSKLSTALRNIGFTQSY 1159
Query: 514 SDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLE 573
D+SLFS G + +L+YVDD+I+AGN LD R K+ L F +KDLG LKYFLGLE
Sbjct: 1160 EDYSLFSLKNGDTIIHVLVYVDDLIVAGNNLDAIDRFKSQLHKCFHMKDLGKLKYFLGLE 1219
Query: 574 VSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGGGA--------TL*G 625
VS G L QRKY LD+V ++G+LG KP + P+ + +L+ G L G
Sbjct: 1220 VSRGPDGFCLSQRKYALDIVKETGLLGCKPSAVPIALNHKLASITGPVFTNPEQYRRLVG 1279
Query: 626 CFFIQKTHRKTVLSYYNQV*YNLCSSAAKSVPLQSYCDSLCCSS*NSKVSERESKKRVIF 685
FI T + LSY + S PL ++ ++ K S + IF
Sbjct: 1280 -RFIYLTITRPDLSYAVHI-----LSQFMQAPLVAHWEAALRLVRYLKGSPAQG----IF 1329
Query: 686 PKEFCSTIVGV**C--RLGGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYR 743
+ S I+ C C TRRS+++Y ++G+S I W++KKQ T+S SS+EA YR
Sbjct: 1330 LRSDSSLIINA-YCDSDYNACPLTRRSLSAYVVYLGDSPISWKTKKQDTVSYSSAEAEYR 1388
Query: 744 ALASATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
A+A EL L LLKDL + P ++CD+++A+HIAANPVFHERTKH+E +C
Sbjct: 1389 AMAYTLKELKWLKALLKDLGVHHSSPMKLHCDSEAAIHIAANPVFHERTKHIESDC 1444
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 390 bits (1002), Expect = e-108
Identities = 230/586 (39%), Positives = 344/586 (58%), Gaps = 41/586 (6%)
Query: 229 LLPETSVPASSSNSLDDLSVSIPSADATSVQVPLPSPESPSTTSVSDHTRRPTRPRHQPS 288
L P S S++ + D + P + S+ LPSP+ +T +S RR T+ P+
Sbjct: 849 LFPLASSQQSATTASDVFTPMDPLSSGNSITSHLPSPQISPSTQISK--RRITK---FPA 903
Query: 289 HLRNYVLHTVSSSCKASQTSSGIKYPISNYMSYSNLSIPHHAYAMSLSLDSEPHTYAEAS 348
HL++Y + V+ +PIS+ +SYS +S H Y ++S P +Y EA
Sbjct: 904 HLQDYHCYFVNKDDS---------HPISSSLSYSQISPSHMLYINNISKIPIPQSYHEAK 954
Query: 349 KHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA* 408
K W A++ EI A+E TW + LPP + KWV+ +K A+G++ER+KAR+VA
Sbjct: 955 DSKEWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWVFTVKFHADGSLERFKARIVAK 1014
Query: 409 GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVP 468
GY Q EG+ Y +TFSP AK+ V+++L +++ W+L+QLD++NAFL GDL E +YMK+P
Sbjct: 1015 GYTQKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQLDISNAFLNGDLEETIYMKLP 1074
Query: 469 EGVSCVDS-----GKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQ 523
+G + + VC+L KS YGLKQASRQW+ KF++ L+ G+++ H DH+LF +
Sbjct: 1075 DGYADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSNSLLALGFEKQHGDHTLFVRCI 1134
Query: 524 GQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISL 583
G F +LL+YVDDI++A + AL +FK+++LG LKYFLGLEV+ +++GISL
Sbjct: 1135 GSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRELGPLKYFLGLEVARTSEGISL 1194
Query: 584 CQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGG--------GATL*GCFFIQKTHRK 635
QRKY L+L+ + +L KP S P+ P+ RLS++ G L G R
Sbjct: 1195 SQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLLEDKEMYRRLVGKLMYLTITRP 1254
Query: 636 TVLSYYNQV*YNLC--SSAAKSVPLQSYCDSLCCSS*NSKVSERESKKRVIFPKEFCSTI 693
+ N+ LC SSA ++ L + L + + + + + E T+
Sbjct: 1255 DITFAVNK----LCQFSSAPRTAHLAAVYKVL-------QYIKGTVGQGLFYSAEDDLTL 1303
Query: 694 VGV**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL* 753
G G C D+RRS T + F+G+SLI WRSKKQ T+S+SS+EA YRALA A+CE+
Sbjct: 1304 KGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYRALALASCEMA 1363
Query: 754 RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
L+ LL L++ P ++Y D+ +A++IA NPVFHERTKH+EI+C
Sbjct: 1364 WLSTLLLALRVHSGVP-ILYSDSTAAVYIATNPVFHERTKHIEIDC 1408
>At4g17450 retrotransposon like protein
Length = 1433
Score = 387 bits (994), Expect = e-107
Identities = 212/500 (42%), Positives = 302/500 (60%), Gaps = 26/500 (5%)
Query: 315 ISNYMSYSNLSIPHHAYAMSLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVP 374
+ ++ Y+N + P HA+ +++ P Y+EA K W DAM EI A+ TWS+V
Sbjct: 891 LQDFHCYNNTTEPFHAFINNITNAVIPQRYSEAKDFKAWCDAMKEEIGAMVRTNTWSVVS 950
Query: 375 LPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMV 434
LPPN I KWV+ IK A+G++ERYKARLVA GY Q EG+ Y +TFSP AKLT VRM+
Sbjct: 951 LPPNKKAIGCKWVFTIKHNADGSIERYKARLVAKGYTQEEGLDYEETFSPVAKLTSVRMM 1010
Query: 435 LALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVS-----CVDSGKVCKLHKSSYGL 489
L LA+ W +HQLD++NAFL GDL E++YMK+P G + + +C+LHKS YGL
Sbjct: 1011 LLLAAKMKWSVHQLDISNAFLNGDLDEEIYMKIPPGYADLVGEALPPHAICRLHKSIYGL 1070
Query: 490 KQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTR 549
KQASRQWY K ++ L G++++++DH+LF K +L+YVDDI++ N D +
Sbjct: 1071 KQASRQWYLKLSNTLKGMGFQKSNADHTLFIKYANGVLMGVLVYVDDIMIVSNSDDAVAQ 1130
Query: 550 IKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLD 609
A L + FK++DLG KYFLG+E++ S KGIS+CQRKY L+L+ +G LGSKP S PLD
Sbjct: 1131 FTAELKSYFKLRDLGAAKYFLGIEIARSEKGISICQRKYILELLSTTGFLGSKPSSIPLD 1190
Query: 610 PSSRLSQDGG--------GATL*GCFFIQKTHRKTVLSYYNQV*YNLC--SSAAKSVPLQ 659
PS +L+++ G L G + R + N LC S A SV L
Sbjct: 1191 PSVKLNKEDGVPLTDSTSYRKLVGKLMYLQITRPDIAYAVN----TLCQFSHAPTSVHLS 1246
Query: 660 SYCDSLCCSS*NSKVSERESKKRVIFPKEFCSTIVGV**CRLGGCVDTRRSVTSYCFFIG 719
+ L + + + + + + + G G C D+RR V +YC FIG
Sbjct: 1247 AVHKVL-------RYLKGTVGQGLFYSADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIG 1299
Query: 720 NSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSA 779
+ L+ W+SKKQ T+S S++EA +RA++ T E+ L+ L D ++ + P+ +YCDN +A
Sbjct: 1300 DYLVSWKSKKQDTVSMSTAEAEFRAMSQGTKEMIWLSRLFDDFKVPFIPPAYLYCDNTAA 1359
Query: 780 LHIAANPVFHERTKHLEIEC 799
LHI N VFHERTK +E++C
Sbjct: 1360 LHIVNNSVFHERTKFVELDC 1379
>At1g70010 hypothetical protein
Length = 1315
Score = 385 bits (990), Expect = e-107
Identities = 232/569 (40%), Positives = 329/569 (57%), Gaps = 47/569 (8%)
Query: 251 PSADATSVQVPLPSPESPSTTSVSDHTRRPTRPRHQPSHLRNYVLHTVSSSCKASQTSSG 310
PS ++SV++ LPS +P+ + R +P++L++Y H+V SS
Sbjct: 722 PSDSSSSVEI-LPSA-NPTNNVPEPSVQTSHRKAKKPAYLQDYYCHSVVSSTP------- 772
Query: 311 IKYPISNYMSYSNLSIPHHAYAMSLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTW 370
+ I ++SY ++ P+ + L EP Y EA K + W DAM E LE TW
Sbjct: 773 --HEIRKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTW 830
Query: 371 SLVPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTI 430
+ LP + I +W++KIK ++G+VERYKARLVA GY Q EGI Y +TFSP AKL
Sbjct: 831 EVCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNS 890
Query: 431 VRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVS-----CVDSGKVCKLHKS 485
V+++L +A+ L QLD++NAFL GDL E++YM++P+G + + VC+L KS
Sbjct: 891 VKLLLGVAARFKLSLTQLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKS 950
Query: 486 SYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNFLD 545
YGLKQASRQWY KF+S L+ G+ Q++ DH+ F K F +L+Y+DDII+A N
Sbjct: 951 LYGLKQASRQWYLKFSSTLLGLGFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDA 1010
Query: 546 EFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKPVS 605
+K+ + + FK++DLG LKYFLGLE+ S KGI + QRKY LDL+ ++G LG KP S
Sbjct: 1011 AVDILKSQMKSFFKLRDLGELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSS 1070
Query: 606 TPLDPSSRLSQDGGG--------ATL*GCFFIQKTHRKTVLSYYNQV*YNLCSSAAKSVP 657
P+DPS + D GG L G R + N++ S A +
Sbjct: 1071 IPMDPSMVFAHDSGGDFVEVGPYRRLIGRLMYLNITRPDITFAVNKL--AQFSMAPRKAH 1128
Query: 658 LQSYCDSLCCSS*N-------SKVSERESKKRVIFPKEFCSTIVGV**CRLGGCVDTRRS 710
LQ+ L S SE + K V ++ S CR D+RRS
Sbjct: 1129 LQAVYKILQYIKGTIGQGLFYSATSELQLK--VYANADYNS-------CR-----DSRRS 1174
Query: 711 VTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEPVKPS 770
+ YC F+G+SLICW+S+KQ +SKSS+EA YR+L+ AT EL LT LK+LQ+ KP+
Sbjct: 1175 TSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSLSVATDELVWLTNFLKELQVPLSKPT 1234
Query: 771 VIYCDNQSALHIAANPVFHERTKHLEIEC 799
+++CDN++A+HIA N VFHERTKH+E +C
Sbjct: 1235 LLFCDNEAAIHIANNHVFHERTKHIESDC 1263
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 375 bits (963), Expect = e-104
Identities = 235/592 (39%), Positives = 330/592 (55%), Gaps = 41/592 (6%)
Query: 230 LPETSVPASSSNSLDDLSVS---IPSADATSVQVPLPSPESPSTTSVSDHTRRPTRPRHQ 286
+P+ + P+S LSVS P+ T + VP+ SP S R+ R H
Sbjct: 877 VPDDTPPSSP------LSVSPSGSPNTPTTPIVVPVASPIPVSPPK----QRKSKRATHP 926
Query: 287 PSHLRNYVL----------HTVSSSCKASQTSSGIK-YPISNYMSYSNLSIPHHAYAMSL 335
P L +YVL H + + S T G +P+++Y+S + S H AY ++
Sbjct: 927 PPKLNDYVLYNAMYTPSSIHALPADPSQSSTVPGKSLFPLTDYVSDAAFSSSHRAYLAAI 986
Query: 336 SLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRAN 395
+ + EP + EA + K W DAM E+ ALE N TW +V LPP V I ++WV+K K ++
Sbjct: 987 TDNVEPKHFKEAVQIKVWNDAMFTEVDALEINKTWDIVDLPPGKVAIGSQWVFKTKYNSD 1046
Query: 396 GTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFL 455
GTVERYKARLV G Q+EG Y +TF+P ++T VR +L + N W ++Q+DV+NAFL
Sbjct: 1047 GTVERYKARLVVQGNKQVEGEDYKETFAPVVRMTTVRTLLRNVAANQWEVYQMDVHNAFL 1106
Query: 456 LGDLYEDVYMKVPEGVSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSD 515
GDL E+VYMK+P G KVC+L KS YGLKQA R W+ K + L+ G+ Q++ D
Sbjct: 1107 HGDLEEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDSLLRFGFVQSYED 1166
Query: 516 HSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVS 575
+SLFS T+ +LIYVDD+++ GN + K L F +KDLG LKYFLG+EVS
Sbjct: 1167 YSLFSYTRNNIELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYFLGIEVS 1226
Query: 576 HSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPL--------DPSSRLSQDGGGATL*GCF 627
+GI L QRKY LD++ DSG LGS+P TPL D LS L G
Sbjct: 1227 RGPEGIFLSQRKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPYRRLVGRL 1286
Query: 628 FIQKTHRKTVLSYYNQV*YNLCSSAAKSVPLQSYCDSLCCSS*NSKVSERESKKRVIFPK 687
+ H + LSY V + P +++ D+ K S + ++
Sbjct: 1287 -LYLLHTRPELSYSVHVLAQFMQN-----PREAHFDAALRVVRYLKGSPGQG---ILLNA 1337
Query: 688 EFCSTIVGV**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALAS 747
+ T+ C TRRS+++Y +G S I W++KKQ T+S SS+EA YRA++
Sbjct: 1338 DPDLTLEVYCDSDWQSCPLTRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSY 1397
Query: 748 ATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
A E+ L LLK+L IE P+ +YCD+++A+HIAANPVFHERTKH+E +C
Sbjct: 1398 ALKEIKWLRKLLKELGIEQSTPARLYCDSKAAIHIAANPVFHERTKHIESDC 1449
>At1g47650 hypothetical protein
Length = 1409
Score = 342 bits (876), Expect = 6e-94
Identities = 187/457 (40%), Positives = 269/457 (57%), Gaps = 31/457 (6%)
Query: 360 EISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEGIYYF 419
EI A+E TW + LP + KWV+ +K A+G++ERYKARLVA GY Q EG+ Y
Sbjct: 750 EIGAMETTHTWEITTLPAGKKAVGCKWVFSLKFLADGSLERYKARLVAKGYTQKEGLDYT 809
Query: 420 DTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVSC-----V 474
DTFSP AK+T ++++L +++ W L QLDV+NAFL G+L E++YMK+PEG + +
Sbjct: 810 DTFSPVAKMTTIKLLLKISASKKWFLKQLDVSNAFLNGELEEEIYMKLPEGYAARKGITL 869
Query: 475 DSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYV 534
VC+L +S YGLKQASRQW+ KF++ L+ G+K+ H DH+LF K F I+L+YV
Sbjct: 870 PPNSVCRLKRSIYGLKQASRQWFKKFSASLLDLGFKKTHGDHTLFIKEYDGEFVIVLVYV 929
Query: 535 DDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQRKYCLDLVH 594
DDI +A ++ L FK++DLG LKYFLGLE++ + GIS+CQRKY L+L+
Sbjct: 930 DDIAIASTSEGAAIQLTQDLQQRFKLRDLGDLKYFLGLEIARTEAGISICQRKYALELLA 989
Query: 595 DSGVLGSKPVSTPLDPSSRLSQDGGGATL*GCFFIQKTHRKTVLSYYNQV*YNLCSSAAK 654
+G++ KPVS P+ P+ ++ + G Y ++ L
Sbjct: 990 STGMVNCKPVSVPMIPNVKMMKTDGDLL-------------DDREQYRRIVGKLMYLTIT 1036
Query: 655 SVPLQSYCDSLCCSS*NSKVSERESKKRVI------------FPKEFCSTIVGV**CRLG 702
+ + + LC S + S ++ RV+ + T+ G
Sbjct: 1037 RLDITFAVNKLCQFSSAPRTSHLQAAYRVLQYIKGSVGQGLFYSASADLTLKGFADSDWA 1096
Query: 703 GCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDL 762
C D+RRS T + F+G+SLI WRSKKQ +S+SS+EA YRALA TCEL L LL L
Sbjct: 1097 SCPDSRRSTTGFSMFVGDSLISWRSKKQHVVSRSSAEAEYRALALVTCELVWLHTLLMSL 1156
Query: 763 QIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
+ + P V+Y D+ +A++IA NPVFHERTKH+E++C
Sbjct: 1157 KASYLVP-VLYSDSTTAIYIATNPVFHERTKHIELDC 1192
>At1g57640
Length = 1444
Score = 335 bits (859), Expect = 6e-92
Identities = 173/390 (44%), Positives = 251/390 (64%), Gaps = 8/390 (2%)
Query: 229 LLPETSVPASSSN---SLDDLSVSIPSADATSVQVPLPSPESPSTTSVSDHTRRPTRPRH 285
++PE + +SS + SL L + S+ + +PL S TT RR +R
Sbjct: 825 IIPEINQESSSPSEFVSLSSLDPFLASSTVQTADLPLSS-----TTPAPIQLRRSSRQTQ 879
Query: 286 QPSHLRNYVLHTVSSSCKASQTSSGIKYPISNYMSYSNLSIPHHAYAMSLSLDSEPHTYA 345
+P L+N+V +TVS + + SS YPI Y+ + H A+ +++ EP TY
Sbjct: 880 KPMKLKNFVTNTVSVESISPEASSSSLYPIEKYVDCHRFTSSHKAFLAAVTAGMEPTTYN 939
Query: 346 EASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERYKARL 405
EA K W +AM+ EI +L N T+S+V LPP + NKWVYKIK R++G +ERYKARL
Sbjct: 940 EAMVDKAWREAMSAEIESLRVNQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARL 999
Query: 406 VA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYM 465
V G Q EG+ Y +TF+P AK++ VR+ L +A+ +WH+HQ+DV+NAFL GDL E+VYM
Sbjct: 1000 VVLGNCQKEGVDYDETFAPVAKMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYM 1059
Query: 466 KVPEGVSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQ 525
K+P+G C D KVC+LHKS YGLKQA R W++K +S L G+ Q+ SD+SLFS
Sbjct: 1060 KLPQGFQCDDPSKVCRLHKSLYGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDG 1119
Query: 526 SFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQ 585
F +L+YVDD+I++G+ D + K+ L++ F +KDLG+LKYFLG+EVS +A+G L Q
Sbjct: 1120 IFVHVLVYVDDLIISGSCPDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQ 1179
Query: 586 RKYCLDLVHDSGVLGSKPVSTPLDPSSRLS 615
RKY LD++ + G+LG++P + PL+ + +LS
Sbjct: 1180 RKYVLDIISEMGLLGARPSAFPLEQNHKLS 1209
Score = 92.4 bits (228), Expect = 9e-19
Identities = 46/96 (47%), Positives = 68/96 (69%)
Query: 704 CVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQ 763
C TRRS+T Y +G++ I W++KKQ T+S+SS+EA YRA+A T EL L +L DL
Sbjct: 1297 CPLTRRSLTGYFVQLGDTPISWKTKKQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLG 1356
Query: 764 IEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
+ V+ I+ D++SA+ ++ NPV HERTKH+E++C
Sbjct: 1357 VSHVQAMRIFSDSKSAIALSVNPVQHERTKHVEVDC 1392
>At2g15920 putative retroelement pol polyprotein
Length = 1100
Score = 334 bits (857), Expect = 1e-91
Identities = 211/550 (38%), Positives = 303/550 (54%), Gaps = 88/550 (16%)
Query: 282 RPRHQPSHLRNYVLHTVSSSCKASQTSSGIKYPISNYMSYSNLSIPHHAYAMSLSLDSEP 341
R R +HL +YV CK +Q+ KYPIS+ +SYS +S H Y +++
Sbjct: 550 RVRKPSTHLSDYV-------CKNTQSDQ--KYPISSTISYSQISPSHMCYIDNITKILIS 600
Query: 342 HTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERY 401
+ EA K W +A+++EI A+E+ W + LP + + KWV+ +K A+G++ERY
Sbjct: 601 TNFPEAQGTKEWCEAVDVEIGAMESTNKWEITTLPKDKKAVGCKWVFTLKILADGSLERY 660
Query: 402 KARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYE 461
KARLVA GY Q EG+ Y +TFSP AK+T ++++L + + W L QLDV+N+FL G+L E
Sbjct: 661 KARLVAKGYTQKEGLDYTNTFSPVAKMTTIKLLLKIYASKKWFLKQLDVSNSFLNGELEE 720
Query: 462 DVYMKVPEGVS-----CVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDH 516
++YMK+PEG + + S VC+L +S YGLKQASRQW+ KF++ L++ G+ + H DH
Sbjct: 721 EIYMKLPEGSAERKGIILPSNAVCRLKRSIYGLKQASRQWFKKFSASLLSLGFFKIHGDH 780
Query: 517 SLFSKTQGQSFTI--LLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEV 574
+LF G + + +L+YVDDI++A D L N FK++DLG LKYFL LE+
Sbjct: 781 TLFLIDCGGEYVVVLVLVYVDDIVIASTNED--------LHNLFKLRDLGDLKYFLCLEI 832
Query: 575 SHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGG--------------- 619
S + GISLC RK +L+ G++ KPVS P+ + +L + G
Sbjct: 833 SRTEVGISLCHRKNAWELLASIGMINCKPVSVPMITNVKLMKADGELLEDREQYRRIVGK 892
Query: 620 ---------GATL*GCFFIQKTHRKTVLSYYNQ-V*YNLCSSAAKSVPLQSYCDSLCCSS 669
TL +Q HR VL Y V L SA+ + L+ + DS S
Sbjct: 893 LMDLTITRPDITL-RTTHLQAAHR--VLQYIKGIVGQGLFYSASSDLTLKGFADSDWVS- 948
Query: 670 *NSKVSERESKKRVIFPKEFCSTIVGV**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKK 729
C D+RRS T + F+G+SLI RSKK
Sbjct: 949 ----------------------------------CADSRRSTTGFTMFVGDSLISLRSKK 974
Query: 730 QQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFH 789
Q+ +S+SS+EA YRALA ATCEL L LL L P +++ ++ +A++IA NPVFH
Sbjct: 975 QRVVSRSSAEAEYRALALATCELVSLHTLLVSLTAATSVP-ILFSNSTTAIYIAINPVFH 1033
Query: 790 ERTKHLEIEC 799
ERTKH EI+C
Sbjct: 1034 ERTKHTEIDC 1043
>At2g07010 putative retroelement pol polyprotein
Length = 1413
Score = 310 bits (793), Expect = 3e-84
Identities = 211/578 (36%), Positives = 296/578 (50%), Gaps = 94/578 (16%)
Query: 236 PASSSNSLDDLSVSIPSADATSVQVPLPSPESPSTTSVSDHT--RRPTRPRHQPSHLRNY 293
P S S S+ S+ +TS + SP TT + ++T R+ R QP+ L++Y
Sbjct: 864 PLSPSTSVTPTQTPTNSSSSTSPSTNV-SPPQQDTTPIIENTPPRQGKRQVQQPARLKDY 922
Query: 294 VLHTVSS--------SCKASQTSSGIK----YPISNYMSYSNLSIPHHAYAMSLSLDSEP 341
+L+ S S SQ+SS I+ YP+++Y+S S H + +++ + EP
Sbjct: 923 ILYNASCTPNTPHVLSPSTSQSSSSIQGNLQYPLTDYISDECFSAGHKVFLAAITANDEP 982
Query: 342 HTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERY 401
+ E K K W DAM E+ ALE N TW +V LP V I ++WVYK K A+GTVERY
Sbjct: 983 KHFKEDVKVKVWNDAMYKEVDALEVNKTWDIVDLPTGKVAIGSQWVYKTKFNADGTVERY 1042
Query: 402 KARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYE 461
KARLV G NQIEG Y +TF+P K+T VR +L L + N W ++Q+DV+NAFL GDL E
Sbjct: 1043 KARLVVQGNNQIEGEDYTETFAPVVKMTTVRTLLRLVAANQWEVYQMDVHNAFLHGDLEE 1102
Query: 462 DVYMKVPEGVSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSK 521
+VYMK+P G KVC+L KS YGLKQA R W+ K + L G+ Q + D+S FS
Sbjct: 1103 EVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDALKRFGFIQGYEDYSFFSY 1162
Query: 522 TQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGI 581
+ +L+YVDD+I+ GN DE+ K K +LG S G
Sbjct: 1163 SCKGIELRVLVYVDDLIICGN--DEYMVQK--------------FKEYLGRCFSMKDLG- 1205
Query: 582 SLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGGGATL*GCFFIQKTHRKTVLSYY 641
+ KY L + G P L+ + R+ + G+ G
Sbjct: 1206 ---KLKYFLGIEVSRGP--DAPREAHLEAAMRIVRYLKGSPGQGILL------------- 1247
Query: 642 NQV*YNLCSSAAKSVPLQSYCDSLCCSS*NSKVSERESKKRVIFPKEFCSTIVGV**CRL 701
SA K + L+ YCDS +F S
Sbjct: 1248 ---------SANKDLTLEVYCDS-----------------------DFQS---------- 1265
Query: 702 GGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKD 761
C TRRS+++Y +G S I W++KKQ T+S SS+EA YRA++ A E+ L LLK+
Sbjct: 1266 --CPLTRRSLSAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSVALKEIKWLNKLLKE 1323
Query: 762 LQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
L I P+ ++CD+++A+ IAANPVFHERTKH+E +C
Sbjct: 1324 LGITLAAPTRLFCDSKAAISIAANPVFHERTKHIERDC 1361
>At4g07810 putative polyprotein
Length = 1366
Score = 304 bits (779), Expect = 1e-82
Identities = 208/606 (34%), Positives = 317/606 (51%), Gaps = 76/606 (12%)
Query: 203 LIISQKQCHGLFS-HLRMRILRALVLILLPETSVPASS-SNSLDDLSVSIPSADATSVQV 260
L IS+KQ F + +L V ++ T+VPA + + SL LS ++ + +
Sbjct: 766 LPISEKQKENRFQIYDYFNVLNLEVCPVIEPTTVPAHTHTRSLAPLSTTVTNDQFGNDM- 824
Query: 261 PLPSPESPSTTSVSDHTRRPTRPRHQPSHLRNYVLHTVSSSCKASQTSSGIKYPISNYMS 320
D+T P + PS+L Y H + + S + G + +S+++S
Sbjct: 825 --------------DNTLMPRKETRAPSYLSQY--HCSNVLKEPSSSLHGTAHSLSSHLS 868
Query: 321 YSNLSIPHHAYAMSLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVV 380
Y LS + + ++ + EP T+ EA+ + W+DAMN+E+ AL + T + L
Sbjct: 869 YDKLSNEYRLFCFAIIAEKEPTTFKEAALLQKWLDAMNVELDALVSTSTREICSLHDGKR 928
Query: 381 PIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASV 440
I KWV+KIK +++GT+ERYKARLVA GY Q EG+ Y DTFSP AKLT VR++LALA++
Sbjct: 929 AIGCKWVFKIKYKSDGTIERYKARLVANGYTQQEGVDYIDTFSPIAKLTSVRLILALAAI 988
Query: 441 NNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVS-----CVDSGKVCKLHKSSYGLKQASRQ 495
+NW + Q+DV NAFL GD E++YM++P+G + + VC+L KS YGLKQASRQ
Sbjct: 989 HNWSISQMDVTNAFLHGDFEEEIYMQLPQGYTPRKGELLPKRPVCRLVKSLYGLKQASRQ 1048
Query: 496 WYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALD 555
W+ KF+ +L+ G+ Q+ D +LF + + +F LL+YVDDI+L N +K L
Sbjct: 1049 WFHKFSGVLIQNGFMQSLFDPTLFVRVREDTFLALLVYVDDIMLVSNKDSAVIEVKQILA 1108
Query: 556 NAFKIKDLGVLKYFLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLS 615
FK+KDLG +YFLGLE++ S +GIS+ QRKY L+L+ + G LG KPV TP++ + +LS
Sbjct: 1109 KEFKLKDLGQKRYFLGLEIARSKEGISISQRKYALELLEEFGFLGCKPVPTPMELNLKLS 1168
Query: 616 QDGGGATL*GCFFIQKTHRKTVLSYYNQV*YNLCSSAAK-----SVPLQSYCDSLCCSS* 670
Q+ G L + + R L Y ++C + K S P + + L +
Sbjct: 1169 QEDGALLLDASHYRKLIGR---LVYLTVTRPDICFAVNKLNQYMSAPREPH---LMAARR 1222
Query: 671 NSKVSERESKKRVIFPKEFCSTIVGV**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKKQ 730
+ + + + V +P T C ++ S+ S++ W
Sbjct: 1223 ILRYLKNDPGQGVFYPASSTLTFRAFADADWSNCPESSISI---------SIVFW----- 1268
Query: 731 QTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEPVKPSVIYC--DNQSALHIAANPVF 788
K S+EA +L+ L P I+ D++SALHIA N VF
Sbjct: 1269 ---LKLSTEA----------------WLVLSL------PDTIFVYYDDESALHIAKNSVF 1303
Query: 789 HERTKH 794
HE TK+
Sbjct: 1304 HESTKN 1309
>At2g24660 putative retroelement pol polyprotein
Length = 1156
Score = 303 bits (775), Expect = 3e-82
Identities = 172/405 (42%), Positives = 238/405 (58%), Gaps = 19/405 (4%)
Query: 232 ETSVPASSSNSLDDLSVSIPSADATSV----QVPLPSPESPSTTSVSDHTRRPTRPRHQP 287
++S P + S D L+ S D S PL +SP + S R+ R Q
Sbjct: 515 DSSTPDKNLASGDTLAQIDDSPDIVSTPNRNNQPLFVVDSPFVEATSPRQRK--RQIRQS 572
Query: 288 SHLRNYVLHTVSSSC--------KASQTSSGIK-----YPISNYMSYSNLSIPHHAYAMS 334
L++YVL+ + S +SQ+SS ++ YP+S+Y+S S H A+ +
Sbjct: 573 VRLQDYVLYNATVSPINPHALPDSSSQSSSMVQGTSSLYPLSDYVSDDCFSAGHKAFLAA 632
Query: 335 LSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRA 394
++ + EP + EA + K W DAM E+ ALE N TW +V LPP V I ++WVYK K A
Sbjct: 633 ITANDEPKHFKEAVRIKVWNDAMFKEVDALEINKTWDIVDLPPGKVAIGSQWVYKTKYNA 692
Query: 395 NGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAF 454
+G++ERYKARLV G Q+EG Y +TF+P K+T VR +L L + N W ++Q+DVNNAF
Sbjct: 693 DGSIERYKARLVVQGNKQVEGEDYNETFAPVVKMTTVRTLLRLVAANQWEVYQMDVNNAF 752
Query: 455 LLGDLYEDVYMKVPEGVSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHS 514
L GDL E+VYMK+P G KVC+L KS YGLKQA R W+ K + L+ G+ Q H
Sbjct: 753 LHGDLDEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDALLRFGFVQGHE 812
Query: 515 DHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEV 574
D+S FS T+ +L+YVDD+++ GN + K L F +KDLG LKYFLG+EV
Sbjct: 813 DYSFFSYTRNGIELRVLVYVDDLLICGNDGYMLQKFKEYLGRCFSMKDLGKLKYFLGIEV 872
Query: 575 SHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGG 619
S ++GI L QRKY LD++ DSG LG +P TPL+ + L+ D G
Sbjct: 873 SRGSEGIFLSQRKYALDIITDSGNLGCRPALTPLEQNHHLATDDG 917
Score = 75.9 bits (185), Expect = 8e-14
Identities = 42/96 (43%), Positives = 56/96 (57%), Gaps = 21/96 (21%)
Query: 704 CVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQ 763
C TRRS+++Y +G S I W++KKQ T+S SS+EA YRA++ A E+ L LLK+L
Sbjct: 1001 CPKTRRSLSAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSVALREIKWLRKLLKEL- 1059
Query: 764 IEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
ANPVFHERTKH+E +C
Sbjct: 1060 --------------------ANPVFHERTKHIESDC 1075
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 301 bits (771), Expect = 9e-82
Identities = 148/304 (48%), Positives = 212/304 (69%), Gaps = 1/304 (0%)
Query: 312 KYPISNYMSYSNLSIPHHAYAMSLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWS 371
++P S MS + + H A+ +++ EP TY EA K W +AM+ EI +L N T+S
Sbjct: 572 EFPYSK-MSCNRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVNQTFS 630
Query: 372 LVPLPPNVVPIDNKWVYKIKRRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIV 431
+V LPP + NKWVYKIK R++G +ERYKARLV G Q EG+ Y +TF+P AK++ V
Sbjct: 631 IVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAKMSTV 690
Query: 432 RMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVSCVDSGKVCKLHKSSYGLKQ 491
R+ L +A+ +WH+HQ+DV+NAFL GDL E+VYMK+P+G C D KVC+LHKS YGLKQ
Sbjct: 691 RLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLYGLKQ 750
Query: 492 ASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIK 551
A R W++K +S L G+ Q+ SD+SLFS F +L+YVDD+I++G+ D + K
Sbjct: 751 APRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGVFVHVLVYVDDLIISGSCPDAVAQFK 810
Query: 552 AALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPS 611
+ L++ F +KDLG+LKYFLG+EVS +A+G L QRKY LD++ + G+LG++P + PL+ +
Sbjct: 811 SYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAFPLEQN 870
Query: 612 SRLS 615
+LS
Sbjct: 871 HKLS 874
Score = 92.8 bits (229), Expect = 7e-19
Identities = 47/96 (48%), Positives = 68/96 (69%)
Query: 704 CVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQ 763
C TRRS+T Y +G++ I W++KKQ TIS+SS+EA YRA+A T EL L +L DL
Sbjct: 962 CPLTRRSLTGYFVQLGDTPISWKTKKQPTISRSSAEAEYRAMAFLTQELMWLKRVLYDLG 1021
Query: 764 IEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
+ V+ I+ D++SA+ ++ NPV HERTKH+E++C
Sbjct: 1022 VSHVQAMRIFSDSKSAIALSVNPVQHERTKHVEVDC 1057
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 290 bits (742), Expect = 2e-78
Identities = 176/474 (37%), Positives = 258/474 (54%), Gaps = 18/474 (3%)
Query: 335 LSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRA 394
L + EP TY EA K W +AMN EI +E N TW LV P I KW+YKIK A
Sbjct: 831 LVANEEPQTYDEARGDKEWEEAMNEEIKVIEKNRTWKLVDKPEKKNVISVKWIYKIKTDA 890
Query: 395 NGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAF 454
+G ++KARLVA G++Q GI Y +TF+P ++ +R +LA A+ W L+Q+DV +AF
Sbjct: 891 SGNHVKHKARLVARGFSQEYGIDYLETFAPVSRYDTIRALLAYAAQMKWRLYQMDVKSAF 950
Query: 455 LLGDLYEDVYMKVPEG-VSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAH 513
L G+L E+VY+ P G V KV +L+K+ YGLKQA R WY + S + G+ ++
Sbjct: 951 LNGELEEEVYVTQPPGFVIEGKEEKVLRLYKALYGLKQAPRAWYERIDSYFIQNGFARSM 1010
Query: 514 SDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLE 573
+D +L+SK +G+ I+ +YVDD+I+ GN K + + F++ DLG+L YFLG+E
Sbjct: 1011 NDAALYSKKKGEDVLIVSLYVDDLIITGNNTHLINTFKKNMKDEFEMTDLGLLNYFLGME 1070
Query: 574 VSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGGGATL*GCFFIQKTH 633
V+ GI L Q KY L+ G+ SK VSTPL P + G F T
Sbjct: 1071 VNQDDSGIFLSQEKYANKLIDKFGMKESKSVSTPLTPQGKRKGVEGDDK----EFADPTK 1126
Query: 634 RKTVLSYYNQV*YNLCSSAAKSVPLQSYCDSLCCSS*NSKVSERESKKR---------VI 684
+ ++ LC+S + SY S E + R V+
Sbjct: 1127 YRRIVGGL----LYLCASRPDVMYASSYLSRYMSSPSIQHYQEAKRVLRYVKGTSNFGVL 1182
Query: 685 FPKEFCSTIVGV**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRA 744
F + +VG GG ++ ++S T Y F +G ++ CW+S KQQT+++S++EA Y A
Sbjct: 1183 FTSKETPRLVGYSDSDWGGSLEDKKSTTGYVFTLGLAMFCWQSCKQQTVAQSTAEAEYIA 1242
Query: 745 LASATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIE 798
+ +AT + L L +D ++ + I CDN+SA+ I NPV H RTKH+EI+
Sbjct: 1243 VCAATNQAIWLQRLFEDFGLKFKEGIPILCDNKSAIAIGRNPVQHRRTKHIEIK 1296
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 281 bits (720), Expect = 8e-76
Identities = 159/413 (38%), Positives = 236/413 (56%), Gaps = 27/413 (6%)
Query: 234 SVPASSSNSLDDLSVSIP-SADATSVQVPLPSPESPSTTSVSD------HTRRPTRPRHQ 286
S+P DD + S SA + S PLPS + T D RP R
Sbjct: 396 SMPLDDDLRADDNNASTSNSASSASSIPPLPSTVNTQNTDALDIDTNSVPIARPKRNAKA 455
Query: 287 PSHLRNYVLHTVSSSCKASQTSSG---------------IKYPISNYMSYSNLSIPHHAY 331
P++L Y ++V S T+S YP+S +SY L+ H+Y
Sbjct: 456 PAYLSEYHCNSVPFLSSLSPTTSTSIETPSSSIPPKKITTPYPMSTAISYDKLTPLFHSY 515
Query: 332 AMSLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIK 391
+ ++++EP + +A K + W A N E+ ALE N TW + L + KWV+ IK
Sbjct: 516 ICAYNVETEPKAFTQAMKSEKWTRAANEELHALEQNKTWIVESLTEGKNVVGCKWVFTIK 575
Query: 392 RRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVN 451
+G++ERYKARLVA G+ Q EGI Y +TFSP AK V+++L LA+ W L Q+DV+
Sbjct: 576 YNPDGSIERYKARLVAQGFTQQEGIDYMETFSPVAKFGSVKLLLGLAAATGWSLTQMDVS 635
Query: 452 NAFLLGDLYEDVYMKVPEGVS-----CVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVT 506
NAFL G+L E++YM +P+G + + S VC+L KS YGLKQASRQWY + +S+ +
Sbjct: 636 NAFLHGELDEEIYMSLPQGYTPPTGISLPSKPVCRLLKSLYGLKQASRQWYKRLSSVFLG 695
Query: 507 CGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVL 566
+ Q+ +D+++F K S ++L+YVDD+++A N +K L + FKIKDLG
Sbjct: 696 ANFIQSPADNTMFVKVSCTSIIVVLVYVDDLMIASNDSSAVENLKELLRSEFKIKDLGPA 755
Query: 567 KYFLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGG 619
++FLGLE++ S++GIS+CQRKY +L+ D G+ G KP S P+DP+ L+++ G
Sbjct: 756 RFFLGLEIARSSEGISVCQRKYAQNLLEDVGLSGCKPSSIPMDPNLHLTKEMG 808
Score = 131 bits (330), Expect = 1e-30
Identities = 58/98 (59%), Positives = 77/98 (78%)
Query: 702 GGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKD 761
G C D+RRSVT +C ++G SLI W+SKKQ +S+SS+E+ YR+LA ATCE+ L LLKD
Sbjct: 890 GTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKD 949
Query: 762 LQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
L + P+ ++CDN+SALH+A NPVFHERTKH+EI+C
Sbjct: 950 LHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEIDC 987
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 260 bits (664), Expect = 2e-69
Identities = 153/392 (39%), Positives = 216/392 (55%), Gaps = 43/392 (10%)
Query: 238 SSSNSLDDLSVSIPSADATSVQVP--------LPSPESPSTTS-------VSDHTRRPTR 282
S+S+ LD V P+ TS P LP + TT+ V R+ TR
Sbjct: 734 STSSPLDTEVVPTPATSTTSASSPSSPTNLQFLPLSRAKPTTAQAVAPPAVPPPRRQSTR 793
Query: 283 PRHQPSHLRNYVLHTVSSSCKASQTSSGIKYPISNYMSYSNLSIPHHAYAMSLSLDSEPH 342
+ P L+++V++T S+ +S I Y + S H Y
Sbjct: 794 NKAPPVTLKDFVVNTTVCQESPSKLNS-ILYQLQKRDDTRRFSASHTTY----------- 841
Query: 343 TYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYKIKRRANGTVERYK 402
+ I A E N TW++ LPP I ++WVYK+K ++G+VERYK
Sbjct: 842 ----------------VAIDAQEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYK 885
Query: 403 ARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYED 462
ARLVA G Q EG Y +TF+P AK+ VR+ L +A NW +HQ+DV+NAFL GDL E+
Sbjct: 886 ARLVALGNKQKEGEDYGETFAPVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREE 945
Query: 463 VYMKVPEGVSCVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKT 522
VYMK+P G KVC+L K+ YGLKQA R W+ K T+ L G++Q+ +D+SLF+
Sbjct: 946 VYMKLPPGFEASHPNKVCRLRKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLV 1005
Query: 523 QGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGIS 582
+G +LIYVDD+I+ GN + K L + F +KDLG LKYFLG+EV+ S GI
Sbjct: 1006 KGSVRIKILIYVDDLIITGNSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTGIY 1065
Query: 583 LCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRL 614
+CQRKY LD++ ++G+LG KP + PL+ + +L
Sbjct: 1066 ICQRKYALDIISETGLLGVKPANFPLEQNHKL 1097
Score = 99.8 bits (247), Expect = 5e-21
Identities = 49/92 (53%), Positives = 67/92 (72%)
Query: 707 TRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEP 766
+RRSVT Y G+S I W++KKQ T+SKSS+EA YRA++ EL L LL L +
Sbjct: 1189 SRRSVTGYFVQFGDSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSH 1248
Query: 767 VKPSVIYCDNQSALHIAANPVFHERTKHLEIE 798
V+P ++ CD++SA++IA NPVFHERTKH+EI+
Sbjct: 1249 VQPMIMCCDSKSAIYIATNPVFHERTKHIEID 1280
>At4g04410 putative polyprotein
Length = 1017
Score = 254 bits (650), Expect = 1e-67
Identities = 159/412 (38%), Positives = 225/412 (54%), Gaps = 34/412 (8%)
Query: 241 NSLDDLSVSIPSAD-ATSVQVPLP----SPESPSTTSVSDHTRRPT-----RPRHQPSHL 290
NSL DLS PS D ATS+Q L +P S T D P + + S+L
Sbjct: 598 NSLADLST--PSTDRATSLQFLLDHLGVTPSSERETKTRDLIEEPITIDQENEQEEASNL 655
Query: 291 R---------------------NYVLHTVSSSCKASQTSSGIKYPISNYMSYSNLSIPHH 329
+ + VL S + + + +PI S + L H
Sbjct: 656 QQDGEINGIQIHDDMDTQNEDGDEVLGLREKSSRLYYNNKAVAHPIQAVCSLALLPQDHQ 715
Query: 330 AYAMSLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDNKWVYK 389
A+ + + TY EA + + W++A+ E A+ N TW LPP + +KWV+
Sbjct: 716 AFIGKIEANFVLQTYEEAKESEEWINAVADETGAMIRNHTWDEEDLPPGKRAVSSKWVFT 775
Query: 390 IKRRANGTVERYKARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLD 449
IK +NG +ER+KARLVA G+ Q G Y +TF+P AKL VR+VL+LA+ +W L Q+D
Sbjct: 776 IKYLSNGEIERHKARLVACGFTQTYGRDYTETFAPVAKLHTVRVVLSLATNLSWDLWQMD 835
Query: 450 VNNAFLLGDLYEDVYMKVPEGVS-CVDSGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCG 508
V NAFL G+L E+VYM P G+ + GKV +L K+ YGLKQ+ R WY ++ L G
Sbjct: 836 VKNAFLQGELEEEVYMTPPPGLEDSIAPGKVLRLRKAIYGLKQSPRAWYHNLSTTLKGKG 895
Query: 509 YKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKY 568
+K++ +DH+LF+ Q + LIYVD+II++G+ + K L + F IKDLG LKY
Sbjct: 896 FKKSEADHTLFTLQSDQGIIVALIYVDNIIISGDNKEGIHDTKLFLKSTFDIKDLGELKY 955
Query: 569 FLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGGG 620
FLG+EV S +G+ L QRKY LDL++++G LGSKP TPL + + GGG
Sbjct: 956 FLGIEVCRSPEGLFLSQRKYTLDLLNETGKLGSKPAKTPLVDGYTVKRTGGG 1007
>At1g59265 polyprotein, putative
Length = 1466
Score = 253 bits (647), Expect = 2e-67
Identities = 161/402 (40%), Positives = 227/402 (56%), Gaps = 28/402 (6%)
Query: 232 ETSVPASSSNSLDDLSVSIPSADATSVQVPLPSPES-PS-TTSVSDHTRRPTRPR---HQ 286
+T +S + S ++ + PS A S+ P S S PS TTS S + PT P H
Sbjct: 849 QTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHP 908
Query: 287 PSHLRNYVLHT----VSSSCKASQTSSGIKYPISNYMSYSNLSIPHHAYAMSLSLDSEPH 342
P L V + +++ ++ +GI P P ++ A+SL+ +SEP
Sbjct: 909 PPPLAQIVNNNNQAPLNTHSMGTRAKAGIIKPN-----------PKYSLAVSLAAESEPR 957
Query: 343 TYAEASKHKCWVDAMNLEISALEANGTWSLVPLPPNVVPIDN-KWVYKIKRRANGTVERY 401
T +A K + W +AM EI+A N TW LVP PP+ V I +W++ K ++G++ RY
Sbjct: 958 TAIQALKDERWRNAMGSEINAQIGNHTWDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRY 1017
Query: 402 KARLVA*GYNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYE 461
KARLVA GYNQ G+ Y +TFSP K T +R+VL +A +W + QLDVNNAFL G L +
Sbjct: 1018 KARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTD 1077
Query: 462 DVYMKVPEGVSCVD-SGKVCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFS 520
DVYM P G D VCKL K+ YGLKQA R WY + + L+T G+ + SD SLF
Sbjct: 1078 DVYMSQPPGFIDKDRPNYVCKLRKALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFV 1137
Query: 521 KTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDN---AFKIKDLGVLKYFLGLEVSHS 577
+G+S +L+YVDDI++ GN + T + LDN F +KD L YFLG+E
Sbjct: 1138 LQRGKSIVYMLVYVDDILITGN---DPTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRV 1194
Query: 578 AKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGG 619
G+ L QR+Y LDL+ + ++ +KPV+TP+ PS +LS G
Sbjct: 1195 PTGLHLSQRRYILDLLARTNMITAKPVTTPMAPSPKLSLYSG 1236
Score = 84.0 bits (206), Expect = 3e-16
Identities = 41/96 (42%), Positives = 60/96 (61%)
Query: 703 GCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDL 762
G D S Y ++G+ I W SKKQ+ + +SS+EA YR++A+ + E+ + LL +L
Sbjct: 1319 GDKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTEL 1378
Query: 763 QIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIE 798
I +P VIYCDN A ++ ANPVFH R KH+ I+
Sbjct: 1379 GIRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAID 1414
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 252 bits (643), Expect = 7e-67
Identities = 152/393 (38%), Positives = 219/393 (55%), Gaps = 24/393 (6%)
Query: 236 PASSSNSLDDLSVSIPSADATSVQVPLPS-----PESPSTTSVSDHTRRPTRPRHQPSHL 290
P S S + + + +P + +S +P PS P SPS++S S P P P +
Sbjct: 842 PNSPSPNSPNQNSPLPQSPISSPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPA--PPII 899
Query: 291 RNYVLHTVSSSCKASQTSSGIKYPISNYMSYSNLSIPHHAYAMSLSLDSEPHTYAEASKH 350
+ V++ A++ GI+ P Y +YA SL+ +SEP T +A K
Sbjct: 900 QVNAQAPVNTHSMATRAKDGIRKPNQKY-----------SYATSLAANSEPRTAIQAMKD 948
Query: 351 KCWVDAMNLEISALEANGTWSLVPLPP-NVVPIDNKWVYKIKRRANGTVERYKARLVA*G 409
W AM EI+A N TW LVP PP +V + +W++ K ++G++ RYKARLVA G
Sbjct: 949 DRWRQAMGSEINAQIGNHTWDLVPPPPPSVTIVGCRWIFTKKFNSDGSLNRYKARLVAKG 1008
Query: 410 YNQIEGIYYFDTFSPTAKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPE 469
YNQ G+ Y +TFSP K T +R+VL +A +W + QLDVNNAFL G L ++VYM P
Sbjct: 1009 YNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQPP 1068
Query: 470 GVSCVDSGK---VCKLHKSSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQS 526
G VD + VC+L K+ YGLKQA R WY + + L+T G+ + SD SLF +G+S
Sbjct: 1069 GF--VDKDRPDYVCRLRKAIYGLKQAPRAWYVELRTYLLTVGFVNSISDTSLFVLQRGRS 1126
Query: 527 FTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQR 586
+L+YVDDI++ GN AL F +K+ L YFLG+E +G+ L QR
Sbjct: 1127 IIYMLVYVDDILITGNDTVLLKHTLDALSQRFSVKEHEDLHYFLGIEAKRVPQGLHLSQR 1186
Query: 587 KYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGG 619
+Y LDL+ + +L +KPV+TP+ S +L+ G
Sbjct: 1187 RYTLDLLARTNMLTAKPVATPMATSPKLTLHSG 1219
Score = 83.6 bits (205), Expect = 4e-16
Identities = 41/96 (42%), Positives = 60/96 (61%)
Query: 703 GCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDL 762
G D S Y ++G+ I W SKKQ+ + +SS+EA YR++A+ + EL + LL +L
Sbjct: 1302 GDTDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSELQWICSLLTEL 1361
Query: 763 QIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIE 798
I+ P VIYCDN A ++ ANPVFH R KH+ ++
Sbjct: 1362 GIQLSHPPVIYCDNVGATYLCANPVFHSRMKHIALD 1397
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.338 0.145 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,338,557
Number of Sequences: 26719
Number of extensions: 660527
Number of successful extensions: 3198
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 100
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 2794
Number of HSP's gapped (non-prelim): 248
length of query: 799
length of database: 11,318,596
effective HSP length: 107
effective length of query: 692
effective length of database: 8,459,663
effective search space: 5854086796
effective search space used: 5854086796
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0171.6


