

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
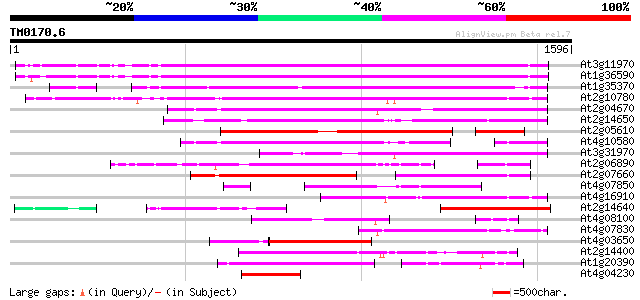
Query= TM0170.6
(1596 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g11970 hypothetical protein 1036 0.0
At1g36590 hypothetical protein 1031 0.0
At1g35370 hypothetical protein 916 0.0
At2g10780 pseudogene 671 0.0
At2g04670 putative retroelement pol polyprotein 623 e-178
At2g14650 putative retroelement pol polyprotein 613 e-175
At2g05610 putative retroelement pol polyprotein 568 e-162
At4g10580 putative reverse-transcriptase -like protein 477 e-134
At3g31970 hypothetical protein 438 e-122
At2g06890 putative retroelement integrase 430 e-120
At2g07660 putative retroelement pol polyprotein 380 e-105
At4g07850 putative polyprotein 369 e-102
At4g16910 retrotransposon like protein 351 2e-96
At2g14640 putative retroelement pol polyprotein 304 3e-82
At4g08100 putative polyprotein 283 4e-76
At4g07830 putative reverse transcriptase 268 2e-71
At4g03650 putative reverse transcriptase 265 2e-70
At2g14400 putative retroelement pol polyprotein 222 1e-57
At1g20390 hypothetical protein 219 1e-56
At4g04230 putative transposon protein 193 6e-49
>At3g11970 hypothetical protein
Length = 1499
Score = 1036 bits (2680), Expect = 0.0
Identities = 597/1530 (39%), Positives = 887/1530 (57%), Gaps = 73/1530 (4%)
Query: 18 VTQMEAKIDAMESELAAIRTALAAVTTAMKDLPTTLGTMVEKAVGKSVGIDVDSGDARPE 77
VT++E + E+ L AV + +T G MV++ +D + +
Sbjct: 26 VTRLETTVAEQHKEMMKQFADLYAVLSR-----STAGKMVDE----QSTLDRSAPRSSQS 76
Query: 78 REPREKTPESVGLRESGSDQPVLQGEALNEFRQSVK--KVELPMFDGKDLAGWISRAEIY 135
E R P+ R++ Q ++ + N + + K++ P FDG L W+ + E +
Sbjct: 77 MENRSGYPDPY--RDARHQQ--VRSDHFNAYNNLTRLGKIDFPRFDGTRLKEWLFKVEEF 132
Query: 136 FKVQETSPEVKVSLAQLSMEG--GTIH--FYNSLLATEEELTWERFRDALLERYGGNGDG 191
F V T ++KV +A + + T H F S + E W+ + L ER+ + D
Sbjct: 133 FGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLEVLYDWKGYVKLLKERFEDDCD- 191
Query: 192 DVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKVRSLVAMG 251
D +L L++ + +Y FE + ++ L E+ +L GL+ + + VR
Sbjct: 192 DPMAELKHLQETDGIIDYHQKFELIKTRV-NLSEEYLVSVYLAGLRTDTQMHVRMFQPQ- 249
Query: 252 GVNRARLLVVTRAVEKEVKGDGVAGQARANRFGGNGSRSGFSGATKGNGTEWIWVKGNKE 311
L + + EK NR G G++ K +G +
Sbjct: 250 --TVRHCLFLGKTYEKAHPKKPANTTWSTNRSAPTG---GYNKYQK---------EGESK 295
Query: 312 TGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQLMERRQKGLCFKCGGPYHRN 371
T N+ N P Q K +S ++ +RR KGLC+ C Y
Sbjct: 296 TDHYGNKGNFKPVSQQPKK----------------MSQQEMSDRRSKGLCYFCDEKYTPE 339
Query: 372 HVCPDKHLRLLILEEDGEELDESKMLAMEVNEDEEETQGELSLMSLCELGMKTGGIPRTM 431
H K +L ++ D EE ++++ E+ D++E ++S+ ++ + +TM
Sbjct: 340 HYLVHKKTQLFRMDVD-EEFEDARE---ELVNDDDEHMPQISVNAVSGIAGY-----KTM 390
Query: 432 KLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPRMT-VKLGDGYKSQAQGRCA 490
+++GT + + +L+DSG+THNF+D +LG +V DT +T V + DG K + +G+
Sbjct: 391 RVKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKV-DTAGLTRVSVADGRKLRVEGKVT 449
Query: 491 GLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQG 550
++ + L L G D+VLG++WL+TLG + M F + + + L G
Sbjct: 450 DFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHG 509
Query: 551 MDTRGEHMEALQSITATGERKPGL----LGTRNEEKAGKQLPGEINSSQLEE---LDKLL 603
+ + Q + E + L + +E G+ +S+L E ++++L
Sbjct: 510 LTSGSVREVKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVL 569
Query: 604 YRFDVVFQEKQGLPPGRGRE-HCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGI 662
+ +F E LPP R + H I + EG PVN RPYRY H KNEI+K V ++L+ G
Sbjct: 570 NEYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGT 629
Query: 663 IRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSK 722
++ S+S Y+SPV+LVKKKD TW +CVDYR LN +TV D FPIP+IE+L+DEL GAV FSK
Sbjct: 630 VQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSK 689
Query: 723 LDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRG 782
+DL++GYHQVR+ +D+ KTAF+TH GH+EY+VMPFGL NAP+TFQ LMN +F+P LR+
Sbjct: 690 IDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKF 749
Query: 783 VLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGV 842
VLVFFDDILVYS + +H HL++V EV++ ++L A KC FA VEYLGH IS QG+
Sbjct: 750 VLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGI 809
Query: 843 AVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAFNWGKQ 902
DP K+ +VK WP+P +K +RGFLGL GYYR+F++++G IA PL LTK DAF W
Sbjct: 810 ETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTAV 869
Query: 903 AQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRN 962
AQQAFEDLK L APVL+LP F ++F +E DA G GIGA+L+ HP+AY S+ L +
Sbjct: 870 AQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQ 929
Query: 963 LAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLG 1022
L S YEKEL+AV A++ WR YLL F++ TDQRSLK L Q++ T QQ W KLL
Sbjct: 930 LHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLE 989
Query: 1023 YNFEIIYKPGKTNQGADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKLCQIVSEV 1082
+++EI Y+ GK N ADALSR++ + L ++ + I D +L I++ +
Sbjct: 990 FDYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLLKDIQAGYANDSQLQDIITAL 1049
Query: 1083 ALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATN 1142
DP + + Q L + ++V+ ++ +L H + GGHSG T++R+
Sbjct: 1050 QRDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVTHQRVKGL 1109
Query: 1143 LYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRG 1202
YW+GM K IQ ++ +C CQ+ K ++ GLLQPLPIP+ +W ++S++FI GLP S G
Sbjct: 1110 FYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVSGG 1169
Query: 1203 FEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFW 1262
I VVVDRL+K +HFI L HPY+A +VA + + +LHG P+SI+SDRD +F S FW
Sbjct: 1170 KTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGCPTSIVSDRDVVFTSEFW 1229
Query: 1263 KEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNT 1322
+E F LQG K++SAYHP+SDGQTE+VNRCLETYLRC D+P+ W+ WL AEYW+NT
Sbjct: 1230 REFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAEYWYNT 1289
Query: 1323 SYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERM 1382
+YHS+++ TPFE VYG+ PPV ++ GE++V V + LQ+R++ L LK HL+ AQ RM
Sbjct: 1290 NYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRM 1349
Query: 1383 RAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRL 1442
+ A+ R +EFE+G++V+VK++ +RQ S+ R + KL+ +YFGPY II R G VAY+L
Sbjct: 1350 KQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKL 1409
Query: 1443 KLPEGARIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRDILRQGESI 1502
LP +++HPVFH+S LK VGN LP + +++ P V+ R+ + RQG+++
Sbjct: 1410 ALPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVM--QDVFEKVPEKVVERKMVNRQGKAV 1467
Query: 1503 SQVLVQWQGKTPEEATWEDLATIRSEFPVF 1532
++VLV+W + EEATWE L ++ FP F
Sbjct: 1468 TKVLVKWSNEPLEEATWEFLFDLQKTFPEF 1497
>At1g36590 hypothetical protein
Length = 1499
Score = 1031 bits (2667), Expect = 0.0
Identities = 598/1537 (38%), Positives = 885/1537 (56%), Gaps = 87/1537 (5%)
Query: 18 VTQMEAKIDAMESELAAIRTALAAVTTAMKDLPTTLGTMVEK-------AVGKSVGIDVD 70
VT++E + E+ L AV + +T G MV++ A S ++
Sbjct: 26 VTRLETTVAEQHKEMMKQFADLYAVLSR-----STAGKMVDEQSTLDRSAPRSSQSMENR 80
Query: 71 SGDARPEREPREKTPESVGLRESGSDQPVLQGEALNEFRQSVK--KVELPMFDGKDLAGW 128
SG P R+ R + S + N + + K++ P FDG L W
Sbjct: 81 SGYLDPYRDARHQQVRS---------------DHFNAYNNLTRLGKIDFPRFDGTRLKEW 125
Query: 129 ISRAEIYFKVQETSPEVKVSLAQLSMEG--GTIH--FYNSLLATEEELTWERFRDALLER 184
+ + E +F V T ++KV +A + + T H F S + E W+ + L ER
Sbjct: 126 LFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLEVLYDWKGYVKLLKER 185
Query: 185 YGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKV 244
+ + D D +L L++ + +Y FE + ++ L E+ +L GL+ + + V
Sbjct: 186 FEDDCD-DPMAELKHLQETDGIIDYHQKFELIKTRV-NLSEEYLVSVYLAGLRTDTQMHV 243
Query: 245 RSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFGGNGSRSGFSGATKGNGTEWI 304
R L + + EK NR G G++ K
Sbjct: 244 RMFQPQ---TVRHCLFLGKTYEKAHLKKPANTTWSTNRSAPTG---GYNKYQK------- 290
Query: 305 WVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQLMERRQKGLCFKC 364
+G +T N+ N P Q K +S ++ +RR KGLC+ C
Sbjct: 291 --EGESKTDHYGNKGNFKPVSQQPKK----------------MSQQEMSDRRSKGLCYFC 332
Query: 365 GGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNEDEEETQGELSLMSLCELGMKT 424
Y H K +L ++ D EE ++++ E+ D++E ++S+ ++ +
Sbjct: 333 DEKYTPEHYLVHKKTQLFRMDVD-EEFEDARE---ELVNDDDEHMPQISVNAVSGIAGY- 387
Query: 425 GGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPRMT-VKLGDGYKS 483
+TM+++GT + + +L+DSG+THNF+D +LG +V DT +T V + DG K
Sbjct: 388 ----KTMRVKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKV-DTAGLTRVSVADGRKL 442
Query: 484 QAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDK 543
+ +G+ ++ + L L G D+VLG++WL+TLG + M F +
Sbjct: 443 RVEGKVTDFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNN 502
Query: 544 KWITLQGMDTRGEHMEALQSITATGERKPGL----LGTRNEEKAGKQLPGEINSSQLEE- 598
+ + L G+ + Q + E + L + +E G+ +S+L E
Sbjct: 503 QKVLLHGLTSGSVREVKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEE 562
Query: 599 --LDKLLYRFDVVFQEKQGLPPGRGRE-HCITIQEGKGPVNVRPYRYPHHHKNEIEKQVR 655
++++L + +F E LPP R + H I + EG PVN RPYRY H KNEI+K V
Sbjct: 563 SVVEEVLNEYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVE 622
Query: 656 EMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELH 715
++L+ G ++ S+S Y+SPV+LVKKKD TW +CVDYR LN +TV D FPIP+IE+L+DEL
Sbjct: 623 DLLTNGTVQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELG 682
Query: 716 GAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVF 775
GAV FSK+DL++GYHQVR+ +D+ KTAF+TH GH+EY+VMPFGL NAP+TFQ LMN +F
Sbjct: 683 GAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIF 742
Query: 776 RPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGH 835
+P LR+ VLVFFDDILVYS + +H HL++V EV++ ++L A KC FA VEYLGH
Sbjct: 743 KPFLRKFVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGH 802
Query: 836 LISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKD 895
IS QG+ DP K+ +VK WP+P +K +RGFLGL GYYR+F++++G IA PL LTK D
Sbjct: 803 FISAQGIETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTD 862
Query: 896 AFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFS 955
AF W AQQAFEDLK L APVL+LP F ++F +E DA G GIGA+L+ HP+AY S
Sbjct: 863 AFEWTAVAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYIS 922
Query: 956 KALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQN 1015
+ L + L S YEKEL+AV A++ WR YLL F++ TDQRSLK L Q++ T QQ
Sbjct: 923 RQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQ 982
Query: 1016 WAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKL 1075
W KLL +++EI Y+ GK N ADALSR++ + L ++ + I D +L
Sbjct: 983 WLPKLLEFDYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLLKDIQAGYANDSQL 1042
Query: 1076 CQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRT 1135
I++ + DP + + Q L + ++V+ ++ +L H + GGHSG T
Sbjct: 1043 QDIITALQRDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVT 1102
Query: 1136 YRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFII 1195
++R+ Y +GM K IQ ++ +C CQ+ K ++ GLLQPLPIP+ +W ++S++FI
Sbjct: 1103 HQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIE 1162
Query: 1196 GLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDP 1255
GLP S G I VVVDRL+K +HFI L HPY+A +VA+ + + +LHG P+SI+SDRD
Sbjct: 1163 GLPVSGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFKLHGCPTSIVSDRDV 1222
Query: 1256 LFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHW 1315
+F S FW+E F LQG K++SAYHP+SDGQTE+VNRCLETYLRC D+P+ W+ WL
Sbjct: 1223 VFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLAL 1282
Query: 1316 AEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHL 1375
AEYW+NT+YHS+++ TPFE VYG+ PPV ++ GE++V V + LQ+R++ L LK HL
Sbjct: 1283 AEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHL 1342
Query: 1376 VAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRV 1435
+ AQ RM+ A+ R +EFE+G++V+VK++ +RQ S+ R + KL+ +YFGPY II R
Sbjct: 1343 MRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRC 1402
Query: 1436 GAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRDI 1495
G VAY+L LP +++HPVFH+S LK VGN LP + +++ P V+ R+ +
Sbjct: 1403 GEVAYKLALPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVM--QDVFEKVPEKVVERKMV 1460
Query: 1496 LRQGESISQVLVQWQGKTPEEATWEDLATIRSEFPVF 1532
RQG+++++VLV+W + EEATWE L ++ FP F
Sbjct: 1461 NRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFPEF 1497
>At1g35370 hypothetical protein
Length = 1447
Score = 916 bits (2367), Expect = 0.0
Identities = 509/1195 (42%), Positives = 718/1195 (59%), Gaps = 54/1195 (4%)
Query: 348 SYPQLMERRQK----GLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNE 403
SY + E QK GLC+ C + H K +L ++ D E D A+EV
Sbjct: 289 SYRPVKEVEQKSDHLGLCYFCDEKFTPEHYLVHKKTQLFRMDVDEEFED-----AVEVLS 343
Query: 404 DEEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRL 463
D++ Q + +S+ + +G +TM ++GT++ + +L+DSG+THNF+D + +L
Sbjct: 344 DDDHEQKPMPQISVNAVSGISGY--KTMGVKGTVDKRDLFILIDSGSTHNFIDSTVAAKL 401
Query: 464 GWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLK 523
G V V + DG K G+ G ++ + L L G D+VLG++WL+
Sbjct: 402 GCHVESAGLTKVAVADGRKLNVDGQIKGFTWKLQSTTFQSDILLIPLQGVDMVLGVQWLE 461
Query: 524 TLGDTIVNWDTQLMSFWSDKKWITLQGMDT---RGEHMEALQSITATGERKPGLLGTRNE 580
TLG + M F+ + + L G+ T R LQ T + + ++ R
Sbjct: 462 TLGRISWEFKKLEMQFFYKNQRVWLHGIITGSVRDIKAHKLQK-TQADQIQLAMVCVREV 520
Query: 581 EKAGKQLPGEIN---SSQLEE--LDKLLYRFDVVFQEKQGLPPGRGR-EHCITIQEGKGP 634
+Q G I+ S +EE + ++ F VF E LPP R + +H I + EG P
Sbjct: 521 VSDEEQEIGSISALTSDVVEESVVQNIVEEFPDVFAEPTDLPPFREKHDHKIKLLEGANP 580
Query: 635 VNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALN 694
VN RPYRY H K+EI+K V++M+ +G I+ S+S ++SPV+LVKKKD TW +CVDY LN
Sbjct: 581 VNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFASPVVLVKKKDGTWRLCVDYTELN 640
Query: 695 SVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYM 754
+TV D+F IP+IE+L+DEL G+V FSK+DL++GYHQVR+ +D+ KTAF+TH GH+EY+
Sbjct: 641 GMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYL 700
Query: 755 VMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHH 814
VM FGL NAP+TFQSLMN VFR LR+ VLVFFDDIL+YS + +H HL V EV++ H
Sbjct: 701 VMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLH 760
Query: 815 QLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYY 874
+L A K E+LGH IS + + DP K+ +VK WP P VK VRGFLG GYY
Sbjct: 761 KLFAKGSK--------EHLGHFISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFAGYY 812
Query: 875 RKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECD 934
R+F++N+G IA PL LTK D F W +AQ AF+ LK L APVLALP F ++F +E D
Sbjct: 813 RRFVRNFGVIAGPLHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETD 872
Query: 935 ASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVS 994
A G GI A+L+ HP+AY S+ L + L S YEKEL+A A++ WR YLL F++
Sbjct: 873 ACGQGIRAVLMQKGHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIK 932
Query: 995 TDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRPLV 1054
TDQRSLK L Q++ T QQ W KLL +++EI Y+ GK N ADALSR++ + L +
Sbjct: 933 TDQRSLKYLLEQRLNTPVQQQWLPKLLEFDYEIQYRQGKENLVADALSRVEGSEVLHMAL 992
Query: 1055 SYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLI 1114
S + I + D L I+S + P A Y Q L + ++V+ N +
Sbjct: 993 SIVECDFLKEIQVAYESDGVLKDIISALQQHPDAKKHYSWSQDILRRKSKIVVPNDVEIT 1052
Query: 1115 PKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAG 1174
KLL+ H + GG SG +++R+ + YW+GM K IQ F+ +C CQ+ K ++ G
Sbjct: 1053 NKLLQWLHCSGMGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPG 1112
Query: 1175 LLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEI 1234
LLQPLPIP+++W D+S++FI GLP S G I VVVDRL+K +HF+ L HPY+A +VA+
Sbjct: 1113 LLQPLPIPDKIWCDVSMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQA 1172
Query: 1235 FAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCL 1294
F + + HG P+SI+SDRD LF S FWKE F+LQG + +MSSAYHP+SDGQTE+VNRCL
Sbjct: 1173 FLDNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCL 1232
Query: 1295 ETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRV 1354
E YLRC +P W WL AEYW+NT+YHS++Q TPFE VYG+ PP+ ++ G+++V
Sbjct: 1233 ENYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKV 1292
Query: 1355 EAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLA 1414
V + LQ+R+ L LK HL+ AQ RM+ A+ R + F++G++V+VK++ +RQ S+
Sbjct: 1293 AVVARSLQERENMLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVV 1352
Query: 1415 NRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGLPE 1474
RV+ KL+ +YFGPY II + G V VGN LP
Sbjct: 1353 LRVNQKLSPKYFGPYKIIEKCGEV-----------------------MVGNVTTSTQLPS 1389
Query: 1475 QLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEF 1529
L ++ P +L R+ + RQG + + VLV+W G+ EEATW+ L + +F
Sbjct: 1390 VL--PDIFEKAPEYILERKLVKRQGRAATMVLVKWIGEPVEEATWKFLFDRQQKF 1442
Score = 49.3 bits (116), Expect = 2e-05
Identities = 35/136 (25%), Positives = 63/136 (45%), Gaps = 6/136 (4%)
Query: 114 KVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSME--GGTIH--FYNSLLATE 169
K++ P FDG + W+ + E +F V T E+KV + + + T H F S + +
Sbjct: 116 KIDFPRFDGSRINEWLFKVEEFFGVDFTPEEMKVKMVAIHFDSHAATWHHSFIQSGIGLD 175
Query: 170 EELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQ 229
W + L +R+ D D +L +L++ + EY FE + ++ L E+
Sbjct: 176 VFFNWPEYVKLLKDRFEDACD-DPMAELKKLQETDGIVEYHQQFELIKVRL-NLSEEYLV 233
Query: 230 GYFLHGLKEEIRGKVR 245
+L GL+ + + VR
Sbjct: 234 SVYLAGLRTDTQMHVR 249
>At2g10780 pseudogene
Length = 1611
Score = 671 bits (1730), Expect = 0.0
Identities = 478/1532 (31%), Positives = 743/1532 (48%), Gaps = 117/1532 (7%)
Query: 46 MKDLPTTLGTMVEKAVGKSVGIDVDSGDARPEREPREKTPESVGLRESGSDQPVLQGEAL 105
MK + L T+ + + V + VD+ + E E P+ R+ G +L+ +
Sbjct: 118 MKGMMAQLATLTKAVIADPVVLKVDN-QKDDDSEVVEVNPQPNHGRKRGDYLSLLEHVSR 176
Query: 106 NEFRQSVKKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSL 165
R + + + D W SR + FK + + +A +EG +++ ++
Sbjct: 177 LGTRHFMGSTDPIVADE-----WRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTV 231
Query: 166 LAT--EEELTWERFRDALLERYGGNGDGDVYE--QLSELRQQGIVEEYITDFEYLTAQIP 221
+E ++ F D ++Y D E L ++ V EY +F L +
Sbjct: 232 EKRRGDEVRSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRYVG 291
Query: 222 K-LPEKQYQ-GYFLHGLKEEIRGK--VRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQ 277
+ L E+Q Q F+ GL+ EIR VR+ ++ + R ++ +E+E +
Sbjct: 292 RELEEEQAQLRRFIRGLRIEIRNHCLVRTFNSVSELVE-RAAMIEEGIEEERYLNREKAP 350
Query: 278 ARANRFGGNGSRSGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRA 337
R N+ + K + T K + +TG+ V N G K G R
Sbjct: 351 IRNNQSTKPADKK--RKFDKVDNT-----KSDAKTGECVTC-GKNHSGTCWKAIGACGRC 402
Query: 338 GPRDRGFNHLSYPQLMERRQKGL------CFKCGGPYHRNHVCPDKHLRLLILEEDGEEL 391
G +D S P++ + K L CF CG H CP
Sbjct: 403 GSKDHAIQ--SCPKMEPGQSKVLGEETRTCFYCGKTGHLKRECPK--------------- 445
Query: 392 DESKMLAMEVNEDEEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGAT 451
L E + + +G G+P K + + V +
Sbjct: 446 -----LTAEKQAGQRDNRGG-------------NGLPPPPKRQAVASRV-----YELSEE 482
Query: 452 HNFVDCFLVRRLGW-EVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDL 510
N F G+ + +T V+ G G G+ + + ++ + L
Sbjct: 483 ANDAGNFRAITGGFRKEPNTDYGMVRAAGGQAMYPTGLVRGISVVVNGVNMPADLIIVPL 542
Query: 511 GGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHM--EALQSITATG 568
D++LG++WL ++ + F D+ + QG+ T + A+Q+ G
Sbjct: 543 KKHDVILGMDWLGKY-KAHIDCHRGRIQFERDEGMLKFQGIRTTSGSLVISAIQAERMLG 601
Query: 569 ERKPGLLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITI 628
+ L T ++ G S++L+++ ++ F VF G+PP R I +
Sbjct: 602 KGCEAYLATITTKEVGA-------SAELKDIP-IVNEFSDVFAAVSGVPPDRSDPFTIEL 653
Query: 629 QEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCV 688
+ G P++ PYR +++KQ+ E+L G IR S+S + +PV+ VKKKD ++ +C+
Sbjct: 654 EPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKDGSFRLCI 713
Query: 689 DYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHE 748
DYR LN VTV +K+P+P I+EL+D+L GA +FSK+DL SGYHQ+ + DV KTAFRT
Sbjct: 714 DYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRY 773
Query: 749 GHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVL 808
H+E++VMPFGL NAP+ F +MN VFR L V++F +DILVYSK+W H HL VL
Sbjct: 774 DHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVL 833
Query: 809 EVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFL 868
E L+ H+L A KC F + V +LGH+IS+QGV+VDP K+ S+K WP P+N +R FL
Sbjct: 834 ERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFL 893
Query: 869 GLTGYYRKFIQNYGKIARPLTELTKKD-AFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQ 927
GL GYYR+F+ ++ +A+PLT LT KD AFNW + +++F +LK LT APVL LP+ +
Sbjct: 894 GLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVLPEEGE 953
Query: 928 EFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLL 987
+ + DAS +G+G +L+ IAY S+ L ++ E+ AV ++ WR YL
Sbjct: 954 PYTVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLY 1013
Query: 988 GRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALS--RID 1045
G K + TD +SLK Q + Q+ W + YN +I Y PGK NQ ADALS R +
Sbjct: 1014 GAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVADYNLDIAYHPGKANQVADALSRRRSE 1073
Query: 1046 EAGELRPLVSYPLWEEQGVIADENQKDP------KLCQIVSEVALDPQAHPGYR------ 1093
E + + V A + +P ++S + L + +
Sbjct: 1074 VEAERSQVDLVNMMGTLHVNALSKEVEPLGLGAADQADLLSRIRLAQERDEEIKGWAQNN 1133
Query: 1094 ------VVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRG 1147
GT++ RV + N +L ++L E H + H G + YR + +W G
Sbjct: 1134 KTEYQTSNNGTIVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVG 1193
Query: 1148 MTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPR--SRGFEA 1205
M K + +V C CQ K P+GLLQ LPIPE W+ I+++F+ GLP A
Sbjct: 1194 MKKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNA 1253
Query: 1206 IFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEM 1265
++VVVDRLTK +HF+ + A +AE + EI+RLHG+P SI+SDRD F S FWK
Sbjct: 1254 VWVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAF 1313
Query: 1266 FRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYH 1325
+ GT+ +S+AYHP++D Q+E + LE LR D W +L E+ +N S+
Sbjct: 1314 QKALGTRVNLSTAYHPQTDEQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQ 1373
Query: 1326 SATQQTPFEAVYGRKPPVLTRWV-LGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRA 1384
++ +P+EA+YGR W +GE R+ + + E ++ LK L AQ+R ++
Sbjct: 1374 ASIGMSPYEALYGRACRTPLCWTPVGERRLFG-PTIVDETTERMKFLKIKLKEAQDRQKS 1432
Query: 1385 QANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKL 1444
AN +RK EF+VG+ V++K ++ KL+ RY GPY +I RVGAVAY+L L
Sbjct: 1433 YANKRRKELEFQVGDLVYLKAMTYKGAGRFTS-RKKLSPRYVGPYKVIERVGAVAYKLDL 1491
Query: 1445 PEGAR-IHPVFHISLLKKAVGNYVEE-----PGLPEQLDGEELPSIQPALVLARRDILRQ 1498
P H VFH+S L+K + + E PGL E + E P ++ R +
Sbjct: 1492 PPKLNAFHNVFHVSQLRKCLSDQEESVEDIPPGLKENMTVE----AWPVRIMDRMTKGTR 1547
Query: 1499 GESISQVLVQWQGKTPEEATWEDLATIRSEFP 1530
G++ + V W + EE TWE +++ FP
Sbjct: 1548 GKARDLLKVLWNCRGREEYTWETENKMKANFP 1579
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 623 bits (1607), Expect = e-178
Identities = 388/1109 (34%), Positives = 574/1109 (50%), Gaps = 74/1109 (6%)
Query: 449 GATHNFVDCFLVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLF 508
G + C + + + + VK+ G GR G+ I++ + +
Sbjct: 339 GGRAGYFSCGSLDHTVADCTERGHLHVKVAGGQFLAVLGRAKGVDIQIAGESMPADLIIS 398
Query: 509 DLGGPDIVLGIEWLKTLGDTIVNWDTQL--MSFWSDKKWITLQGMDTRGEHMEALQSITA 566
+ D++LG++WL V+ D +SF + + QG+ + + ++ A
Sbjct: 399 PVELYDVILGMDWLDHYR---VHLDCHRGRVSFERPEGRLVYQGVRPTSGSL-VISAVQA 454
Query: 567 TGERKPG----LLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGR 622
+ G L+ E G+ +I +++ F+ VFQ QGLPP R
Sbjct: 455 KKMIEKGCEAYLVTISMPESLGQVAVSDI---------RVIQEFEDVFQSLQGLPPSRSD 505
Query: 623 EHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQ 682
I ++ G P++ PYR E++KQ+ ++L G IR STS + +PV+ VKKKD
Sbjct: 506 PFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKDG 565
Query: 683 TW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKT 742
++ +C+DYR LN VTV +K+P+P I+ELLD+L GA FSK+DL SGYHQ+ + E DV KT
Sbjct: 566 SFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKT 625
Query: 743 AFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHML 802
AFRT GH+E++VMPF L NAP+ F LMN VF+ L V++F DDILVYSK+ +H +
Sbjct: 626 AFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEV 685
Query: 803 HLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVK 862
HL RV+E L+ +L A KC F + + +LGH++S +GV+VDP K+ +++ WP P N
Sbjct: 686 HLRRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNAT 745
Query: 863 GVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDA-FNWGKQAQQAFEDLKEKLTTAPVLA 921
+R FL LTGYYR+F++ + +A+P+T+LT KD F W + ++ F LKE LT+ PVLA
Sbjct: 746 EIRSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLA 805
Query: 922 LPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQH 981
LP+ Q + + DAS +G+G +L+ IAY S+ L ++ E+ V A++
Sbjct: 806 LPEHGQPYMVYTDASRVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKI 865
Query: 982 WRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADAL 1041
WR YL G K V TD +SLK +Q + Q W + Y+ EI Y PGK N ADAL
Sbjct: 866 WRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQMRWMELVADYDLEIAYHPGKANVVADAL 925
Query: 1042 SR--------------IDEAGELRPLV--SYPLWEEQGVIADENQKDPKLCQIVSEVALD 1085
SR + E G LR V PL E AD + +L Q E +
Sbjct: 926 SRKRVGAAPGQSVEALVSEIGALRLCVVAREPLGLEAVDRADLLTR-ARLAQEKDEGLIA 984
Query: 1086 PQAHPGYR---VVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATN 1142
G GT+ RV + L ++L E H + H G + YR +
Sbjct: 985 ASKAEGSEYQFAANGTIFVYGRVCVPKDEELRREILSEAHASMFSIHPGATKMYRDLKRY 1044
Query: 1143 LYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRG 1202
W GM + + +V CDVCQ K P
Sbjct: 1045 YQWVGMKRDVANWVAECDVCQLVKAEHQVP------------------------------ 1074
Query: 1203 FEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFW 1262
+AI+V++DRLTK +HF+ ++ A +A+ + EI++LHGVP SI+SDRD F FW
Sbjct: 1075 -DAIWVIMDRLTKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFW 1133
Query: 1263 KEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNT 1322
+ GT+ +MS+AYHP++DGQ+E + LE LR D WA L E+ +N
Sbjct: 1134 RAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNN 1193
Query: 1323 SYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERM 1382
SY ++ PFEA+YGR RW E R +Q+ E +R LK ++ AQ R
Sbjct: 1194 SYQASIGMAPFEALYGRPCWTPLRWTQVEERSIYGADYVQETTERIRVLKLNMKEAQARQ 1253
Query: 1383 RAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRL 1442
R+ A+ +R+ EFEVG+ V++K+ R + + + KL+ RY GP+ I+ RVG VAYRL
Sbjct: 1254 RSYADKRRRELEFEVGDRVYLKMAMLRGPN-RSILETKLSPRYMGPFRIVERVGPVAYRL 1312
Query: 1443 KLPEGAR-IHPVFHISLLKKAVGNYVEE-PGLPEQLDGEELPSIQPALVLARRDILRQGE 1500
+LP+ R H VFH+ +L+K + E +PE L +P VL RR + +
Sbjct: 1313 ELPDVMRAFHKVFHVLMLRKCLHKDDEVLVKIPEDLQPNMTLEARPVRVLERRIKELRRK 1372
Query: 1501 SISQVLVQWQGKTPEEATWEDLATIRSEF 1529
I + V W E TWE A +++ F
Sbjct: 1373 KIPLIKVLWDCDGVTEETWEPEARMKARF 1401
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 613 bits (1580), Expect = e-175
Identities = 388/1101 (35%), Positives = 578/1101 (52%), Gaps = 95/1101 (8%)
Query: 437 INGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPRM-TVKLGDGYKSQAQGRCAGLKIE 495
+ GV VL DSGA+H F+ R ++ VK+ G GR G+ I+
Sbjct: 308 VGGVEAHVLFDSGASHCFITPESASRGNIRGDPGEQLGAVKVAGGQFVAVLGRTKGVDIQ 367
Query: 496 MGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRG 555
+ + + + D++LG++WL D
Sbjct: 368 IAGESMPADLIISPVELYDVILGMDWL-----------------------------DHYR 398
Query: 556 EHMEALQSITATGERKPGLLGTRNEEKAGKQLPGEINSSQLEELDKLLY-RFDVVFQEKQ 614
H++ + + + L+ +G + + + ++ E Y F+ VFQ Q
Sbjct: 399 VHLDCHRGRVSFERPEGSLVYQGVRPTSGSLVISAVQAEKMIEKGCEAYLEFEDVFQSLQ 458
Query: 615 GLPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPV 674
GLPP R I ++ G P++ PYR E++KQ+ ++L A PV
Sbjct: 459 GLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGA------------PV 506
Query: 675 ILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRV 734
+ VKKKD ++ +C+DYR LN VTV +K+P+P I+ELLD+L GA FSK+DL SGYH + +
Sbjct: 507 LFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPI 566
Query: 735 REEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYS 794
E DV KTAFRT GH+E++VMPFGL NAP+ F LMN VF+ +L V++F DDILVYS
Sbjct: 567 AEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYS 626
Query: 795 KTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKS 854
K+ +H +HL RV+E L+ +L A KC F + + +LGH++S +GV+VDP K+ +++
Sbjct: 627 KSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRD 686
Query: 855 WPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDA-FNWGKQAQQAFEDLKEK 913
W P N +R FLGL GYYR+F++ + +A+P+T+LT KD F W + ++ F LKE
Sbjct: 687 WHTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEM 746
Query: 914 LTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELM 973
LT+ PVLALP+ + + + DASG+G+G +L+ IAY S+ L ++ E+
Sbjct: 747 LTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMA 806
Query: 974 AVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGK 1033
AV A++ WR YL G V TD +SLK Q + Q+ W + Y+ EI Y PGK
Sbjct: 807 AVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLEIAYHPGK 866
Query: 1034 TNQGADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYR 1093
N ADALS G L E G + +LC + E L +A
Sbjct: 867 ANVVADALSH-KRVGAAPGQSVEALVSEIGAL--------RLCAVARE-PLGLEA----- 911
Query: 1094 VVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQ 1153
V + LL + R+ LI Y ++ +
Sbjct: 912 VDRADLLTRVRLAQEKDEGLIA---------------------------AYKAEGSEDVA 944
Query: 1154 EFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRL 1213
+V CDVCQ K P G+LQ LPIPE W+ I+++F++GLP SR +AI+V+VDRL
Sbjct: 945 NWVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTKDAIWVIVDRL 1004
Query: 1214 TKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQF 1273
TK +HF+ ++ A +A+ + EI++LHGVP SI+SDRD F S FW+ GT+
Sbjct: 1005 TKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKV 1064
Query: 1274 KMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPF 1333
+MS+AYHP++ GQ+E + LE LR D WA L E+ +N SY ++ PF
Sbjct: 1065 QMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASIGMAPF 1124
Query: 1334 EAVYG---RKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKR 1390
EA+Y R P LT+ +GE + + +Q+ E +R LK ++ AQ+R R+ A+ +R
Sbjct: 1125 EALYERPCRTPLCLTQ--VGERSIYGADY-VQETTERIRVLKLNMKEAQDRQRSYADKRR 1181
Query: 1391 KHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGAR- 1449
+ EFEVG+ V++K+ R + + KL+ RY GP+ I+ RVG VAYRL+LP+ R
Sbjct: 1182 RELEFEVGDRVYLKMAMLRGPN-RSISETKLSPRYMGPFRIVERVGPVAYRLELPDVMRA 1240
Query: 1450 IHPVFHISLLKKAVGNYVEE-PGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQ 1508
H VFH+S+L+K + E +PE L +P VL RR + + I + V
Sbjct: 1241 FHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVL 1300
Query: 1509 WQGKTPEEATWEDLATIRSEF 1529
W + TWE A +++ F
Sbjct: 1301 WDCDGVTKETWEPEARMKARF 1321
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 568 bits (1465), Expect = e-162
Identities = 299/663 (45%), Positives = 406/663 (61%), Gaps = 52/663 (7%)
Query: 599 LDKLLYRFDVVFQEKQGLPPGRG-REHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREM 657
++K++ F +F E LPP R +H I + E PVN RPYRY H KNEI+K V +M
Sbjct: 7 VEKVVTEFPDIFVEPTDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDM 66
Query: 658 LSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGA 717
L++G I+ S+S Y+SPV+LVKKKD TW +CVDYR LN +TV D+FPIP+IE+L+DEL G+
Sbjct: 67 LASGTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGS 126
Query: 718 VFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRP 777
+SK+DL++GYHQVR+ D+HKTAF+TH GHYEY+VMPFGL NAP++FQSLMN F+P
Sbjct: 127 NVYSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKP 186
Query: 778 MLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLI 837
LR+ VLVFFDDIL+YS + +H HLE V EV++ H L A KC FA VEYLGH I
Sbjct: 187 FLRKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFI 246
Query: 838 SEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAF 897
S +G+A DP K+ +V+ WP P N+K + GFLGLTGYYR+F
Sbjct: 247 SGEGIATDPAKIKAVQDWPVPVNLKQLCGFLGLTGYYRRF-------------------- 286
Query: 898 NWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKA 957
F +E DA G GIGA+L+ HP+A+ S+
Sbjct: 287 -------------------------------FVVEMDACGHGIGAVLMQEGHPLAFISRQ 315
Query: 958 LGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWA 1017
L + L S YEKEL+AV ++ WR YL+ F++ TDQRSLK L Q++ T QQ W
Sbjct: 316 LKGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRLNTPIQQQWL 375
Query: 1018 AKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKLCQ 1077
KLL +++EI YK GK N ADALSR++ + L +S + I D +
Sbjct: 376 PKLLEFDYEIQYKQGKENLVADALSRVEGSEVLHMALSVVECDLLKEIQAGYVTDGDIQG 435
Query: 1078 IVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYR 1137
I++ + + Y QG L ++++V+ N S + +L H + GGHSG T++
Sbjct: 436 IITILQQQADSKKHYTWSQGVLRRKNKIVVPNNSGIKDTILRWLHCSGMGGHSGKEVTHQ 495
Query: 1138 RIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGL 1197
R+ YW+ M K IQ F+ +C CQ+ K ++ GLLQPLPIP+R+W D+S++FI GL
Sbjct: 496 RVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPIPDRIWSDVSMDFIDGL 555
Query: 1198 PRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLF 1257
P S G I VVVDRL+K +HFI L HPY+A +VA+ + + +LHG PSSI+ +
Sbjct: 556 PLSNGKTVIMVVVDRLSKAAHFIALAHPYSAMTVAQAYLDNVFKLHGCPSSIVVSSRRVL 615
Query: 1258 VSH 1260
V H
Sbjct: 616 VQH 618
Score = 132 bits (333), Expect = 1e-30
Identities = 63/139 (45%), Positives = 96/139 (68%)
Query: 1326 SATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQ 1385
S + TP+EAVYG+ PP+ ++ GE++V V + +Q+R+ + LK HL+ AQ RM+
Sbjct: 621 SHSTMTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQL 680
Query: 1386 ANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLP 1445
A+ +EFEVG++VFVK++ +RQ S+ R KL+ +YFGPY +I R G VAY+L+LP
Sbjct: 681 ADQHITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLP 740
Query: 1446 EGARIHPVFHISLLKKAVG 1464
+++HPVFH+S L+ VG
Sbjct: 741 ANSQVHPVFHVSQLRVLVG 759
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 477 bits (1227), Expect = e-134
Identities = 281/773 (36%), Positives = 421/773 (54%), Gaps = 48/773 (6%)
Query: 487 GRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWI 546
GR G+ I++ + + + D++LG++WL ++W + F + +
Sbjct: 351 GRAKGVDIQIAGESMPADLIISPVELYDVILGMDWLDYYR-VHLDWHRGRVFFERPEGRL 409
Query: 547 TLQGMDTRGEHMEALQSITATGERKPG----LLGTRNEEKAGKQLPGEINSSQLEELDKL 602
QG+ + + ++ A + G L+ E G+ +I ++
Sbjct: 410 VYQGVRPISGSL-VISAVQAEKMIEKGCEAYLVTISMPESVGQVAVSDI---------RV 459
Query: 603 LYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGI 662
+ F VFQ QGLPP + I ++ G P++ PYR E++KQ++++L G
Sbjct: 460 VQEFQDVFQSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGF 519
Query: 663 IRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSK 722
IR STS + +PV+ VKKKD ++ +C+DYR LN VTV +++P+P I+ELLD+L GA FSK
Sbjct: 520 IRPSTSPWGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSK 579
Query: 723 LDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRG 782
+DL SGYHQ+ + E DV KTAFRT GH+E++VMPFGL NAP+ F LMN VF+ L
Sbjct: 580 IDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEF 639
Query: 783 VLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGV 842
V++F DDILVYSK+ + +HL RV+E L+ +L A KC F + + +LGH++S +GV
Sbjct: 640 VIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGV 699
Query: 843 AVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDA-FNWGK 901
+VDP K+ +++ WP P N +R FLG GYYR+F++ + +A+P+T+LT KD F W +
Sbjct: 700 SVDPEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQ 759
Query: 902 QAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPR 961
+ ++ F LKE LT+ PVLALP+ Q + + DAS +G+G +L+ IAY S+ L
Sbjct: 760 ECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLMQHGKVIAYASRQLMKH 819
Query: 962 NLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLL 1021
++ E+ AV A++ WR YL G K V TD +SLK Q + Q+ W +
Sbjct: 820 EGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVA 879
Query: 1022 GYNFEIIYKPGKTNQGADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKLCQIVSE 1081
Y+ EI Y PGK N DALSR L V L E G + +LC + E
Sbjct: 880 DYDLEIAYHPGKANVVVDALSRKRVGAALGQSVEV-LVSEIGAL--------RLCAVARE 930
Query: 1082 VALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIAT 1141
G V DR + + L K E T + YR +
Sbjct: 931 PL-------GLEAV-------DRADLLTRVRLAQKKDEGLRAT---------KMYRDLKR 967
Query: 1142 NLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSR 1201
W GM + +V CDVCQ K G+LQ LPIPE W+ I+++ ++GL SR
Sbjct: 968 YYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGLRVSR 1027
Query: 1202 GFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRD 1254
+AI+V+VDRLTK +HF+ ++ A +A+ F EI++LHGVP ++ +D
Sbjct: 1028 TKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNMKEAQD 1080
Score = 87.0 bits (214), Expect = 8e-17
Identities = 56/154 (36%), Positives = 84/154 (54%), Gaps = 3/154 (1%)
Query: 1378 AQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGA 1437
AQ+R R+ A+ +R+ EFEVG+ V++K+ R + + KL+ RY GP+ I+ RV
Sbjct: 1078 AQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSIS-ETKLSPRYMGPFKIVERVEP 1136
Query: 1438 VAYRLKLPEGAR-IHPVFHISLLKKAVGNYVEE-PGLPEQLDGEELPSIQPALVLARRDI 1495
VAYRL+LP+ R H VFH+S+L+K + E +PE L +P VL RR
Sbjct: 1137 VAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEALAKIPEDLQPNMTLEARPVRVLERRIK 1196
Query: 1496 LRQGESISQVLVQWQGKTPEEATWEDLATIRSEF 1529
+ + I + V W E TWE A +++ F
Sbjct: 1197 ELRQKKIPLIKVLWDCDGVTEETWEPEARMKARF 1230
>At3g31970 hypothetical protein
Length = 1329
Score = 438 bits (1127), Expect = e-122
Identities = 285/837 (34%), Positives = 422/837 (50%), Gaps = 87/837 (10%)
Query: 711 LDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSL 770
+ E G V S + + + + + E +V KTAFRT GH+E++VMPFGL N P+ F L
Sbjct: 552 MPESVGQVAVSDIRVVHEFEDIPIAEANVRKTAFRTRYGHFEFVVMPFGLTNIPTAFMRL 611
Query: 771 MNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFV 830
MN VF+ L V++F DDILVYSK+ +H + E
Sbjct: 612 MNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVQSEE---------------------SDG 650
Query: 831 EYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTE 890
E G +E GV+VD K+ +++ WP P N +R FLGL GYY++F++ + +A+P+T+
Sbjct: 651 EAAG---AEVGVSVDLEKIEAIRDWPRPTNATEIRSFLGLAGYYKRFVKVFASMAQPMTK 707
Query: 891 LTKKDA-FNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKH 949
LT KD F W + + F LKE LT+ PVLALP+ + + + DAS +G+ +L+
Sbjct: 708 LTGKDVPFVWSPECDEGFMSLKEMLTSTPVLALPEHGEPYMVYTDASRVGLDCVLM---- 763
Query: 950 PIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIV 1009
++ V TD +SLK Q +
Sbjct: 764 ---------------------------------------QRGKVFTDHKSLKYIFTQPEL 784
Query: 1010 TGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRPLVSYPLWEEQGVIADE- 1068
Q+ W + Y+ EI Y GK N ADALSR G + LVS V+A E
Sbjct: 785 NLRQRRWMELVADYDLEIAYHSGKANVVADALSRKRVGGSVEALVSEIGALRLCVMAQEP 844
Query: 1069 -NQKDPKLCQIVSEVALDPQAHPG-----------YR-VVQGTLLYQDRVVISNKSSLIP 1115
+ +++ V L + G Y+ GT+L RV + L
Sbjct: 845 LGLEAVDRADLLTRVRLAQEKDEGLIAASKADGSEYQFAANGTILVHGRVCVPKDEELRR 904
Query: 1116 KLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGL 1175
++L E H + H + YR + W GM + + +V CDVCQ K P GL
Sbjct: 905 EILSEAHASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGL 964
Query: 1176 LQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIF 1235
LQ LPI E W+ I+++F++GLP SR +AI+V+VDRLTK +HF+ ++ A +A+ +
Sbjct: 965 LQSLPILEWKWDFITMDFVVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAVLLAKKY 1024
Query: 1236 AKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLE 1295
EI+ LHGVP SI+SDRD F S FW+ GT+ +MS+AYHP++DGQ+E + LE
Sbjct: 1025 VSEIVELHGVPVSIVSDRDSKFTSAFWRAFQGEMGTKVQMSTAYHPQTDGQSERTIQTLE 1084
Query: 1296 TYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWV-LGETRV 1354
LR D+ WA L E+ +N SY ++ + PFEA+YGR W +GE +
Sbjct: 1085 DMLRMCVLDRGGHWADHLSLVEFAYNNSYQASIRMAPFEALYGRPCRTPLCWTQVGERSI 1144
Query: 1355 EAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLA 1414
+ L + E +R LK ++ AQ+R R+ A+ +R+ EFEVG+ V++K+ R +
Sbjct: 1145 YGADYVL-ETTERIRVLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPN-R 1202
Query: 1415 NRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGAR-IHPVFHISLLKKAVGNYVEE-PGL 1472
+ KL RY GP+ I+ RVG VAYRL+LP+ R H VFH+S+L+K + E +
Sbjct: 1203 SISETKLTPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKI 1262
Query: 1473 PEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEF 1529
E L +P +L RR + + I + V W E TWE A +++ F
Sbjct: 1263 LEDLQPNMTLEARPVRILERRIKELRRKKIPLIKVLWNCDGVTEETWEPEARMKASF 1319
>At2g06890 putative retroelement integrase
Length = 1215
Score = 430 bits (1106), Expect = e-120
Identities = 315/952 (33%), Positives = 468/952 (49%), Gaps = 90/952 (9%)
Query: 286 NGSRSGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFN 345
+ SRS + T T + + T N+P +PR + A +G
Sbjct: 97 SSSRSSYGSGTIAKPT----YQREERTSSYHNKPIVSPRAESKPYA-----AVQDHKGKA 147
Query: 346 HLSYPQLMERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNEDE 405
+S ++ + R C+KC G H + CP+K + +L+ + E +E + E+E
Sbjct: 148 EISTSRVRDVR----CYKCQGKGHYANECPNKRVMILLDNGEIEPEEEIPDSPSSLKENE 203
Query: 406 E-ETQGELSLMSLCELGMKTGGIPRTMK-----LRGTINGVPVVVLVDSGATHNFVDCFL 459
E QGEL L++ L ++T + + R ++G +++D G+ N +
Sbjct: 204 ELPAQGEL-LVARRTLSVQTKTDEQEQRKNLFHTRCHVHGKVCSLIIDGGSCTNVASETM 262
Query: 460 VRRLGWEVV-DTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLG 518
V++LG + + D+ +M VK + I +G+Y + + I+LG
Sbjct: 263 VKKLGLKWLNDSGKMRVK-----------NQVVVPIVIGKYEDEILCDVLPMEAGHILLG 311
Query: 519 IEWL---KTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHMEALQSITATGERKPGLL 575
W K + D N + F K + + L+ ++P
Sbjct: 312 RPWQSDRKVMHDGFTNRHS--FEFKGGKTILVSMTPHEVYQDQIHLKQKKEQVVKQPNFF 369
Query: 576 GTRNEEKAG-------------KQLPGEINSSQL--EELDKLLYRFDVVFQEK--QGLPP 618
E K+ + L N + + E+ LL + VF E +GLPP
Sbjct: 370 AKSGEVKSAYSSKQPMLLFVFKEALTSLTNFAPVLPSEMTSLLQDYKDVFPEDNPKGLPP 429
Query: 619 GRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVK 678
RG EH I G N YR E+++Q
Sbjct: 430 IRGIEHQIDFVPGASLPNRPAYRTNPVETKELQRQ------------------------- 464
Query: 679 KKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREED 738
KD +W MC D RA+N+VTV PIP ++++LDELHG+ FSK+DLKSGYHQ+R+ E D
Sbjct: 465 -KDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKIDLKSGYHQIRMNEGD 523
Query: 739 VHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWS 798
KTAF+T G YE++VMPFGL +APSTF LMN V R + V+V+FDDILVYS++
Sbjct: 524 EWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFVIVYFDDILVYSESLR 583
Query: 799 DHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEP 858
+H+ HL+ VL VL+ +L AN KKC F T + +LG ++S GV VD KV +++ WP P
Sbjct: 584 EHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVKVDEEKVKAIRDWPSP 643
Query: 859 KNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKD-AFNWGKQAQQAFEDLKEKLTTA 917
K V VR F GL G+YR+F +++ I PLTE+ KKD F W K ++AF+ LK+KLT A
Sbjct: 644 KTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKAQEEAFQSLKDKLTNA 703
Query: 918 PVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVAL 977
PVL L +F + F +ECDASGIGIGA+L+ + IA+FS+ LG L Y+KEL A+
Sbjct: 704 PVLILSEFLKTFEIECDASGIGIGAVLMQDQKLIAFFSEKLGGATLNYPTYDKELYALVR 763
Query: 978 AIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQG 1037
A+Q W+ YL + FV+ TD SLK Q+ + W + + + I YK GK N
Sbjct: 764 ALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLNKRHARWVEFIETFAYVIKYKKGKDNVV 823
Query: 1038 ADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQG 1097
ADALS+ L L + EQ I + + D ++ A + A Y
Sbjct: 824 ADALSQ--RYTLLSTLNVKLMGFEQ--IKEVYETDHDFQEVYK--ACEKFASGRYFRQDK 877
Query: 1098 TLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV* 1157
L Y++R+ + N SL + E H GH G +T + + W M ++
Sbjct: 878 FLFYENRLCVPN-CSLRDLFVREAHGGGLMGHFGIAKTLEVMTEHFRWPHMKCDVKRICG 936
Query: 1158 ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVV 1209
C+ C++ K P GL PLPIP+ W DIS++F++GLPR+ G ++IFVV
Sbjct: 937 RCNTCKQAK-SKIQPNGLYTPLPIPKHPWNDISMDFVMGLPRT-GKDSIFVV 986
Score = 48.1 bits (113), Expect = 4e-05
Identities = 44/158 (27%), Positives = 78/158 (48%), Gaps = 12/158 (7%)
Query: 1331 TPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHL---VAAQERMRA-QA 1386
+PF+ VYG P + ++ E V D + + E ++Q+ ++ + + ++ A QA
Sbjct: 988 SPFQIVYGFNP-ISPFDLIPLPLSERVSIDGKKKAELVQQIHENARRNIEEKTKLYAKQA 1046
Query: 1387 NSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPE 1446
N R+ Q FEVG+ V++ +R R + +KL R GP+ II R+ AY+L L +
Sbjct: 1047 NKGRREQIFEVGDMVWIHLRKER---FPAQRKSKLMPRIDGPFKIIKRINDNAYQLDLQD 1103
Query: 1447 --GARIHPVFH--ISLLKKAVGNYVEEPGLPEQLDGEE 1480
R +P ++ ++ + EP L +L EE
Sbjct: 1104 EPDLRSNPFQEGGDDMIMDSINDMEHEPELERELVAEE 1141
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 380 bits (976), Expect = e-105
Identities = 198/478 (41%), Positives = 296/478 (61%), Gaps = 14/478 (2%)
Query: 514 DIVLGIEWL-KTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHM--EALQSITATGER 570
D++LG++WL K G ++ + + F D+ + QG+ T + A+Q+ G+
Sbjct: 41 DVILGMDWLGKYKGH--IDCHRERIQFERDEGMLKFQGIRTTSGSLVISAIQAERMLGKG 98
Query: 571 KPGLLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQE 630
L T ++ G S++L+++ ++ F VF G+PP R I ++
Sbjct: 99 CEAYLATITTKEVGA-------SAELKDI-LIVNEFSDVFAAVSGVPPDRSDPFTIELEP 150
Query: 631 GKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDY 690
G P++ PYR E++KQ+ E+L+ G IR S+S + +PV+ VKKKD ++ +C+DY
Sbjct: 151 GTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGSFRLCIDY 210
Query: 691 RALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGH 750
R LN VTV +K+P+P I+EL+D+L GA +FSK+DL SGYHQ+ + DV KTAFRT GH
Sbjct: 211 RGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGH 270
Query: 751 YEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEV 810
+E++VMPFGL NAP+ F +MN VFR L V++F DDILV+SK+W H HL VLE
Sbjct: 271 FEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLER 330
Query: 811 LQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGL 870
L+ H+L A K F + V +LGH+IS+QGV+VDP K+ S+K WP P+N +R FLGL
Sbjct: 331 LREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGL 390
Query: 871 TGYYRKFIQNYGKIARPLTELTKKD-AFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEF 929
GYYR+F+ ++ +A+PLT LT KD AFNW + +++F +LK L APVL LP+ + +
Sbjct: 391 AGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLPEEGEPY 450
Query: 930 RLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLL 987
+ DAS +G+G +L+ IAY S+ L ++ E+ AV A++ WR YL+
Sbjct: 451 TVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIWRSYLI 508
Score = 252 bits (643), Expect = 1e-66
Identities = 147/395 (37%), Positives = 217/395 (54%), Gaps = 11/395 (2%)
Query: 1097 GTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV 1156
GT++ RV + N +L ++L E H + H G + YR + +W GM K + +V
Sbjct: 535 GTIVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMRKDVARWV 594
Query: 1157 *ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRS--RGFEAIFVVVDRLT 1214
C CQ K P+GLLQ LPI E W+ I+++F+ LP A++VVVDRLT
Sbjct: 595 AKCPTCQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLT 654
Query: 1215 KYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFK 1274
K +HF+ + A +AE + EI+RLHG+P SI+SDRD F S FW + GT+
Sbjct: 655 KSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVN 714
Query: 1275 MSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFE 1334
+S+AYHP++DGQ+E + LE LR D W +L E+ +N S+ ++ +P+E
Sbjct: 715 LSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYE 774
Query: 1335 AVYGRKPPVLTRWV-LGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQ 1393
A+YGR W +GE R+ + + E ++ LK L AQ+R ++ AN +RK
Sbjct: 775 ALYGRACRTPLCWTPVGERRLFG-PTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKEL 833
Query: 1394 EFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARI-HP 1452
EF+VG+ V++K ++ KL+ RY GPY +I RVGAVAY+L LP + H
Sbjct: 834 EFQVGDLVYLKAMTYKGAGRFTS-RKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNVFHN 892
Query: 1453 VFHISLLKKAVGNYVEE-----PGLPEQLDGEELP 1482
VFH+S L+K + + E PGL E + E P
Sbjct: 893 VFHVSQLRKYLSDQEESVEDIPPGLKENMTVEAWP 927
>At4g07850 putative polyprotein
Length = 1138
Score = 369 bits (947), Expect = e-102
Identities = 204/506 (40%), Positives = 283/506 (55%), Gaps = 54/506 (10%)
Query: 838 SEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDA- 896
S GV VD KV +++ WP PK+V VR F GL G+YR+F++++ +A PLTE+ KK+
Sbjct: 533 STDGVKVDEEKVKAIREWPSPKSVGKVRSFHGLAGFYRRFVRDFSTLAAPLTEVIKKNVG 592
Query: 897 FNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSK 956
F W + + AF+ LKEKLT APVL+LPDF + F +ECDA G+GIGA+L+ K PIAYFS+
Sbjct: 593 FKWEQAPEDAFQALKEKLTHAPVLSLPDFLKTFEIECDAPGVGIGAVLMQDKKPIAYFSE 652
Query: 957 ALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNW 1016
LG L Y+KEL A+ A+Q W+ YL ++FV+ TD SLK Q+ + W
Sbjct: 653 KLGGATLNYPTYDKELYALVRALQTWQHYLWPKEFVIHTDHESLKHLKGQQKLNKRHARW 712
Query: 1017 AAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKLC 1076
+ + + I YK GK N ADALSR E
Sbjct: 713 VEFIETFPYVIKYKKGKDNVVADALSRRHE------------------------------ 742
Query: 1077 QIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTY 1136
G L Y +R+ I N SSL + E H GH G +T
Sbjct: 743 --------------------GFLFYDNRLCIPN-SSLRELFIREAHGGGLMGHFGVSKTL 781
Query: 1137 RRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIG 1196
+ + + +W M + ++ C C++ K S P GL PLPIP W DIS++F++G
Sbjct: 782 KVMQDHFHWPHMKRDVERMCERCTTCKQAK-AKSQPHGLCTPLPIPLHPWNDISMDFVVG 840
Query: 1197 LPRSR-GFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDP 1255
LPR+R G ++IFVVVDR +K +HFIP A +A +F +E++RLHG+P +I+SDRD
Sbjct: 841 LPRTRTGKDSIFVVVDRFSKMAHFIPCHKTDDAMHIANLFFREVVRLHGMPKTIVSDRDT 900
Query: 1256 LFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHW 1315
F+S+FWK ++ GT+ S+ HP++DGQTE+VNR L T LR KTW L
Sbjct: 901 KFLSYFWKTLWSKLGTKLLFSTTCHPQTDGQTEVVNRTLSTLLRALIKKNLKTWEDCLPH 960
Query: 1316 AEYWFNTSYHSATQQTPFEAVYGRKP 1341
E+ +N S HSAT+ +PF+ VYG P
Sbjct: 961 VEFAYNHSVHSATKFSPFQIVYGFNP 986
Score = 55.8 bits (133), Expect = 2e-07
Identities = 34/78 (43%), Positives = 43/78 (54%), Gaps = 2/78 (2%)
Query: 609 VFQEK--QGLPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQS 666
VF E+ +GLPP RG EH I G N YR E+EKQV E++ G I +S
Sbjct: 451 VFPEENPEGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELEKQVNELMERGHICES 510
Query: 667 TSAYSSPVILVKKKDQTW 684
S + PV+LV KKD +W
Sbjct: 511 MSPCAVPVLLVPKKDGSW 528
Score = 42.4 bits (98), Expect = 0.002
Identities = 76/390 (19%), Positives = 148/390 (37%), Gaps = 57/390 (14%)
Query: 146 KVSLAQLSMEGGTIHFYNSLLATEEEL------TWERFRDALLERY-GGNGDGDVYEQLS 198
KV +A + +++ ++ T+ L TW + ++ + R+ + +++++L
Sbjct: 95 KVKVAATEFYDYALSWWDQVVTTKRRLGDDSIETWNQLKNIMKRRFVPSHYHRELHQRLR 154
Query: 199 ELRQQG-IVEEYITDFEYLTAQIPKLPE-KQYQGYFLHGLKEEIRGKVRSLVAMGGVNRA 256
L Q VEEY + E L + E + F+ GL +I + + N
Sbjct: 155 NLVQGNRTVEEYFKEMETLMLRADVQEECEATMSRFMGGLNRDILDRFE---VIHYENLE 211
Query: 257 RLLVVTRAVEKEVKGDGVAGQARANRFGGNGSRSGFSGATKGNGTEWIWVKGNKETGQTV 316
L EK++K R+ + N S+ + + K Q
Sbjct: 212 ELFHKAVMFEKQIK-------RRSAKPSYNSSKPSYQ-------------REEKSGFQKE 251
Query: 317 NRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQLMERRQKGLCFKCGGPYHRNHVCPD 376
+P P+ ++ + G + + R + CFKC G H C +
Sbjct: 252 YKPFVKPKVEEISSKGKEKE----------------VTRTRDLKCFKCHGLGHYASECSN 295
Query: 377 KHLRLLILEEDGEELDESKMLAMEVNEDEEETQGEL----SLMSLCELGMKTGGIPRTMK 432
K R++I+ + GE ES+ E ++ EE +GEL ++S+ +
Sbjct: 296 K--RIMIIRDSGEV--ESEDEKPEESDVEEAPKGELLVTMRVLSVLNKAEEQAQRENLFH 351
Query: 433 LRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPR-MTVKLGDGYKSQAQGRCAG 491
R I G +++D G+ N +V++LG E P+ ++ + A R
Sbjct: 352 TRCLIKGKVCSLIIDGGSCTNVASETMVQKLGLEEFPHPKPYKLQWLNESGEMAVTRQVQ 411
Query: 492 LKIEMGEYHLRCSPQLFDLGGPDIVLGIEW 521
+ + +G+Y + L ++LG W
Sbjct: 412 VPLAIGKYEDEILCDILPLEASHVLLGRPW 441
>At4g16910 retrotransposon like protein
Length = 687
Score = 351 bits (900), Expect = 2e-96
Identities = 232/678 (34%), Positives = 349/678 (51%), Gaps = 46/678 (6%)
Query: 884 IARPLTELTKKD-AFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGA 942
+A+P+T+LT KD AFNW ++ +++F +LK LT APVL LP+ + + + DAS +G+G
Sbjct: 1 MAQPMTQLTGKDTAFNWSEECERSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGC 60
Query: 943 ILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKE 1002
+L+ IAY S+ L + E+ AV A++ WR YL G K + TD +SLK
Sbjct: 61 VLIQKGSVIAYASRQLRKHEKNYPTNDLEMAAVVFALKIWRSYLYGAKVQIFTDHKSLKY 120
Query: 1003 FLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRP----LVSYPL 1058
Q + Q+ W + Y+ I Y PGK NQ DALSR+ E LV+
Sbjct: 121 IFTQPELNLRQRRWIKLVADYDLNIAYHPGKANQVVDALSRLRTVVEAERNQVNLVNMMG 180
Query: 1059 WEEQGVIADE---------NQKDPKLCQIVSEVALDPQ----AHPGYRVVQ----GTLLY 1101
++ E NQ D L +I S D + A Q GT++
Sbjct: 181 TLHLNALSKEVEPLGLRAANQAD-LLSRIRSAQERDEEIKGWAQNNKTEYQSSNNGTIVV 239
Query: 1102 QDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDV 1161
RV N +L ++L+E H + H G + YR + +W GM K + +V
Sbjct: 240 NGRVCGPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV----A 295
Query: 1162 CQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRS--RGFEAIFVVVDRLTKYSHF 1219
+ H+ P+G+LQ LPIPE W+ I ++F+ GLP A++VVVDRLTK +HF
Sbjct: 296 KEEHQV----PSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHF 351
Query: 1220 IPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAY 1279
+ + A +AE + EI+RLHG+P SI+SDRD F S FWK ++ GT+ +S+AY
Sbjct: 352 MAISDKDAAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVNLSTAY 411
Query: 1280 HPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGR 1339
HP++DGQ+E + LE LR D W +L E+ +N S+ ++ +P+EA+YGR
Sbjct: 412 HPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGR 471
Query: 1340 KPPVLTRWV-LGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVG 1398
W +GE R+ + + + ++ LK L AQ+R ++ AN +RK EF+VG
Sbjct: 472 AGRTPLCWTPVGERRLFG-PAVVDETTKKMKFLKIKLKEAQDRQKSYANKRRKELEFQVG 530
Query: 1399 EWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGA-RIHPVFHIS 1457
+ V++K ++ KL RY GPY +I RVGAVAY+L LP H VFH+S
Sbjct: 531 DLVYLKAMTYKGAGRFTS-RKKLRPRYVGPYKVIERVGAVAYKLDLPPKLDAFHNVFHVS 589
Query: 1458 LLKKAVGNYVEE-----PGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGK 1512
L+K + E PGL E + E P ++ + +G+S+ + + W
Sbjct: 590 QLRKCLSEQEESMEDVPPGLKENMTVE----AWPVRIMDQMKKGTRGKSMDLLKILWNCG 645
Query: 1513 TPEEATWEDLATIRSEFP 1530
EE TWE +++ FP
Sbjct: 646 GREEYTWETETKMKANFP 663
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 304 bits (778), Expect = 3e-82
Identities = 148/317 (46%), Positives = 214/317 (66%), Gaps = 4/317 (1%)
Query: 1225 PYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESD 1284
P +RSVA F +++LHG+P SI+SD DP+F+S FW+E ++L T+ MS+AYHP++D
Sbjct: 628 PLLSRSVAAKFVSHVVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTD 687
Query: 1285 GQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVL 1344
GQTE+VNRC+E +LRCF PK W+S++ WAEYW+NT++H++T TPF+A+YGR P +
Sbjct: 688 GQTEVVNRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGRPPSPI 747
Query: 1345 TRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVK 1404
+ LG + + + RDE L +LKQHLV A M+ QA+S+ + F+VG+WV ++
Sbjct: 748 PAYELGSVVCGELNEQMAARDELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLLR 807
Query: 1405 IRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVG 1464
I+ +RQ +L R KL+ R++GP+ + + G VAYRL LPEG RIHPVFH+SLLK VG
Sbjct: 808 IQPYRQKTLFRRSSQKLSHRFYGPFQVASKHGEVAYRLTLPEGTRIHPVFHVSLLKPWVG 867
Query: 1465 NYVEEPG-LPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLA 1523
+ + G LP + EL +QP VL R + + ++ +LVQW+G E+ATWE+
Sbjct: 868 DGEPDMGQLPPLRNNGEL-KLQPTAVLEVRWRSQDKKRVADLLVQWEGLHIEDATWEEYD 926
Query: 1524 TIRSEFP--VFNLEDKV 1538
+ + FP V NLEDKV
Sbjct: 927 QLAASFPEFVLNLEDKV 943
Score = 225 bits (573), Expect = 2e-58
Identities = 143/403 (35%), Positives = 213/403 (52%), Gaps = 51/403 (12%)
Query: 388 GEELDESKMLAMEVNEDEEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVD 447
GE DE A+E E E + E++L +L G+ T T+++R TI+ ++ L++
Sbjct: 284 GENDDE----AVEEESTEAEGKPEITLYAL--EGVDTTS---TIRVRATIHRNRLIALIN 334
Query: 448 SGATHNFVDCFLVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQL 507
SG+THNF+ VR + + T TV++ +G + R + + MG +
Sbjct: 335 SGSTHNFIGEKAVRGMNLKATTTKPFTVRVVNGMPLVCRSRYEAIPVVMGGVVFPVTLYA 394
Query: 508 FDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHMEALQSITAT 567
L G D+ +G++WL TLG T+ NW Q + F + L G+ G + ++ T T
Sbjct: 395 LPLMGLDLAMGVQWLSTLGPTLCNWKEQTLQFHWAGDEVRLMGIKPTG--LRGVEHKTIT 452
Query: 568 GERKPG---LLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREH 624
+ + G T + P E+ KL+ F +F+ LP R H
Sbjct: 453 KKARMGHTIFAITMAHNSSDPNTPDG-------EIKKLIEEFAGLFETPTQLPSERPIVH 505
Query: 625 CITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW 684
I +++G PVNVRPYRY + K+E+ + AGIIR S+S + SPV+L
Sbjct: 506 RIALKKGTDPVNVRPYRYAFYQKDEMSR-------AGIIRPSSSPFLSPVLL-------- 550
Query: 685 *MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAF 744
++FPIP ++++LDEL+GAV+F+KLDL +GY QVR+ D+ K AF
Sbjct: 551 ---------------ERFPIPTVDDMLDELNGAVYFTKLDLTAGYQQVRMHSPDIPKIAF 595
Query: 745 RTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFF 787
+TH GHYEY+VMPFGL NAPSTFQ+LMNE+F P+L R V F
Sbjct: 596 QTHNGHYEYLVMPFGLCNAPSTFQALMNEIFWPLLSRSVAAKF 638
Score = 46.6 bits (109), Expect = 1e-04
Identities = 48/237 (20%), Positives = 87/237 (36%), Gaps = 35/237 (14%)
Query: 13 TTRVAVTQMEAKIDAMESELAAIRTALAAVTTAMKD----LPTTLGTMVEKAVGKSVGID 68
T + + Q+E D M+ E+ + A+ + +D L L ++E + G
Sbjct: 3 TNKDQLEQLETSQDMMQDEMQRMTVAMVDLKGDNEDRHRRLEEALAHLIEAVTQRESG-- 60
Query: 69 VDSGDARPEREPREKTPESVGLRESGSDQPVLQGEALNEFRQSVKKVELPMFDGKDLAGW 128
S +R R +S G R + N K++ P F G DL W
Sbjct: 61 -GSAASRGSRRTERNQFQSTGNRGEPDNPFAAVTANNNNAIARPSKLDFPRFHGGDLTKW 119
Query: 129 ISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEELTWERFRDALLERYGGN 188
+S+A+ YF+ E E V ++G ++ + L
Sbjct: 120 LSKAKQYFEYHEGPVEQAVRFTTFHLDGVANGWWQATL---------------------- 157
Query: 189 GDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKVR 245
+ + + Q G + EY +FE+L ++ ++ G F++GL I +R
Sbjct: 158 ------RRSATIGQTGSLSEYQHEFEWLQNKVYGWTQEVLVGAFMNGLYYSISNGIR 208
>At4g08100 putative polyprotein
Length = 1054
Score = 283 bits (725), Expect = 4e-76
Identities = 160/403 (39%), Positives = 227/403 (55%), Gaps = 42/403 (10%)
Query: 689 DYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHE 748
D RA+N++TV + PIP +++ LD+LHG+ FSK+DLKSGYHQ R++E D KTA +T +
Sbjct: 527 DCRAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQ 586
Query: 749 GHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVL 808
YE++VMPFGL NAP+TF LMN V R + V+V+FDDILVYSK H++HL+ VL
Sbjct: 587 RLYEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVL 646
Query: 809 EVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFL 868
++L+ +L AN KKC F T + +LG ++S G+ VD KV +++ WP PKNV
Sbjct: 647 DLLRKEKLYANLKKCTFCTDNLVFLGFVVSADGIKVDEEKVKAIREWPNPKNVS------ 700
Query: 869 GLTGYYRKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQE 928
K F W + AF+ LKEKLT + V LP+F +
Sbjct: 701 -----------------------EKDIGFKWEDAQENAFQALKEKLTNSSVPILPNFMKS 737
Query: 929 FRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLG 988
F +ECDASG+GIGA+L+ PIAYFS+ LG L Y+KEL A+ A+Q W+ YL
Sbjct: 738 FEIECDASGLGIGAVLMQDHKPIAYFSEKLGGATLNYPTYDKELYALVRALQTWQHYLWP 797
Query: 989 RKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQG------ADALS 1042
+KFV+ TD SLK Q+ + W + + + I YK + + G A S
Sbjct: 798 KKFVIHTDHESLKHLKGQQKLNKRHARWVEFIETFPYVIKYKKREAHGGGLMRHSATKFS 857
Query: 1043 RIDEAGELRP-----LVSYPLWEEQGVIADENQKDPKLCQIVS 1080
+ +P L+ PL E + D +KD + Q+++
Sbjct: 858 PFEIVYGFKPTSPLDLIPLPLSERSSL--DGKKKDDLVQQVLN 898
Score = 50.8 bits (120), Expect = 6e-06
Identities = 38/122 (31%), Positives = 64/122 (52%), Gaps = 5/122 (4%)
Query: 1325 HSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRA 1384
HSAT+ +PFE VYG K P ++ E D + +D+ ++Q+ +L A ++ +
Sbjct: 851 HSATKFSPFEIVYGFK-PTSPLDLIPLPLSERSSLDGKKKDDLVQQV-LNLEARTKQYKK 908
Query: 1385 QANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKL 1444
AN RK F G+ V+V +R R + + +KL R GP+ ++ R+ AY+L L
Sbjct: 909 YANKGRKEVIFNEGDQVWVHLRKKRFPEVRS---SKLMPRIDGPFKVLKRINNNAYKLDL 965
Query: 1445 PE 1446
+
Sbjct: 966 QD 967
>At4g07830 putative reverse transcriptase
Length = 611
Score = 268 bits (684), Expect = 2e-71
Identities = 190/560 (33%), Positives = 276/560 (48%), Gaps = 48/560 (8%)
Query: 993 VSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSR--------- 1043
V TD +SLK Q + + W + Y+ EI Y PGK N DALSR
Sbjct: 31 VFTDHKSLKYIFTQLELNLRLRRWMKLVADYDLEIAYHPGKANVVTDALSRKRVGAAPGQ 90
Query: 1044 -----IDEAGELR--PLVSYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYR--- 1093
+ E G LR + PL E V + +L Q E + G+
Sbjct: 91 SVETLVIEIGALRLCAVAREPLGLE-AVDQTDLLSRVQLAQEKDEGLIAASKAEGFEYQF 149
Query: 1094 VVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQ 1153
GT+L RV + L ++L E H + H G + YR + W GM + +
Sbjct: 150 AANGTILVHGRVCVPKDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVG 209
Query: 1154 EFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRL 1213
+V CDVCQ K LLQ LPIPE W+ I+++F++GLP SR +AI+V+VDRL
Sbjct: 210 NWVEECDVCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRTKDAIWVIVDRL 269
Query: 1214 TKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQF 1273
TK +HF+ ++ A +A+ + EI++LHGVP SI+SDRD F S FW+ GT+
Sbjct: 270 TKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKV 329
Query: 1274 KMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPF 1333
+MS+AYHP++DGQ+E + LE LR D WA L E+ +N SY ++ PF
Sbjct: 330 QMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWRGHWADHLSLVEFAYNNSYQASIGMAPF 389
Query: 1334 EAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQ 1393
E +YGR L +GE + + +Q+ E +R LK ++ AQ R R+ A+ +RK
Sbjct: 390 EVLYGRPCRTLCWTQVGERSIYGADY-VQEITERIRVLKLNMKEAQNRQRSYADKRRKEL 448
Query: 1394 EFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGAR-IHP 1452
EFEVG+ S++ H + + RVG VA+RL+L + R H
Sbjct: 449 EFEVGD------------SVSQDGHVARSEQ---------RVGPVAFRLELSDVMRAFHK 487
Query: 1453 VFHISLLKKAVGNYVEE-PGLPEQLDGEELPSIQPALVLARR--DILRQGESISQVLVQW 1509
VFH+S+L+K + E +PE L +P VL RR ++ R+ + +VL
Sbjct: 488 VFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLRNC 547
Query: 1510 QGKTPEEATWEDLATIRSEF 1529
G T E TWE A +++ F
Sbjct: 548 DGVT--EETWEPEARLKARF 565
>At4g03650 putative reverse transcriptase
Length = 839
Score = 265 bits (677), Expect = 2e-70
Identities = 131/295 (44%), Positives = 185/295 (62%), Gaps = 1/295 (0%)
Query: 736 EEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSK 795
E DV KTAFRT GH+E++VMPFGL NAP+ F LMN VF+ L V++F DDILVYSK
Sbjct: 518 EADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSK 577
Query: 796 TWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSW 855
+ +H +HL RV+E L+ +L A KC F + + +LGH++S +GV+VDP K+ +++ W
Sbjct: 578 SPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDW 637
Query: 856 PEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDA-FNWGKQAQQAFEDLKEKL 914
P P N +R FLGL GYYR+FI+ + +A+P+T+LT KD F W + ++ F LKE L
Sbjct: 638 PRPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEML 697
Query: 915 TTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMA 974
T+ PVLALP+ + + + DASG+G+G L+ IAY S+ L ++ E+ A
Sbjct: 698 TSTPVLALPEHGEPYMVYTDASGVGLGCALMQRGKVIAYASRQLRKHEGNYPTHDLEMAA 757
Query: 975 VALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIY 1029
V A++ WR YL G K V TD +SLK Q + Q+ W + Y+ EI Y
Sbjct: 758 VIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAY 812
Score = 50.1 bits (118), Expect = 1e-05
Identities = 48/178 (26%), Positives = 80/178 (43%), Gaps = 16/178 (8%)
Query: 569 ERKPGLLGTRNEEKAGKQLPGEINSS----QLEELD-KLLYRFDVVFQEKQGLPPGRGRE 623
ER G G N+ + + LPG+ + Q+ D +++ F VFQ QGLPP R
Sbjct: 422 ERPEGSAGRENDREGLRGLPGDDIYAGVCGQVAVSDIRVVQEFQYVFQSLQGLPPSRSDP 481
Query: 624 HCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQST--SAYSSPVILVKKKD 681
I ++ P++ PYR E++KQ+ ++L+ +R++ + Y +V
Sbjct: 482 FTIELEPKTAPLSKAPYRMAPAKMAELKKQLEDLLAEADVRKTAFRTRYGHFEFVVMPFG 541
Query: 682 QTW*MCVDYRALNSV--TVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREE 737
T R +NSV D+F I I+++L +SK + H RV E+
Sbjct: 542 LTNAPAAFMRLMNSVFQEFLDEFVIIFIDDIL-------VYSKSPEEHEVHLRRVMEK 592
Score = 32.3 bits (72), Expect = 2.3
Identities = 16/40 (40%), Positives = 28/40 (70%), Gaps = 5/40 (12%)
Query: 1249 IISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTE 1288
+++D D L +++ EM GT+ +MS+ YHP++DGQ+E
Sbjct: 802 LVADYD-LEIAYHLAEM----GTKVQMSTTYHPQTDGQSE 836
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 222 bits (566), Expect = 1e-57
Identities = 225/849 (26%), Positives = 360/849 (41%), Gaps = 118/849 (13%)
Query: 650 IEKQVREMLSAGIIRQ-STSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIE 708
+ +V ++L G IR+ + + ++VKKK+ +C+D+ LN D FP+P I+
Sbjct: 480 VNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHID 539
Query: 709 ELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQ 768
L++ G S +D SGY+Q+ + ED KT F T G Y Y VMPFGL NA +T+
Sbjct: 540 RLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNAGATYP 599
Query: 769 SLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATK 828
L+N++F + + + V+ DD+L+ S DH+ HLE +L +Q+ N KC F
Sbjct: 600 RLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAKCTFGVP 659
Query: 829 FVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPL 888
E+LG++++++G+ +PN++ + + P PKN K V+ G +FI + P
Sbjct: 660 SGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRFISRSTDKSLPF 719
Query: 889 TELTK-KDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLY- 946
++ K F W ++ ++AF LK LT+ PVL P+ ++ L S + +L+
Sbjct: 720 YQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSNHAVSGVLIRE 779
Query: 947 ---GKHPIAYF--SKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLK 1001
+ PI Y ++ G S E L V +++R + L R + Q +
Sbjct: 780 DRGEQKPIYYIIVNQFNGDYEAKDSRMEAYLHVVKDLAKNFRKFELIR---IPRGQNTTA 836
Query: 1002 EFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGK-----TNQGADALSRIDEAGELRPL--- 1053
+ L T D + + + E K K T A A R D A EL P+
Sbjct: 837 DALAALASTSDPEVNSIIPVECISERSIKEEKEAFVVTRSRAAARDRGDTAVELPPVKRR 896
Query: 1054 ------VSYPLWEE-------QGVIAD---ENQKDPKLCQIVSEVALDPQAHPGYRVVQG 1097
V P +E + VI+D E + +P V E L P HP +
Sbjct: 897 KSKEATVPKPAVQENIGIELIEEVISDAKIEAETEP----WVDEDILAPNEHPEPEALNS 952
Query: 1098 TLLYQ----DRVVISNKSSLIPKLLEEFHTTPHGGH-SGFLRTYRRIATNLYWRGMTKTI 1152
LY+ D ++S ++ ++ E H G H SG ++ YW M
Sbjct: 953 --LYRRGVSDPYLLSIFGPVVEIVMSEVHEGLCGSHSSGRAMAFKIKKMCYYWPTMITDC 1010
Query: 1153 QEFV*ACDVCQRHKYLASSPAGLLQPL--PIPERVWEDISLNFIIGLPRS-RGFEAIFVV 1209
++ C CQ H L P+ L + P P W S++ I L RS RG + + V+
Sbjct: 1011 VKYAQRCKRCQLHAPLIHQPSELFSSISAPYPFMRW---SMDIIGPLHRSTRGVQYLLVL 1067
Query: 1210 VDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQ 1269
D +K+ E I L+ W
Sbjct: 1068 TDYFSKW------------------IEAEAILLN------------------W------- 1084
Query: 1270 GTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQ 1329
G + S+ +P+ +GQ E N+ + + L+ W L + + T+ +T
Sbjct: 1085 GIKVSYSTLRYPQGNGQAEAANKTILSNLKKRLNHLKGGWYDELQPVLWAYRTTPRRSTG 1144
Query: 1330 QTPFEAVYGRKPPV----------LTRWVLGETRVEAVEKDLQD-----RDEALRQLKQH 1374
+TPF VYG K V T L E A+ D D RD+AL Q++ +
Sbjct: 1145 ETPFSLVYGMKAVVPAELNVPGLRRTEAPLNEKENSAMLDDSLDTINERRDQALIQIQNY 1204
Query: 1375 LVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGR 1434
AA NSK K + F VG++V ++ +++ A KL + GPY +
Sbjct: 1205 QHAAAR----YYNSKVKSRPFFVGDYVLKRVFDNKKEEGA----GKLGINWEGPYIVTEV 1256
Query: 1435 VGAVAYRLK 1443
V YRLK
Sbjct: 1257 VRNGVYRLK 1265
>At1g20390 hypothetical protein
Length = 1791
Score = 219 bits (558), Expect = 1e-56
Identities = 138/457 (30%), Positives = 226/457 (49%), Gaps = 12/457 (2%)
Query: 590 EINSSQLEELDKLLYRFDVVF----QEKQGLPPGRGREHCITIQEGKGPVNVRPYRYPHH 645
EI+ S EL LL R F ++ +G+ P H + + PV + +
Sbjct: 730 EISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAI-TAHELNVDPTFKPVKQKRRKLGPE 788
Query: 646 HKNEIEKQVREMLSAG-IIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPI 704
+ ++V ++L AG II + + ++VKKK+ W +CVDY LN D +P+
Sbjct: 789 RARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKDSYPL 848
Query: 705 PVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAP 764
P I+ L++ G S +D SGY+Q+ + ++D KT+F T G Y Y VM FGL NA
Sbjct: 849 PHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAG 908
Query: 765 STFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQ 824
+T+Q +N++ + R V V+ DD+LV S DH+ HL + +VL + + N KC
Sbjct: 909 ATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCT 968
Query: 825 FATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKI 884
F E+LG++++++G+ +P ++ ++ P P+N + V+ G +FI
Sbjct: 969 FGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTDK 1028
Query: 885 ARPLTELTKKDA-FNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAI 943
P L K+ A F+W K +++AFE LK+ L+T P+L P+ + L S + ++
Sbjct: 1029 CLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVGETLYLYIAVSDHAVSSV 1088
Query: 944 LLY----GKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRS 999
L+ + PI Y SK+L EK +AV + + RPY V TDQ
Sbjct: 1089 LVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSARKLRPYFQSHTIAVLTDQ-P 1147
Query: 1000 LKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQ 1036
L+ LH +G WA +L Y+ + +P +Q
Sbjct: 1148 LRVALHSPSQSGRMTKWAVELSEYDIDFRPRPAMKSQ 1184
Score = 91.7 bits (226), Expect = 3e-18
Identities = 91/366 (24%), Positives = 152/366 (40%), Gaps = 30/366 (8%)
Query: 1114 IPKLLEEFHTTPHGGHSGFLRTYRRIAT-NLYWRGMTKTIQEFV*ACDVCQRHKYLASSP 1172
I ++++E H G HSG ++ YW M + F C+ CQRH P
Sbjct: 1435 INEIMKETHEGAAGNHSGGRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQP 1494
Query: 1173 AGLLQP--LPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARS 1230
LL+ P P W +++ + +P SR I V+ D TK+ A
Sbjct: 1495 TELLRAGVAPYPFMRW---AMDIVGPMPASRQKRFILVMTDYFTKWVEAESYA-TIRAND 1550
Query: 1231 VAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIV 1290
V K II HG+P II+D F+S ++ + S+ +P+ +GQ E
Sbjct: 1551 VQNFVWKFIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEAT 1610
Query: 1291 NRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYG------------ 1338
N+ + + L+ ++ WA L + + T+ SAT QTPF YG
Sbjct: 1611 NKTILSGLKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYS 1670
Query: 1339 --RKPPVLTRWVLGETRVEAVEKDLQD-RDEALRQLKQHLVAAQERMRAQANSKRKHQEF 1395
R+ ++ L + + DL++ R+ AL +++ + +AA + N K ++ F
Sbjct: 1671 SLRRSMMVKNPELNDRMMLDRLDDLEEIRNAALCRIQNYQLAAAKHY----NQKVHNRHF 1726
Query: 1396 EVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFH 1455
+VG+ V K+ + A KL A + G Y + V Y L G + ++
Sbjct: 1727 DVGDLVLRKVFE----NTAEINAGKLGANWEGSYQVSKIVRPGDYELLTMSGTAVPRTWN 1782
Query: 1456 ISLLKK 1461
LK+
Sbjct: 1783 SMHLKR 1788
>At4g04230 putative transposon protein
Length = 315
Score = 193 bits (491), Expect = 6e-49
Identities = 93/167 (55%), Positives = 126/167 (74%)
Query: 661 GIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFF 720
G IR+S S + PV+LV KKD TW MC+D RA+N++T+ + PIP + ++LDEL GA+ F
Sbjct: 128 GYIRESLSPCAVPVLLVPKKDGTWRMCLDCRAINNITIKYRHPIPRLYDMLDELSGAIIF 187
Query: 721 SKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLR 780
SK+DL+SGYHQVR+RE D KTAF+T +G YE +VMPFGL NAPSTF LMN+V R +
Sbjct: 188 SKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQVLRSFIG 247
Query: 781 RGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFAT 827
+ V+V+FDDIL+Y+K++SDH+ LE +L+ L+ L AN KKC F +
Sbjct: 248 KFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTFCS 294
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,935,600
Number of Sequences: 26719
Number of extensions: 1681143
Number of successful extensions: 4971
Number of sequences better than 10.0: 109
Number of HSP's better than 10.0 without gapping: 72
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 4627
Number of HSP's gapped (non-prelim): 226
length of query: 1596
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1483
effective length of database: 8,299,349
effective search space: 12307934567
effective search space used: 12307934567
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0170.6


