

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0170.5
(603 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
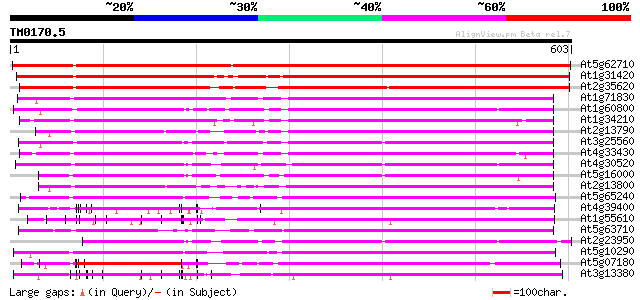
Score E
Sequences producing significant alignments: (bits) Value
At5g62710 receptor - like protein kinase - like 895 0.0
At1g31420 unknown protein 592 e-169
At2g35620 putative receptor-like protein kinase 566 e-162
At1g71830 protein kinase, putative 363 e-100
At1g60800 Putative Serine/Threonine protein kinase 362 e-100
At1g34210 putative protein 358 4e-99
At2g13790 putative receptor protein kinase (F13J11.14/At2g13790) 334 7e-92
At3g25560 unknown protein 331 6e-91
At4g33430 somatic embryogenesis receptor-like kinase 3 (SERK3) 330 1e-90
At4g30520 receptor-like kinase homolog 329 2e-90
At5g16000 receptor protein kinase-like protein 319 3e-87
At2g13800 receptor-like protein kinase 318 4e-87
At5g65240 receptor-like protein kinase 314 8e-86
At4g39400 brassinosteroid insensitive 1 gene (BRI1) 314 1e-85
At1g55610 receptor kinase, putative 311 9e-85
At5g63710 receptor-like protein kinase 310 1e-84
At2g23950 putative LRR receptor protein kinase 309 3e-84
At5g10290 protein serine/threonine kinase-like protein 308 6e-84
At5g07180 receptor-like protein kinase 308 6e-84
At3g13380 brassinosteroid receptor kinase, putative 305 5e-83
>At5g62710 receptor - like protein kinase - like
Length = 604
Score = 895 bits (2313), Expect = 0.0
Identities = 452/605 (74%), Positives = 513/605 (84%), Gaps = 9/605 (1%)
Query: 4 GFPIWVFILI-FTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGI 62
G WVF +I T+F S ALT DG LLE+K NDT+N L NW++ DESPC+WTG+
Sbjct: 2 GISNWVFSVISVATLFVSCSFALTLDGFALLELKSGFNDTRNSLENWKDSDESPCSWTGV 61
Query: 63 TCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELR 122
+C+P D QRV SINLPY QLGGIISPSIGKLSRLQRLALHQNSLHG IPNEITNCTELR
Sbjct: 62 SCNPQD--QRVVSINLPYMQLGGIISPSIGKLSRLQRLALHQNSLHGNIPNEITNCTELR 119
Query: 123 ALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGE 182
A+YLRAN+ QGGIP D+GNL FL ILDLSSN+ KGAIPSS+ RL L+ LNLSTNFFSGE
Sbjct: 120 AMYLRANFLQGGIPPDLGNLTFLTILDLSSNTLKGAIPSSISRLTRLRSLNLSTNFFSGE 179
Query: 183 IPDIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYM 242
IPDIGVLS F +F GNLDLCGRQI+KPCR+S GFPVV+PHAES + + KRSS +
Sbjct: 180 IPDIGVLSRFGVETFTGNLDLCGRQIRKPCRSSMGFPVVLPHAESADESDSPKRSS-RLI 238
Query: 243 KVVLIGAMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDP-EASTKLITFHGDL 301
K +LIGAM+T+ L +V FLWI +LSKKER V +YT+VKKQ DP E S KLITFHGDL
Sbjct: 239 KGILIGAMSTMALAFIVIFVFLWIWMLSKKERKVKKYTEVKKQKDPSETSKKLITFHGDL 298
Query: 302 PYTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELE 361
PY+S+E+IEKLESL+EEDIVGSGGFGTVYRMVMND GTFAVK+IDRSR+G D+VFERE+E
Sbjct: 299 PYSSTELIEKLESLDEEDIVGSGGFGTVYRMVMNDLGTFAVKKIDRSRQGSDRVFEREVE 358
Query: 362 ILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQP--LNWNDRLNIALG 419
ILGS+KHINLVNLRGYCRLPS+RLLIYDYL +GSLDDLLHE ++ LNWN RL IALG
Sbjct: 359 ILGSVKHINLVNLRGYCRLPSSRLLIYDYLTLGSLDDLLHERAQEDGLLNWNARLKIALG 418
Query: 420 SARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTF 479
SARGLAYLHH+C PKIVHRDIKSSNILLN+ +EP +SDFGLAKLLVDEDAHVTTVVAGTF
Sbjct: 419 SARGLAYLHHDCSPKIVHRDIKSSNILLNDKLEPRVSDFGLAKLLVDEDAHVTTVVAGTF 478
Query: 480 GYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRL 539
GYLAPEYLQ+GRATEKSDVYSFGVLLLELVTGKRPTDP F RGLNVVGWMNT+ KENRL
Sbjct: 479 GYLAPEYLQNGRATEKSDVYSFGVLLLELVTGKRPTDPIFVKRGLNVVGWMNTVLKENRL 538
Query: 540 EDVVDRRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVMSPCPS-DFY-E 597
EDV+D+RCTD D ++E +LE+A RCTDAN ++RP+MNQV QLLEQEVMSP D+Y +
Sbjct: 539 EDVIDKRCTDVDEESVEALLEIAERCTDANPENRPAMNQVAQLLEQEVMSPSSGIDYYDD 598
Query: 598 SHSDH 602
SHSD+
Sbjct: 599 SHSDY 603
>At1g31420 unknown protein
Length = 592
Score = 592 bits (1527), Expect = e-169
Identities = 301/594 (50%), Positives = 409/594 (68%), Gaps = 14/594 (2%)
Query: 8 WVFILIFTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHPG 67
W+ ++ + S A++ DG LL + A+ + + + W+ D PC W G+TC
Sbjct: 13 WLLLISLLCSLSNESQAISPDGEALLSFRNAVTRSDSFIHQWRPEDPDPCNWNGVTCDAK 72
Query: 68 DGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLR 127
+RV ++NL Y ++ G + P IGKL L+ L LH N+L+G IP + NCT L ++L+
Sbjct: 73 T--KRVITLNLTYHKIMGPLPPDIGKLDHLRLLMLHNNALYGAIPTALGNCTALEEIHLQ 130
Query: 128 ANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIG 187
+NYF G IP+++G+LP L LD+SSN+ G IP+SLG+L L N+S NF G+IP G
Sbjct: 131 SNYFTGPIPAEMGDLPGLQKLDMSSNTLSGPIPASLGQLKKLSNFNVSNNFLVGQIPSDG 190
Query: 188 VLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLI 247
VLS F KNSFIGNL+LCG+ + C+ G P H++S + K++S +LI
Sbjct: 191 VLSGFSKNSFIGNLNLCGKHVDVVCQDDSGNPS--SHSQSGQNQ---KKNSGK----LLI 241
Query: 248 GAMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSE 307
A T+G LLLV L W L KK V + K V AS ++ FHGDLPY+S +
Sbjct: 242 SASATVGALLLVALMCFWGCFLYKKLGKV-EIKSLAKDVGGGAS--IVMFHGDLPYSSKD 298
Query: 308 IIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIK 367
II+KLE L EE I+G GGFGTVY++ M+D FA+KRI + EG D+ FERELEILGSIK
Sbjct: 299 IIKKLEMLNEEHIIGCGGFGTVYKLAMDDGKVFALKRILKLNEGFDRFFERELEILGSIK 358
Query: 368 HINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYL 427
H LVNLRGYC P+++LL+YDYL GSLD+ LH + L+W+ R+NI +G+A+GL+YL
Sbjct: 359 HRYLVNLRGYCNSPTSKLLLYDYLPGGSLDEALHVERGEQLDWDSRVNIIIGAAKGLSYL 418
Query: 428 HHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYL 487
HH+C P+I+HRDIKSSNILL+ N+E +SDFGLAKLL DE++H+TT+VAGTFGYLAPEY+
Sbjct: 419 HHDCSPRIIHRDIKSSNILLDGNLEARVSDFGLAKLLEDEESHITTIVAGTFGYLAPEYM 478
Query: 488 QSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRC 547
QSGRATEK+DVYSFGVL+LE+++GKRPTD SF +GLNVVGW+ L E R D+VD C
Sbjct: 479 QSGRATEKTDVYSFGVLVLEVLSGKRPTDASFIEKGLNVVGWLKFLISEKRPRDIVDPNC 538
Query: 548 TDADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVMSPCPSDFYESHSD 601
+L+ +L +A +C + ++RP+M++V+QLLE EVM+PCPS+FY+S SD
Sbjct: 539 EGMQMESLDALLSIATQCVSPSPEERPTMHRVVQLLESEVMTPCPSEFYDSSSD 592
>At2g35620 putative receptor-like protein kinase
Length = 567
Score = 567 bits (1460), Expect = e-162
Identities = 294/591 (49%), Positives = 397/591 (66%), Gaps = 25/591 (4%)
Query: 11 ILIFTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHPGDGE 70
I I++ + G LL + + + V+ W+ D PC W G+TC
Sbjct: 2 ITIYSAQIAEGIKSFFSPGEALLSFRNGVLASDGVIGLWRPEDPDPCNWKGVTCDAKT-- 59
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
+RV +++L Y +L G + P +GKL +L+ L LH N+L+ IP + NCT L +YL+ NY
Sbjct: 60 KRVIALSLTYHKLRGPLPPELGKLDQLRLLMLHNNALYQSIPASLGNCTALEGIYLQNNY 119
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLS 190
G IPS+IGNL L LDLS+N+ GAIP+SLG+L L N+S NF G+IP G+L+
Sbjct: 120 ITGTIPSEIGNLSGLKNLDLSNNNLNGAIPASLGQLKRLTKFNVSNNFLVGKIPSDGLLA 179
Query: 191 TFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAM 250
++SF GN +LCG+QI C S S + PT + ++ K +LI A
Sbjct: 180 RLSRDSFNGNRNLCGKQIDIVCNDS---------GNSTASGSPTGQGGNN-PKRLLISAS 229
Query: 251 TTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEIIE 310
T+G LLLV L W L KK V E+ + +I GDLPY S +II+
Sbjct: 230 ATVGGLLLVALMCFWGCFLYKKLGRV------------ESKSLVIDVGGDLPYASKDIIK 277
Query: 311 KLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHIN 370
KLESL EE I+G GGFGTVY++ M+D FA+KRI + EG D+ FERELEILGSIKH
Sbjct: 278 KLESLNEEHIIGCGGFGTVYKLSMDDGNVFALKRIVKLNEGFDRFFERELEILGSIKHRY 337
Query: 371 LVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHE 430
LVNLRGYC P+++LL+YDYL GSLD+ LH+ EQ L+W+ R+NI +G+A+GLAYLHH+
Sbjct: 338 LVNLRGYCNSPTSKLLLYDYLPGGSLDEALHKRGEQ-LDWDSRVNIIIGAAKGLAYLHHD 396
Query: 431 CCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSG 490
C P+I+HRDIKSSNILL+ N+E +SDFGLAKLL DE++H+TT+VAGTFGYLAPEY+QSG
Sbjct: 397 CSPRIIHRDIKSSNILLDGNLEARVSDFGLAKLLEDEESHITTIVAGTFGYLAPEYMQSG 456
Query: 491 RATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRCTDA 550
RATEK+DVYSFGVL+LE+++GK PTD SF +G N+VGW+N L ENR +++VD C
Sbjct: 457 RATEKTDVYSFGVLVLEVLSGKLPTDASFIEKGFNIVGWLNFLISENRAKEIVDLSCEGV 516
Query: 551 DAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVMSPCPSDFYESHSD 601
+ +L+ +L +A +C ++ D+RP+M++V+QLLE EVM+PCPSDFY+S SD
Sbjct: 517 ERESLDALLSIATKCVSSSPDERPTMHRVVQLLESEVMTPCPSDFYDSSSD 567
>At1g71830 protein kinase, putative
Length = 625
Score = 363 bits (933), Expect = e-100
Identities = 224/588 (38%), Positives = 330/588 (56%), Gaps = 29/588 (4%)
Query: 9 VFILIFTTVFTPSSLALTQ---DGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCH 65
VFIL+ + SL L +G L ++ L D NVL +W +PC W +TC+
Sbjct: 7 VFILLSLILLPNHSLWLASANLEGDALHTLRVTLVDPNNVLQSWDPTLVNPCTWFHVTCN 66
Query: 66 PGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALY 125
E V ++L ++L G + P +G L LQ L L+ N++ G IP+ + N T L +L
Sbjct: 67 ---NENSVIRVDLGNAELSGHLVPELGVLKNLQYLELYSNNITGPIPSNLGNLTNLVSLD 123
Query: 126 LRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPD 185
L N F G IP +G L L L L++NS G+IP SL + LQVL+LS N SG +PD
Sbjct: 124 LYLNSFSGPIPESLGKLSKLRFLRLNNNSLTGSIPMSLTNITTLQVLDLSNNRLSGSVPD 183
Query: 186 IGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVV 245
G S F SF NLDLCG PC S F P + + P S + +
Sbjct: 184 NGSFSLFTPISFANNLDLCGPVTSHPCPGSPPFSPPPPFIQPPPVSTP----SGYGITGA 239
Query: 246 LIGAMTTLGLLLLVT--LSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLP- 302
+ G + LL ++F W R + + + + DV + DPE G L
Sbjct: 240 IAGGVAAGAALLFAAPAIAFAWWR----RRKPLDIFFDVPAEEDPEVHL------GQLKR 289
Query: 303 YTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSRE-GCDQVFERELE 361
++ E+ + ++I+G GGFG VY+ + D AVKR+ R G + F+ E+E
Sbjct: 290 FSLRELQVASDGFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTPGGELQFQTEVE 349
Query: 362 ILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHEN--TEQPLNWNDRLNIALG 419
++ H NL+ LRG+C P+ RLL+Y Y+A GS+ L E ++ PL+W R IALG
Sbjct: 350 MISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPPSQPPLDWPTRKRIALG 409
Query: 420 SARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTF 479
SARGL+YLH C PKI+HRD+K++NILL+E E + DFGLAKL+ +D HVTT V GT
Sbjct: 410 SARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTI 469
Query: 480 GYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTD-PSFAN-RGLNVVGWMNTLQKEN 537
G++APEYL +G+++EK+DV+ +G++LLEL+TG+R D AN + ++ W+ L KE
Sbjct: 470 GHIAPEYLSTGKSSEKTDVFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEK 529
Query: 538 RLEDVVDRRC-TDADAGTLEVILELAARCTDANADDRPSMNQVLQLLE 584
+LE +VD T+ + LE ++++A CT + +RP M++V+++LE
Sbjct: 530 KLEMLVDPDLQTNYEERELEQVIQVALLCTQGSPMERPKMSEVVRMLE 577
>At1g60800 Putative Serine/Threonine protein kinase
Length = 632
Score = 362 bits (929), Expect = e-100
Identities = 219/593 (36%), Positives = 335/593 (55%), Gaps = 36/593 (6%)
Query: 5 FPIW-VFILIFTTVFTPSSLALTQDGLT-----LLEIKGALNDTKNVLSNWQEFDESPCA 58
F +W + L+F F SS L+ G+ L+ +K LND VL NW PC+
Sbjct: 6 FVVWRLGFLVFVWFFDISSATLSPTGVNYEVTALVAVKNELNDPYKVLENWDVNSVDPCS 65
Query: 59 WTGITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNC 118
W ++C G V S++LP L G +SP IG L+ LQ + L N++ G IP I
Sbjct: 66 WRMVSCTDG----YVSSLDLPSQSLSGTLSPRIGNLTYLQSVVLQNNAITGPIPETIGRL 121
Query: 119 TELRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNF 178
+L++L L N F G IP+ +G L LN L L++NS G P SL ++ L ++++S N
Sbjct: 122 EKLQSLDLSNNSFTGEIPASLGELKNLNYLRLNNNSLIGTCPESLSKIEGLTLVDISYNN 181
Query: 179 FSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSS 238
SG +P + TF+ IGN +CG + C ++ P+ +P DE+ R++
Sbjct: 182 LSGSLPKVSA-RTFK---VIGNALICGPKAVSNC-SAVPEPLTLPQDGPDESGT---RTN 233
Query: 239 SHYMKVVLIGAMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFH 298
H++ + + + + + FLW R K+ + DV +Q DPE S
Sbjct: 234 GHHVALAFAASFSAAFFVFFTSGMFLWWRYRRNKQI----FFDVNEQYDPEVSL------ 283
Query: 299 GDLP-YTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRI-DRSREGCDQVF 356
G L YT E+ ++I+G GG+G VY+ +ND AVKR+ D + G + F
Sbjct: 284 GHLKRYTFKELRSATNHFNSKNILGRGGYGIVYKGHLNDGTLVAVKRLKDCNIAGGEVQF 343
Query: 357 ERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENT--EQPLNWNDRL 414
+ E+E + H NL+ LRG+C R+L+Y Y+ GS+ L +N E L+W+ R
Sbjct: 344 QTEVETISLALHRNLLRLRGFCSSNQERILVYPYMPNGSVASRLKDNIRGEPALDWSRRK 403
Query: 415 NIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTV 474
IA+G+ARGL YLH +C PKI+HRD+K++NILL+E+ E + DFGLAKLL D+HVTT
Sbjct: 404 KIAVGTARGLVYLHEQCDPKIIHRDVKAANILLDEDFEAVVGDFGLAKLLDHRDSHVTTA 463
Query: 475 VAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTD--PSFANRGLNVVGWMNT 532
V GT G++APEYL +G+++EK+DV+ FG+LLLEL+TG++ D S +G+ ++ W+
Sbjct: 464 VRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLELITGQKALDFGRSAHQKGV-MLDWVKK 522
Query: 533 LQKENRLEDVVDRRCTDA-DAGTLEVILELAARCTDANADDRPSMNQVLQLLE 584
L +E +L+ ++D+ D D LE I+++A CT N RP M++V+++LE
Sbjct: 523 LHQEGKLKQLIDKDLNDKFDRVELEEIVQVALLCTQFNPSHRPKMSEVMKMLE 575
>At1g34210 putative protein
Length = 628
Score = 358 bits (920), Expect = 4e-99
Identities = 227/590 (38%), Positives = 332/590 (55%), Gaps = 43/590 (7%)
Query: 11 ILIFTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHPGDGE 70
+L+F +++ SS +G L ++ L D NVL +W +PC W +TC+ E
Sbjct: 18 LLLFNSLWLASS---NMEGDALHSLRANLVDPNNVLQSWDPTLVNPCTWFHVTCN---NE 71
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
V ++L + L G + P +G+L LQ L L+ N++ G +P+++ N T L +L L N
Sbjct: 72 NSVIRVDLGNADLSGQLVPQLGQLKNLQYLELYSNNITGPVPSDLGNLTNLVSLDLYLNS 131
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLS 190
F G IP +G L L L L++NS G IP SL + LQVL+LS N SG +PD G S
Sbjct: 132 FTGPIPDSLGKLFKLRFLRLNNNSLTGPIPMSLTNIMTLQVLDLSNNRLSGSVPDNGSFS 191
Query: 191 TFQKNSFIGNLDLCGRQIQKPCRTSFGF---PVVIPHAESDEAAVPTKRSSSHYMKVVLI 247
F SF NLDLCG +PC S F P IP VPT Y I
Sbjct: 192 LFTPISFANNLDLCGPVTSRPCPGSPPFSPPPPFIP-----PPIVPTPGG---YSATGAI 243
Query: 248 GAMTTLGLLLLVT---LSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLP-Y 303
G LL L+F W R +E + DV + DPE G L +
Sbjct: 244 AGGVAAGAALLFAAPALAFAWWRRRKPQEF----FFDVPAEEDPEVHL------GQLKRF 293
Query: 304 TSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSRE-GCDQVFERELEI 362
+ E+ +S ++I+G GGFG VY+ + D AVKR+ R G + F+ E+E+
Sbjct: 294 SLRELQVATDSFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTPGGELQFQTEVEM 353
Query: 363 LGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHEN--TEQPLNWNDRLNIALGS 420
+ H NL+ LRG+C P+ RLL+Y Y+A GS+ L E ++ PL W+ R IALGS
Sbjct: 354 ISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPPSQLPLAWSIRQQIALGS 413
Query: 421 ARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFG 480
ARGL+YLH C PKI+HRD+K++NILL+E E + DFGLA+L+ +D HVTT V GT G
Sbjct: 414 ARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLARLMDYKDTHVTTAVRGTIG 473
Query: 481 YLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTD-PSFAN-RGLNVVGWMNTLQKENR 538
++APEYL +G+++EK+DV+ +G++LLEL+TG+R D AN + ++ W+ L KE +
Sbjct: 474 HIAPEYLSTGKSSEKTDVFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKK 533
Query: 539 LEDVVD----RRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLLE 584
LE +VD T+A+ +E ++++A CT ++ +RP M++V+++LE
Sbjct: 534 LEMLVDPDLQSNYTEAE---VEQLIQVALLCTQSSPMERPKMSEVVRMLE 580
>At2g13790 putative receptor protein kinase (F13J11.14/At2g13790)
Length = 615
Score = 334 bits (857), Expect = 7e-92
Identities = 212/568 (37%), Positives = 317/568 (55%), Gaps = 41/568 (7%)
Query: 28 DGLTLLEIKGALND---TKNVLSNWQEFDESPCAWTGITCHPGDGEQRVRSINLPYSQLG 84
+G L ++K +L+ NVL +W +PC W +TC+P E +V ++L ++L
Sbjct: 27 EGDALTQLKNSLSSGDPANNVLQSWDATLVTPCTWFHVTCNP---ENKVTRVDLGNAKLS 83
Query: 85 GIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPF 144
G + P +G+L LQ L L+ N++ G IP E+ + EL +L L AN G IPS +G L
Sbjct: 84 GKLVPELGQLLNLQYLELYSNNITGEIPEELGDLVELVSLDLYANSISGPIPSSLGKLGK 143
Query: 145 LNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNSFIGNLDLC 204
L L L++NS G IP +L + LQVL++S N SG+IP G S F SF N L
Sbjct: 144 LRFLRLNNNSLSGEIPMTLTSV-QLQVLDISNNRLSGDIPVNGSFSLFTPISFANN-SLT 201
Query: 205 GRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLV-TLSF 263
P TS P S + G LL V ++F
Sbjct: 202 DLPEPPPTSTS---------------PTPPPPSGGQMTAAIAGGVAAGAALLFAVPAIAF 246
Query: 264 LWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLP-YTSSEIIEKLESLEEEDIVG 322
W L +K + + DV + DPE G L +T E++ ++ ++++G
Sbjct: 247 AW--WLRRKPQD--HFFDVPAEEDPEVHL------GQLKRFTLRELLVATDNFSNKNVLG 296
Query: 323 SGGFGTVYRMVMNDCGTFAVKRIDRSR-EGCDQVFERELEILGSIKHINLVNLRGYCRLP 381
GGFG VY+ + D AVKR+ R +G + F+ E+E++ H NL+ LRG+C P
Sbjct: 297 RGGFGKVYKGRLADGNLVAVKRLKEERTKGGELQFQTEVEMISMAVHRNLLRLRGFCMTP 356
Query: 382 SARLLIYDYLAIGSLDDLLHENTE--QPLNWNDRLNIALGSARGLAYLHHECCPKIVHRD 439
+ RLL+Y Y+A GS+ L E E L+W R +IALGSARGLAYLH C KI+HRD
Sbjct: 357 TERLLVYPYMANGSVASCLRERPEGNPALDWPKRKHIALGSARGLAYLHDHCDQKIIHRD 416
Query: 440 IKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVY 499
+K++NILL+E E + DFGLAKL+ D+HVTT V GT G++APEYL +G+++EK+DV+
Sbjct: 417 VKAANILLDEEFEAVVGDFGLAKLMNYNDSHVTTAVRGTIGHIAPEYLSTGKSSEKTDVF 476
Query: 500 SFGVLLLELVTGKRPTD-PSFAN-RGLNVVGWMNTLQKENRLEDVVDRRCTDADAGT-LE 556
+GV+LLEL+TG++ D AN + ++ W+ + KE +LE +VD T +E
Sbjct: 477 GYGVMLLELITGQKAFDLARLANDDDIMLLDWVKEVLKEKKLESLVDAELEGKYVETEVE 536
Query: 557 VILELAARCTDANADDRPSMNQVLQLLE 584
++++A CT ++A +RP M++V+++LE
Sbjct: 537 QLIQMALLCTQSSAMERPKMSEVVRMLE 564
>At3g25560 unknown protein
Length = 635
Score = 331 bits (849), Expect = 6e-91
Identities = 211/585 (36%), Positives = 316/585 (53%), Gaps = 31/585 (5%)
Query: 10 FILIFTTVFTPSSLALTQDGLT-----LLEIKGALNDTKNVLSNWQEFDESPCAWTGITC 64
F F + SS LT G+ L+ IK +L D VL NW + PC+W ITC
Sbjct: 19 FFFFFICFLSSSSAELTDKGVNFEVVALIGIKSSLTDPHGVLMNWDDTAVDPCSWNMITC 78
Query: 65 HPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRAL 124
G V + P L G +S SIG L+ LQ + L N + G IP+EI +L+ L
Sbjct: 79 SDGF----VIRLEAPSQNLSGTLSSSIGNLTNLQTVLLQNNYITGNIPHEIGKLMKLKTL 134
Query: 125 YLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
L N F G IP + L L +++NS G IPSSL + L L+LS N SG +P
Sbjct: 135 DLSTNNFTGQIPFTLSYSKNLQYLRVNNNSLTGTIPSSLANMTQLTFLDLSYNNLSGPVP 194
Query: 185 DIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKV 244
+ TF + +GN +C +K C + P+ I S + + + +
Sbjct: 195 R-SLAKTF---NVMGNSQICPTGTEKDCNGTQPKPMSITLNSSQNKS--SDGGTKNRKIA 248
Query: 245 VLIGAMTTLGLLLLVTLSFL-WIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLP- 302
V+ G T LL++ FL W R K+ + + D+ +Q E G+L
Sbjct: 249 VVFGVSLTCVCLLIIGFGFLLWWRRRHNKQ---VLFFDINEQNKEEMCL------GNLRR 299
Query: 303 YTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQV-FERELE 361
+ E+ + +++VG GGFG VY+ ++D AVKR+ G +V F+ ELE
Sbjct: 300 FNFKELQSATSNFSSKNLVGKGGFGNVYKGCLHDGSIIAVKRLKDINNGGGEVQFQTELE 359
Query: 362 ILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSA 421
++ H NL+ L G+C S RLL+Y Y++ GS+ L + L+W R IALG+
Sbjct: 360 MISLAVHRNLLRLYGFCTTSSERLLVYPYMSNGSVASRL--KAKPVLDWGTRKRIALGAG 417
Query: 422 RGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGY 481
RGL YLH +C PKI+HRD+K++NILL++ E + DFGLAKLL E++HVTT V GT G+
Sbjct: 418 RGLLYLHEQCDPKIIHRDVKAANILLDDYFEAVVGDFGLAKLLDHEESHVTTAVRGTVGH 477
Query: 482 LAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTD-PSFANRGLNVVGWMNTLQKENRLE 540
+APEYL +G+++EK+DV+ FG+LLLEL+TG R + AN+ ++ W+ LQ+E +LE
Sbjct: 478 IAPEYLSTGQSSEKTDVFGFGILLLELITGLRALEFGKAANQRGAILDWVKKLQQEKKLE 537
Query: 541 DVVDRRC-TDADAGTLEVILELAARCTDANADDRPSMNQVLQLLE 584
+VD+ ++ D +E ++++A CT RP M++V+++LE
Sbjct: 538 QIVDKDLKSNYDRIEVEEMVQVALLCTQYLPIHRPKMSEVVRMLE 582
>At4g33430 somatic embryogenesis receptor-like kinase 3 (SERK3)
Length = 615
Score = 330 bits (847), Expect = 1e-90
Identities = 216/585 (36%), Positives = 318/585 (53%), Gaps = 45/585 (7%)
Query: 11 ILIFTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHPGDGE 70
IL+ V S A +G L +K +L D VL +W +PC W +TC+ +
Sbjct: 14 ILVLDLVLRVSGNA---EGDALSALKNSLADPNKVLQSWDATLVTPCTWFHVTCN---SD 67
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
V ++L + L G + +G+L LQ L L+ N++ G IP ++ N TEL +L L N
Sbjct: 68 NSVTRVDLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNN 127
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLS 190
G IPS +G L L L L++NS G IP SL + LQVL+LS N +G+IP G S
Sbjct: 128 LSGPIPSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIPVNGSFS 187
Query: 191 TFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAM 250
F SF N L P P + P P+ S+ + G
Sbjct: 188 LFTPISF-ANTKLTPLPASPP-------PPISP-------TPPSPAGSNRITGAIAGGVA 232
Query: 251 TTLGLLLLV-TLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLP-YTSSEI 308
LL V ++ W R +++ + DV + DPE G L ++ E+
Sbjct: 233 AGAALLFAVPAIALAWWR----RKKPQDHFFDVPAEEDPEVHL------GQLKRFSLREL 282
Query: 309 IEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSR-EGCDQVFERELEILGSIK 367
++ ++I+G GGFG VY+ + D AVKR+ R +G + F+ E+E++
Sbjct: 283 QVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVEMISMAV 342
Query: 368 HINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQ--PLNWNDRLNIALGSARGLA 425
H NL+ LRG+C P+ RLL+Y Y+A GS+ L E E PL+W R IALGSARGLA
Sbjct: 343 HRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALGSARGLA 402
Query: 426 YLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPE 485
YLH C PKI+HRD+K++NILL+E E + DFGLAKL+ +D HVTT V GT G++APE
Sbjct: 403 YLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPE 462
Query: 486 YLQSGRATEKSDVYSFGVLLLELVTGKRPTD-PSFAN-RGLNVVGWMNTLQKENRLEDVV 543
YL +G+++EK+DV+ +GV+LLEL+TG+R D AN + ++ W+ L KE +LE +V
Sbjct: 463 YLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEALV 522
Query: 544 DRRCTDADAG----TLEVILELAARCTDANADDRPSMNQVLQLLE 584
D D +E ++++A CT ++ +RP M++V+++LE
Sbjct: 523 D---VDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLE 564
>At4g30520 receptor-like kinase homolog
Length = 648
Score = 329 bits (844), Expect = 2e-90
Identities = 209/585 (35%), Positives = 319/585 (53%), Gaps = 34/585 (5%)
Query: 7 IWVFILIFTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHP 66
++ F+ + + T SS + L+ I+ L+D L+NW EF PC+W ITC P
Sbjct: 16 LYSFLFLCFSTLTLSSEPRNPEVEALISIRNNLHDPHGALNNWDEFSVDPCSWAMITCSP 75
Query: 67 GDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYL 126
+ V + P L G +S SIG L+ L++++L N++ G IP E+ +L+ L L
Sbjct: 76 ---DNLVIGLGAPSQSLSGGLSESIGNLTNLRQVSLQNNNISGKIPPELGFLPKLQTLDL 132
Query: 127 RANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDI 186
N F G IP I L L L L++NS G P+SL ++PHL L+LS N SG +P
Sbjct: 133 SNNRFSGDIPVSIDQLSSLQYLRLNNNSLSGPFPASLSQIPHLSFLDLSYNNLSGPVPKF 192
Query: 187 GVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVL 246
TF + GN +C + C S + +V SS +
Sbjct: 193 PA-RTF---NVAGNPLICRSNPPEICSGSIN---------ASPLSVSLSSSSGRRSNRLA 239
Query: 247 IGAMTTLGLLLLVTL---SFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPY 303
I +LG ++++ L SF W R K+ R ++ + K++ + L +F
Sbjct: 240 IALSVSLGSVVILVLALGSFCWYR--KKQRRLLILNLNDKQEEGLQGLGNLRSF------ 291
Query: 304 TSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRI-DRSREGCDQVFERELEI 362
T E+ + ++I+G+GGFG VYR + D AVKR+ D + D F ELE+
Sbjct: 292 TFRELHVYTDGFSSKNILGAGGFGNVYRGKLGDGTMVAVKRLKDINGTSGDSQFRMELEM 351
Query: 363 LGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSAR 422
+ H NL+ L GYC RLL+Y Y+ GS+ L ++ L+WN R IA+G+AR
Sbjct: 352 ISLAVHKNLLRLIGYCATSGERLLVYPYMPNGSVASKL--KSKPALDWNMRKRIAIGAAR 409
Query: 423 GLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYL 482
GL YLH +C PKI+HRD+K++NILL+E E + DFGLAKLL D+HVTT V GT G++
Sbjct: 410 GLLYLHEQCDPKIIHRDVKAANILLDECFEAVVGDFGLAKLLNHADSHVTTAVRGTVGHI 469
Query: 483 APEYLQSGRATEKSDVYSFGVLLLELVTGKRPTD--PSFANRGLNVVGWMNTLQKENRLE 540
APEYL +G+++EK+DV+ FG+LLLEL+TG R + + + +G ++ W+ L +E ++E
Sbjct: 470 APEYLSTGQSSEKTDVFGFGILLLELITGLRALEFGKTVSQKGA-MLEWVRKLHEEMKVE 528
Query: 541 DVVDRRC-TDADAGTLEVILELAARCTDANADDRPSMNQVLQLLE 584
+++DR T+ D + +L++A CT RP M++V+ +LE
Sbjct: 529 ELLDRELGTNYDKIEVGEMLQVALLCTQYLPAHRPKMSEVVLMLE 573
>At5g16000 receptor protein kinase-like protein
Length = 638
Score = 319 bits (817), Expect = 3e-87
Identities = 196/560 (35%), Positives = 306/560 (54%), Gaps = 27/560 (4%)
Query: 32 LLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHPGDGEQRVRSINLPYSQLGGIISPSI 91
L++IK +L+D VL NW PC+WT +TC E V + P L G +SPSI
Sbjct: 45 LMDIKASLHDPHGVLDNWDRDAVDPCSWTMVTC---SSENFVIGLGTPSQNLSGTLSPSI 101
Query: 92 GKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNILDLS 151
L+ L+ + L N++ G IP EI T L L L N+F G IP +G L L L L+
Sbjct: 102 TNLTNLRIVLLQNNNIKGKIPAEIGRLTRLETLDLSDNFFHGEIPFSVGYLQSLQYLRLN 161
Query: 152 SNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKP 211
+NS G P SL + L L+LS N SG +P TF S +GN +C +
Sbjct: 162 NNSLSGVFPLSLSNMTQLAFLDLSYNNLSGPVPRFAA-KTF---SIVGNPLICPTGTEPD 217
Query: 212 CRTSFGFPVVIPHAESDEAAVPTKRSSS--HYMKVVLIGAMTTLGLLLLVTLSFLWIRLL 269
C + P+ + ++ VP S H M + + ++ T+ L+ + FLW R
Sbjct: 218 CNGTTLIPMSM---NLNQTGVPLYAGGSRNHKMAIAVGSSVGTVSLIFIAVGLFLWWRQR 274
Query: 270 SKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEIIEKLESLEEEDIVGSGGFGTV 329
+ + DVK E ++ + E+ + ++++G GG+G V
Sbjct: 275 HNQNT----FFDVKDGNHHEE----VSLGNLRRFGFRELQIATNNFSSKNLLGKGGYGNV 326
Query: 330 YRMVMNDCGTFAVKRI-DRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIY 388
Y+ ++ D AVKR+ D G + F+ E+E++ H NL+ L G+C + +LL+Y
Sbjct: 327 YKGILGDSTVVAVKRLKDGGALGGEIQFQTEVEMISLAVHRNLLRLYGFCITQTEKLLVY 386
Query: 389 DYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLN 448
Y++ GS+ + + L+W+ R IA+G+ARGL YLH +C PKI+HRD+K++NILL+
Sbjct: 387 PYMSNGSVASRM--KAKPVLDWSIRKRIAIGAARGLVYLHEQCDPKIIHRDVKAANILLD 444
Query: 449 ENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLEL 508
+ E + DFGLAKLL +D+HVTT V GT G++APEYL +G+++EK+DV+ FG+LLLEL
Sbjct: 445 DYCEAVVGDFGLAKLLDHQDSHVTTAVRGTVGHIAPEYLSTGQSSEKTDVFGFGILLLEL 504
Query: 509 VTGKRPTD-PSFANRGLNVVGWMNTLQKENRLEDVVDR---RCTDADAGTLEVILELAAR 564
VTG+R + AN+ ++ W+ + +E +LE +VD+ + D L+ ++ +A
Sbjct: 505 VTGQRAFEFGKAANQKGVMLDWVKKIHQEKKLELLVDKELLKKKSYDEIELDEMVRVALL 564
Query: 565 CTDANADDRPSMNQVLQLLE 584
CT RP M++V+++LE
Sbjct: 565 CTQYLPGHRPKMSEVVRMLE 584
>At2g13800 receptor-like protein kinase
Length = 601
Score = 318 bits (816), Expect = 4e-87
Identities = 206/563 (36%), Positives = 312/563 (54%), Gaps = 53/563 (9%)
Query: 32 LLEIKGALND---TKNVLSNWQEFDESPCAWTGITCHPGDGEQRVRSINLPYSQLGGIIS 88
L+ ++ +L+ T N+L +W +PC+W +TC+ E V ++L + L G +
Sbjct: 31 LIALRSSLSSGDHTNNILQSWNATHVTPCSWFHVTCNT---ENSVTRLDLGSANLSGELV 87
Query: 89 PSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNIL 148
P + +L LQ L L N++ G IP E+ + EL +L L AN G IPS +G L L L
Sbjct: 88 PQLAQLPNLQYLELFNNNITGEIPEELGDLMELVSLDLFANNISGPIPSSLGKLGKLRFL 147
Query: 149 DLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNSFIGNLDLCGRQI 208
L +NS G IP SL LP L VL++S N SG+IP G S F SF N +
Sbjct: 148 RLYNNSLSGEIPRSLTALP-LDVLDISNNRLSGDIPVNGSFSQFTSMSFANN-----KLR 201
Query: 209 QKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSFLWIRL 268
+P S P AA+ VV + A G LL L++ W+R
Sbjct: 202 PRPASPS-------PSPSGTSAAI-----------VVGVAA----GAALLFALAW-WLR- 237
Query: 269 LSKKERAVMRYTDVKKQVDPEASTKLITFHGDLP-YTSSEIIEKLESLEEEDIVGSGGFG 327
+ + DV + DPE + G ++ E++ E + +++G G FG
Sbjct: 238 ----RKLQGHFLDVPAEEDPEV------YLGQFKRFSLRELLVATEKFSKRNVLGKGRFG 287
Query: 328 TVYRMVMNDCGTFAVKRIDRSR-EGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLL 386
+Y+ + D AVKR++ R +G + F+ E+E++ H NL+ LRG+C P+ RLL
Sbjct: 288 ILYKGRLADDTLVAVKRLNEERTKGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLL 347
Query: 387 IYDYLAIGSLDDLLHENTE--QPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSN 444
+Y Y+A GS+ L E E L+W R +IALGSARGLAYLH C KI+H D+K++N
Sbjct: 348 VYPYMANGSVASCLRERPEGNPALDWPKRKHIALGSARGLAYLHDHCDQKIIHLDVKAAN 407
Query: 445 ILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVL 504
ILL+E E + DFGLAKL+ D+HVTT V GT G++APEYL +G+++EK+DV+ +GV+
Sbjct: 408 ILLDEEFEAVVGDFGLAKLMNYNDSHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVM 467
Query: 505 LLELVTGKRPTD-PSFAN-RGLNVVGWMNTLQKENRLEDVVDRRCTDADAGT-LEVILEL 561
LLEL+TG++ D AN + ++ W+ + KE +LE +VD T +E ++++
Sbjct: 468 LLELITGQKAFDLARLANDDDIMLLDWVKEVLKEKKLESLVDAELEGKYVETEVEQLIQM 527
Query: 562 AARCTDANADDRPSMNQVLQLLE 584
A CT ++A +RP M++V+++LE
Sbjct: 528 ALLCTQSSAMERPKMSEVVRMLE 550
>At5g65240 receptor-like protein kinase
Length = 607
Score = 314 bits (805), Expect = 8e-86
Identities = 203/584 (34%), Positives = 322/584 (54%), Gaps = 40/584 (6%)
Query: 12 LIFTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHPGDGEQ 71
L+F++++ SS++ G L ++ +L + LS+W + PC W+ + C D ++
Sbjct: 9 LVFSSLW--SSVSPDAQGDALFALRSSLRASPEQLSDWNQNQVDPCTWSQVIC---DDKK 63
Query: 72 RVRSINLPYSQLG-GIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
V S+ L Y G +S IG L+ L+ L L N + G IP I N + L +L L N+
Sbjct: 64 HVTSVTLSYMNFSSGTLSSGIGILTTLKTLTLKGNGIMGGIPESIGNLSSLTSLDLEDNH 123
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLS 190
IPS +GNL L L LS N+ G+IP SL L L + L +N SGEIP L
Sbjct: 124 LTDRIPSTLGNLKNLQFLTLSRNNLNGSIPDSLTGLSKLINILLDSNNLSGEIPQS--LF 181
Query: 191 TFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAM 250
K +F N CG +PC T ES + + R + ++ G +
Sbjct: 182 KIPKYNFTANNLSCGGTFPQPCVT-----------ESSPSGDSSSRKTG-----IIAGVV 225
Query: 251 TTLGLLLLVTLSFLWIRLLSKKERAVMR--YTDVKKQVDPEASTKLITFHGDLPYTSSEI 308
+ + ++LL F + K + R + DV +VD + I F + E+
Sbjct: 226 SGIAVILL---GFFFFFFCKDKHKGYKRDVFVDVAGEVD-----RRIAFGQLRRFAWREL 277
Query: 309 IEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRI-DRSREGCDQVFERELEILGSIK 367
+ E++++G GGFG VY+ +++D AVKR+ D R G D+ F+RE+E++
Sbjct: 278 QLATDEFSEKNVLGQGGFGKVYKGLLSDGTKVAVKRLTDFERPGGDEAFQREVEMISVAV 337
Query: 368 HINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENT--EQPLNWNDRLNIALGSARGLA 425
H NL+ L G+C + RLL+Y ++ S+ L E + L+W R IALG+ARGL
Sbjct: 338 HRNLLRLIGFCTTQTERLLVYPFMQNLSVAYCLREIKPGDPVLDWFRRKQIALGAARGLE 397
Query: 426 YLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPE 485
YLH C PKI+HRD+K++N+LL+E+ E + DFGLAKL+ +VTT V GT G++APE
Sbjct: 398 YLHEHCNPKIIHRDVKAANVLLDEDFEAVVGDFGLAKLVDVRRTNVTTQVRGTMGHIAPE 457
Query: 486 YLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNV--VGWMNTLQKENRLEDVV 543
+ +G+++EK+DV+ +G++LLELVTG+R D S +V + + L++E RLED+V
Sbjct: 458 CISTGKSSEKTDVFGYGIMLLELVTGQRAIDFSRLEEEDDVLLLDHVKKLEREKRLEDIV 517
Query: 544 DRRC-TDADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQE 586
D++ D +E+++++A CT A ++RP+M++V+++LE E
Sbjct: 518 DKKLDEDYIKEEVEMMIQVALLCTQAAPEERPAMSEVVRMLEGE 561
>At4g39400 brassinosteroid insensitive 1 gene (BRI1)
Length = 1196
Score = 314 bits (804), Expect = 1e-85
Identities = 198/540 (36%), Positives = 288/540 (52%), Gaps = 49/540 (9%)
Query: 77 NLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIP 136
N+ GG SP+ + L + N L G IP EI + L L L N G IP
Sbjct: 636 NITSRVYGGHTSPTFDNNGSMMFLDMSYNMLSGYIPKEIGSMPYLFILNLGHNDISGSIP 695
Query: 137 SDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNS 196
++G+L LNILDLSSN G IP ++ L L ++LS N SG IP++G TF
Sbjct: 696 DEVGDLRGLNILDLSSNKLDGRIPQAMSALTMLTEIDLSNNNLSGPIPEMGQFETFPPAK 755
Query: 197 FIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLL 256
F+ N LCG +P +P + A SH + + +GLL
Sbjct: 756 FLNNPGLCG------------YP--LPRCDPSNADGYAHHQRSHGRRPASLAGSVAMGLL 801
Query: 257 LLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEA-----------------------STK 293
F I L+ ++ R R + + ++ E S
Sbjct: 802 FSFVCIFGLI-LVGREMRKRRRKKEAELEMYAEGHGNSGDRTANNTNWKLTGVKEALSIN 860
Query: 294 LITFHGDL-PYTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGC 352
L F L T +++++ + ++GSGGFG VY+ ++ D A+K++
Sbjct: 861 LAAFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQG 920
Query: 353 DQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQ--PLNW 410
D+ F E+E +G IKH NLV L GYC++ RLL+Y+++ GSL+D+LH+ + LNW
Sbjct: 921 DREFMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNW 980
Query: 411 NDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAH 470
+ R IA+GSARGLA+LHH C P I+HRD+KSSN+LL+EN+E +SDFG+A+L+ D H
Sbjct: 981 STRRKIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTH 1040
Query: 471 VT-TVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTD-PSFANRGLNVVG 528
++ + +AGT GY+ PEY QS R + K DVYS+GV+LLEL+TGKRPTD P F + N+VG
Sbjct: 1041 LSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDFGDN--NLVG 1098
Query: 529 WMNTLQKENRLEDVVDRRCTDAD-AGTLEVI--LELAARCTDANADDRPSMNQVLQLLEQ 585
W+ K R+ DV D D A +E++ L++A C D A RP+M QV+ + ++
Sbjct: 1099 WVKQHAK-LRISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAMFKE 1157
Score = 84.3 bits (207), Expect = 2e-16
Identities = 48/110 (43%), Positives = 62/110 (55%)
Query: 75 SINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGG 134
S++L ++ L G I S+G LS+L+ L L N L G IP E+ L L L N G
Sbjct: 444 SLHLSFNYLSGTIPSSLGSLSKLRDLKLWLNMLEGEIPQELMYVKTLETLILDFNDLTGE 503
Query: 135 IPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
IPS + N LN + LS+N G IP +GRL +L +L LS N FSG IP
Sbjct: 504 IPSGLSNCTNLNWISLSNNRLTGEIPKWIGRLENLAILKLSNNSFSGNIP 553
Score = 69.3 bits (168), Expect = 6e-12
Identities = 65/211 (30%), Positives = 106/211 (49%), Gaps = 19/211 (9%)
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITN-CTELRALYLRANYF 131
++ +N+ +Q G I P L LQ L+L +N G IP+ ++ C L L L N+F
Sbjct: 271 LKLLNISSNQFVGPIPPL--PLKSLQYLSLAENKFTGEIPDFLSGACDTLTGLDLSGNHF 328
Query: 132 QGGIPSDIGNLPFLNILDLSSNSFKGAIP-SSLGRLPHLQVLNLSTNFFSGEIPD-IGVL 189
G +P G+ L L LSSN+F G +P +L ++ L+VL+LS N FSGE+P+ + L
Sbjct: 329 YGAVPPFFGSCSLLESLALSSNNFSGELPMDTLLKMRGLKVLDLSFNEFSGELPESLTNL 388
Query: 190 ST------FQKNSFIGNL--DLC---GRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSS 238
S N+F G + +LC +Q+ + GF IP S+ + + + S
Sbjct: 389 SASLLTLDLSSNNFSGPILPNLCQNPKNTLQELYLQNNGFTGKIPPTLSNCSELVSLHLS 448
Query: 239 SHYMKVVLIGAMTTLGLLLLVTLSFLWIRLL 269
+Y+ + ++LG L + LW+ +L
Sbjct: 449 FNYLSGTI---PSSLGSLSKLRDLKLWLNML 476
Score = 68.6 bits (166), Expect = 1e-11
Identities = 42/112 (37%), Positives = 60/112 (53%), Gaps = 2/112 (1%)
Query: 75 SINLPYSQLGGIISPSI--GKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
+++L + G I P++ + LQ L L N G IP ++NC+EL +L+L NY
Sbjct: 394 TLDLSSNNFSGPILPNLCQNPKNTLQELYLQNNGFTGKIPPTLSNCSELVSLHLSFNYLS 453
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
G IPS +G+L L L L N +G IP L + L+ L L N +GEIP
Sbjct: 454 GTIPSSLGSLSKLRDLKLWLNMLEGEIPQELMYVKTLETLILDFNDLTGEIP 505
Score = 62.4 bits (150), Expect = 7e-10
Identities = 42/119 (35%), Positives = 57/119 (47%), Gaps = 23/119 (19%)
Query: 89 PSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPS-DIGNLPFLNI 147
P +G S LQ L + N L G I+ CTEL+ L + +N F G IP + +L +L++
Sbjct: 239 PFLGDCSALQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPIPPLPLKSLQYLSL 298
Query: 148 ----------------------LDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
LDLS N F GA+P G L+ L LS+N FSGE+P
Sbjct: 299 AENKFTGEIPDFLSGACDTLTGLDLSGNHFYGAVPPFFGSCSLLESLALSSNNFSGELP 357
Score = 55.5 bits (132), Expect = 9e-08
Identities = 34/110 (30%), Positives = 57/110 (50%), Gaps = 4/110 (3%)
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
+ ++ L ++ L G I + + L ++L N L G IP I L L L N F
Sbjct: 490 LETLILDFNDLTGEIPSGLSNCTNLNWISLSNNRLTGEIPKWIGRLENLAILKLSNNSFS 549
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGE 182
G IP+++G+ L LDL++N F G IP+++ + Q ++ NF +G+
Sbjct: 550 GNIPAELGDCRSLIWLDLNTNLFNGTIPAAMFK----QSGKIAANFIAGK 595
Score = 54.7 bits (130), Expect = 1e-07
Identities = 36/123 (29%), Positives = 63/123 (50%), Gaps = 10/123 (8%)
Query: 83 LGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNL 142
+G ++S G+L + LA+ N + G + +++ C L L + +N F GIP +G+
Sbjct: 191 VGWVLSDGCGEL---KHLAISGNKISGDV--DVSRCVNLEFLDVSSNNFSTGIPF-LGDC 244
Query: 143 PFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLS----TFQKNSFI 198
L LD+S N G ++ L++LN+S+N F G IP + + S + +N F
Sbjct: 245 SALQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPIPPLPLKSLQYLSLAENKFT 304
Query: 199 GNL 201
G +
Sbjct: 305 GEI 307
Score = 53.5 bits (127), Expect = 3e-07
Identities = 63/247 (25%), Positives = 102/247 (40%), Gaps = 63/247 (25%)
Query: 10 FILIFTTVFTPS-SLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHPGD 68
F F+ F S S +L ++ L+ K L D KN+L +W +++PC + G+TC
Sbjct: 16 FFSFFSLSFQASPSQSLYREIHQLISFKDVLPD-KNLLPDWSS-NKNPCTFDGVTCR--- 70
Query: 69 GEQRVRSINLPYSQLG---GIISPSIGKLSRLQRLALHQNSLHGIIPN------------ 113
+ +V SI+L L +S S+ L+ L+ L L + ++G +
Sbjct: 71 -DDKVTSIDLSSKPLNVGFSAVSSSLLSLTGLESLFLSNSHINGSVSGFKCSASLTSLDL 129
Query: 114 -------------EITNCTELRALYLRANY--FQGGIPSDIGNLPFLNILDLSSNSFKGA 158
+ +C+ L+ L + +N F G + + L L +LDLS+NS GA
Sbjct: 130 SRNSLSGPVTTLTSLGSCSGLKFLNVSSNTLDFPGKVSGGL-KLNSLEVLDLSANSISGA 188
Query: 159 ------IPSSLGRLPHLQV-------------------LNLSTNFFSGEIPDIGVLSTFQ 193
+ G L HL + L++S+N FS IP +G S Q
Sbjct: 189 NVVGWVLSDGCGELKHLAISGNKISGDVDVSRCVNLEFLDVSSNNFSTGIPFLGDCSALQ 248
Query: 194 KNSFIGN 200
GN
Sbjct: 249 HLDISGN 255
>At1g55610 receptor kinase, putative
Length = 1166
Score = 311 bits (796), Expect = 9e-85
Identities = 183/500 (36%), Positives = 277/500 (54%), Gaps = 31/500 (6%)
Query: 105 NSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLG 164
N++ G IP N L+ L L N G IP G L + +LDLS N+ +G +P SLG
Sbjct: 649 NAVSGFIPPGYGNMGYLQVLNLGHNRITGTIPDSFGGLKAIGVLDLSHNNLQGYLPGSLG 708
Query: 165 RLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIP- 223
L L L++S N +G IP G L+TF + + N LCG + +PC ++ P+
Sbjct: 709 SLSFLSDLDVSNNNLTGPIPFGGQLTTFPVSRYANNSGLCGVPL-RPCGSAPRRPITSRI 767
Query: 224 HAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVK 283
HA+ A V+ G + +++ ++ +R + KKE+ +Y +
Sbjct: 768 HAKKQTVAT-----------AVIAGIAFSFMCFVMLVMALYRVRKVQKKEQKREKYIESL 816
Query: 284 ----------KQVDPEASTKLITFHGDL-PYTSSEIIEKLESLEEEDIVGSGGFGTVYRM 332
V S + TF L T + ++E E +VGSGGFG VY+
Sbjct: 817 PTSGSCSWKLSSVPEPLSINVATFEKPLRKLTFAHLLEATNGFSAETMVGSGGFGEVYKA 876
Query: 333 VMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLA 392
+ D A+K++ R D+ F E+E +G IKH NLV L GYC++ RLL+Y+Y+
Sbjct: 877 QLRDGSVVAIKKLIRITGQGDREFMAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMK 936
Query: 393 IGSLDDLLHENTEQP----LNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLN 448
GSL+ +LHE + + LNW R IA+G+ARGLA+LHH C P I+HRD+KSSN+LL+
Sbjct: 937 WGSLETVLHEKSSKKGGIYLNWAARKKIAIGAARGLAFLHHSCIPHIIHRDMKSSNVLLD 996
Query: 449 ENMEPHISDFGLAKLLVDEDAHVT-TVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLE 507
E+ E +SDFG+A+L+ D H++ + +AGT GY+ PEY QS R T K DVYS+GV+LLE
Sbjct: 997 EDFEARVSDFGMARLVSALDTHLSVSTLAGTPGYVPPEYYQSFRCTAKGDVYSYGVILLE 1056
Query: 508 LVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRCTDADAGTLEVI--LELAARC 565
L++GK+P DP N+VGW L +E R +++D +G +E+ L++A++C
Sbjct: 1057 LLSGKKPIDPGEFGEDNNLVGWAKQLYREKRGAEILDPELVTDKSGDVELFHYLKIASQC 1116
Query: 566 TDANADDRPSMNQVLQLLEQ 585
D RP+M Q++ + ++
Sbjct: 1117 LDDRPFKRPTMIQLMAMFKE 1136
Score = 68.6 bits (166), Expect = 1e-11
Identities = 38/92 (41%), Positives = 52/92 (56%)
Query: 93 KLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNILDLSS 152
K L+ L L+ N L G IP I+ CT + + L +N G IPS IGNL L IL L +
Sbjct: 473 KGGNLETLILNNNLLTGSIPESISRCTNMIWISLSSNRLTGKIPSGIGNLSKLAILQLGN 532
Query: 153 NSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
NS G +P LG L L+L++N +G++P
Sbjct: 533 NSLSGNVPRQLGNCKSLIWLDLNSNNLTGDLP 564
Score = 60.5 bits (145), Expect = 3e-09
Identities = 47/194 (24%), Positives = 79/194 (40%), Gaps = 47/194 (24%)
Query: 20 PSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTGITCHPGDGEQRVRSINLP 79
P L+L L +L++ G N S + C W ++++NL
Sbjct: 294 PPELSLLCKTLVILDLSG------NTFSGELPSQFTACVW-------------LQNLNLG 334
Query: 80 YSQLGG-IISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRAN--------- 129
+ L G ++ + K++ + L + N++ G +P +TNC+ LR L L +N
Sbjct: 335 NNYLSGDFLNTVVSKITGITYLYVAYNNISGSVPISLTNCSNLRVLDLSSNGFTGNVPSG 394
Query: 130 ------------------YFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQV 171
Y G +P ++G L +DLS N G IP + LP+L
Sbjct: 395 FCSLQSSPVLEKILIANNYLSGTVPMELGKCKSLKTIDLSFNELTGPIPKEIWMLPNLSD 454
Query: 172 LNLSTNFFSGEIPD 185
L + N +G IP+
Sbjct: 455 LVMWANNLTGTIPE 468
Score = 57.4 bits (137), Expect = 2e-08
Identities = 46/139 (33%), Positives = 67/139 (48%), Gaps = 13/139 (9%)
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
+ I + + L G + +GK L+ + L N L G IP EI L L + AN
Sbjct: 404 LEKILIANNYLSGTVPMELGKCKSLKTIDLSFNELTGPIPKEIWMLPNLSDLVMWANNLT 463
Query: 133 GGIPSDI----GNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPD-IG 187
G IP + GN L L L++N G+IP S+ R ++ ++LS+N +G+IP IG
Sbjct: 464 GTIPEGVCVKGGN---LETLILNNNLLTGSIPESISRCTNMIWISLSSNRLTGKIPSGIG 520
Query: 188 VLSTFQ-----KNSFIGNL 201
LS NS GN+
Sbjct: 521 NLSKLAILQLGNNSLSGNV 539
Score = 54.7 bits (130), Expect = 1e-07
Identities = 46/138 (33%), Positives = 65/138 (46%), Gaps = 5/138 (3%)
Query: 73 VRSINLPYSQLGGIISP-SIGKLSRLQRLALHQNSLHGI-IPNEITNCTELRALYLRANY 130
++ ++L ++ L G S S G L +L QN+L G P + NC L L + N
Sbjct: 203 LKYLDLTHNNLSGDFSDLSFGICGNLTFFSLSQNNLSGDKFPITLPNCKFLETLNISRNN 262
Query: 131 FQGGIPSDI--GNLPFLNILDLSSNSFKGAIPSSLGRL-PHLQVLNLSTNFFSGEIPDIG 187
G IP+ G+ L L L+ N G IP L L L +L+LS N FSGE+P
Sbjct: 263 LAGKIPNGEYWGSFQNLKQLSLAHNRLSGEIPPELSLLCKTLVILDLSGNTFSGELPSQF 322
Query: 188 VLSTFQKNSFIGNLDLCG 205
+ +N +GN L G
Sbjct: 323 TACVWLQNLNLGNNYLSG 340
Score = 53.1 bits (126), Expect = 4e-07
Identities = 35/103 (33%), Positives = 52/103 (49%), Gaps = 4/103 (3%)
Query: 61 GITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTE 120
G+ G+ E + + NL L G I SI + + + ++L N L G IP+ I N ++
Sbjct: 469 GVCVKGGNLETLILNNNL----LTGSIPESISRCTNMIWISLSSNRLTGKIPSGIGNLSK 524
Query: 121 LRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSL 163
L L L N G +P +GN L LDL+SN+ G +P L
Sbjct: 525 LAILQLGNNSLSGNVPRQLGNCKSLIWLDLNSNNLTGDLPGEL 567
Score = 46.2 bits (108), Expect = 5e-05
Identities = 25/66 (37%), Positives = 37/66 (55%)
Query: 76 INLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGI 135
I+L ++L G I IG LS+L L L NSL G +P ++ NC L L L +N G +
Sbjct: 504 ISLSSNRLTGKIPSGIGNLSKLAILQLGNNSLSGNVPRQLGNCKSLIWLDLNSNNLTGDL 563
Query: 136 PSDIGN 141
P ++ +
Sbjct: 564 PGELAS 569
Score = 46.2 bits (108), Expect = 5e-05
Identities = 47/176 (26%), Positives = 75/176 (41%), Gaps = 32/176 (18%)
Query: 40 NDTKNVLSNWQ-EFDESPCAWTGITCHPGDGEQRVRSINLPYSQLGGIIS---------- 88
+D NVL NW+ E C+W G++C DG R+ ++L S L G ++
Sbjct: 48 SDPNNVLGNWKYESGRGSCSWRGVSCSD-DG--RIVGLDLRNSGLTGTLNLVNLTALPNL 104
Query: 89 --------------PSIGKLSRLQRLALHQNSL--HGIIPNEITNCTELRALYLRANYFQ 132
S G LQ L L NS+ + ++ + C+ L ++ + N
Sbjct: 105 QNLYLQGNYFSSGGDSSGSDCYLQVLDLSSNSISDYSMVDYVFSKCSNLVSVNISNNKLV 164
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSS-LGRLP-HLQVLNLSTNFFSGEIPDI 186
G + +L L +DLS N IP S + P L+ L+L+ N SG+ D+
Sbjct: 165 GKLGFAPSSLQSLTTVDLSYNILSDKIPESFISDFPASLKYLDLTHNNLSGDFSDL 220
>At5g63710 receptor-like protein kinase
Length = 614
Score = 310 bits (794), Expect = 1e-84
Identities = 205/581 (35%), Positives = 306/581 (52%), Gaps = 58/581 (9%)
Query: 10 FILIFTTVFTPSSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPC-AWTGITCHPGD 68
F+ + T S+ +G LL+++ +LND+ N L W SPC +W+ +TC
Sbjct: 35 FMALAFVGITSSTTQPDIEGGALLQLRDSLNDSSNRLK-WTRDFVSPCYSWSYVTCRG-- 91
Query: 69 GEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRA 128
Q V ++NL S G +SP+I KL L L L NSL G +P+ + N L+ L L
Sbjct: 92 --QSVVALNLASSGFTGTLSPAITKLKFLVTLELQNNSLSGALPDSLGNMVNLQTLNLSV 149
Query: 129 NYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGV 188
N F G IP+ L L LDLSSN+ G+IP T FFS
Sbjct: 150 NSFSGSIPASWSQLSNLKHLDLSSNNLTGSIP---------------TQFFS-------- 186
Query: 189 LSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIG 248
+ TF F G +CG+ + +PC +S PV SS ++ + +
Sbjct: 187 IPTF---DFSGTQLICGKSLNQPCSSSSRLPVT---------------SSKKKLRDITLT 228
Query: 249 AMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEI 308
A ++L FL ++ R D+ V E K I+F ++ EI
Sbjct: 229 ASCVASIIL-----FLGAMVMYHHHRVRRTKYDIFFDVAGEDDRK-ISFGQLKRFSLREI 282
Query: 309 IEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRI-DRSREGCDQVFERELEILGSIK 367
+S E +++G GGFG VYR ++ D AVKR+ D G + F+RE++++
Sbjct: 283 QLATDSFNESNLIGQGGFGKVYRGLLPDKTKVAVKRLADYFSPGGEAAFQREIQLISVAV 342
Query: 368 HINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHE--NTEQPLNWNDRLNIALGSARGLA 425
H NL+ L G+C S R+L+Y Y+ S+ L + E+ L+W R +A GSA GL
Sbjct: 343 HKNLLRLIGFCTTSSERILVYPYMENLSVAYRLRDLKAGEEGLDWPTRKRVAFGSAHGLE 402
Query: 426 YLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPE 485
YLH C PKI+HRD+K++NILL+ N EP + DFGLAKL+ HVTT V GT G++APE
Sbjct: 403 YLHEHCNPKIIHRDLKAANILLDNNFEPVLGDFGLAKLVDTSLTHVTTQVRGTMGHIAPE 462
Query: 486 YLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNV--VGWMNTLQKENRLEDVV 543
YL +G+++EK+DV+ +G+ LLELVTG+R D S N+ + + L +E RL D+V
Sbjct: 463 YLCTGKSSEKTDVFGYGITLLELVTGQRAIDFSRLEEEENILLLDHIKKLLREQRLRDIV 522
Query: 544 DRRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLLE 584
D T D+ +E I+++A CT + +DRP+M++V+++L+
Sbjct: 523 DSNLTTYDSKEVETIVQVALLCTQGSPEDRPAMSEVVKMLQ 563
>At2g23950 putative LRR receptor protein kinase
Length = 607
Score = 309 bits (791), Expect = 3e-84
Identities = 196/533 (36%), Positives = 298/533 (55%), Gaps = 38/533 (7%)
Query: 79 PYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSD 138
P L G +S SIG L+ L++++L N++ G IP EI + +L+ L L N F G IP
Sbjct: 55 PSQSLSGTLSGSIGNLTNLRQVSLQNNNISGKIPPEICSLPKLQTLDLSNNRFSGEIPGS 114
Query: 139 IGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNSFI 198
+ L L L L++NS G P+SL ++PHL L+LS N G +P TF +
Sbjct: 115 VNQLSNLQYLRLNNNSLSGPFPASLSQIPHLSFLDLSYNNLRGPVPKFPA-RTF---NVA 170
Query: 199 GNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLL 258
GN +C + + C S + +V + SS ++ + +LG +
Sbjct: 171 GNPLICKNSLPEICSGSIS---------ASPLSVSLRSSSGRRTNILAVALGVSLGFAVS 221
Query: 259 VTLS--FLWIRLLSKKER--AVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEIIEKLES 314
V LS F+W R KK+R ++R +D +++ L+ +T E+ +
Sbjct: 222 VILSLGFIWYR---KKQRRLTMLRISDKQEE-------GLLGLGNLRSFTFRELHVATDG 271
Query: 315 LEEEDIVGSGGFGTVYRMVMNDCGTFAVKRI-DRSREGCDQVFERELEILGSIKHINLVN 373
+ I+G+GGFG VYR D AVKR+ D + + F ELE++ H NL+
Sbjct: 272 FSSKSILGAGGFGNVYRGKFGDGTVVAVKRLKDVNGTSGNSQFRTELEMISLAVHRNLLR 331
Query: 374 LRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHECCP 433
L GYC S RLL+Y Y++ GS+ L + L+WN R IA+G+ARGL YLH +C P
Sbjct: 332 LIGYCASSSERLLVYPYMSNGSVASRL--KAKPALDWNTRKKIAIGAARGLFYLHEQCDP 389
Query: 434 KIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRAT 493
KI+HRD+K++NILL+E E + DFGLAKLL ED+HVTT V GT G++APEYL +G+++
Sbjct: 390 KIIHRDVKAANILLDEYFEAVVGDFGLAKLLNHEDSHVTTAVRGTVGHIAPEYLSTGQSS 449
Query: 494 EKSDVYSFGVLLLELVTGKRPTD--PSFANRGLNVVGWMNTLQKENRLEDVVDRRC-TDA 550
EK+DV+ FG+LLLEL+TG R + S + +G ++ W+ L KE ++E++VDR T
Sbjct: 450 EKTDVFGFGILLLELITGMRALEFGKSVSQKGA-MLEWVRKLHKEMKVEELVDRELGTTY 508
Query: 551 DAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVMSPCPSDFYESHSDHS 603
D + +L++A CT RP M++V+Q+LE + + ++ + + DHS
Sbjct: 509 DRIEVGEMLQVALLCTQFLPAHRPKMSEVVQMLEGDGL----AERWAASHDHS 557
>At5g10290 protein serine/threonine kinase-like protein
Length = 613
Score = 308 bits (789), Expect = 6e-84
Identities = 205/591 (34%), Positives = 312/591 (52%), Gaps = 36/591 (6%)
Query: 5 FPIWVFILIFTTVFTP---SSLALTQDGLTLLEIKGALNDTKNVLSNWQEFDESPCAWTG 61
F + + FT +F S ++ G L ++ +L N LS+W + +PC W+
Sbjct: 4 FSLQKMAMAFTLLFFACLCSFVSPDAQGDALFALRISLRALPNQLSDWNQNQVNPCTWSQ 63
Query: 62 ITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTEL 121
+ C D + V S+ L G +S +G L L+ L L N + G IP + N T L
Sbjct: 64 VIC---DDKNFVTSLTLSDMNFSGTLSSRVGILENLKTLTLKGNGITGEIPEDFGNLTSL 120
Query: 122 RALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSG 181
+L L N G IPS IGNL L L LS N G IP SL LP+L L L +N SG
Sbjct: 121 TSLDLEDNQLTGRIPSTIGNLKKLQFLTLSRNKLNGTIPESLTGLPNLLNLLLDSNSLSG 180
Query: 182 EIPDIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHY 241
+IP L K +F N CG + PC +AV SS
Sbjct: 181 QIPQS--LFEIPKYNFTSNNLNCGGRQPHPC----------------VSAVAHSGDSSKP 222
Query: 242 MKVVLIGAMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDL 301
++ G + + ++L L FL+ + K R + + DV +VD + I F
Sbjct: 223 KTGIIAGVVAGVTVVLFGILLFLFCKDRHKGYRRDV-FVDVAGEVD-----RRIAFGQLK 276
Query: 302 PYTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRI-DRSREGCDQVFEREL 360
+ E+ ++ E++++G GGFG VY+ V+ D AVKR+ D G D F+RE+
Sbjct: 277 RFAWRELQLATDNFSEKNVLGQGGFGKVYKGVLPDNTKVAVKRLTDFESPGGDAAFQREV 336
Query: 361 EILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENT--EQPLNWNDRLNIAL 418
E++ H NL+ L G+C + RLL+Y ++ SL L E + L+W R IAL
Sbjct: 337 EMISVAVHRNLLRLIGFCTTQTERLLVYPFMQNLSLAHRLREIKAGDPVLDWETRKRIAL 396
Query: 419 GSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGT 478
G+ARG YLH C PKI+HRD+K++N+LL+E+ E + DFGLAKL+ +VTT V GT
Sbjct: 397 GAARGFEYLHEHCNPKIIHRDVKAANVLLDEDFEAVVGDFGLAKLVDVRRTNVTTQVRGT 456
Query: 479 FGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNV--VGWMNTLQKE 536
G++APEYL +G+++E++DV+ +G++LLELVTG+R D S +V + + L++E
Sbjct: 457 MGHIAPEYLSTGKSSERTDVFGYGIMLLELVTGQRAIDFSRLEEEDDVLLLDHVKKLERE 516
Query: 537 NRLEDVVDRRCT-DADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQE 586
RL +VD+ + +E+++++A CT + +DRP M++V+++LE E
Sbjct: 517 KRLGAIVDKNLDGEYIKEEVEMMIQVALLCTQGSPEDRPVMSEVVRMLEGE 567
>At5g07180 receptor-like protein kinase
Length = 932
Score = 308 bits (789), Expect = 6e-84
Identities = 191/522 (36%), Positives = 285/522 (54%), Gaps = 35/522 (6%)
Query: 75 SINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGG 134
+++L + G I ++G L L L L +N L+G +P E N ++ + + N+ G
Sbjct: 401 TLDLSGNNFSGSIPLTLGDLEHLLILNLSRNHLNGTLPAEFGNLRSIQIIDVSFNFLAGV 460
Query: 135 IPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQK 194
IP+++G L +N L L++N G IP L L LN+S N SG IP + + F
Sbjct: 461 IPTELGQLQNINSLILNNNKIHGKIPDQLTNCFSLANLNISFNNLSGIIPPMKNFTRFSP 520
Query: 195 NSFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLG 254
SF GN LCG + C P+ S + +V +I LG
Sbjct: 521 ASFFGNPFLCGNWVGSICG-------------------PSLPKSQVFTRVAVI--CMVLG 559
Query: 255 LLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLP-YTSSEIIEKLE 313
+ L+ + F+ + SK+++ V++ + + PE STKL+ H D+ +T +I+ E
Sbjct: 560 FITLICMIFIAV-YKSKQQKPVLKGSSKQ----PEGSTKLVILHMDMAIHTFDDIMRVTE 614
Query: 314 SLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVN 373
+L+E+ I+G G TVY+ A+KRI + FE ELE +GSI+H N+V+
Sbjct: 615 NLDEKYIIGYGASSTVYKCTSKTSRPIAIKRIYNQYPSNFREFETELETIGSIRHRNIVS 674
Query: 374 LRGYCRLPSARLLIYDYLAIGSLDDLLH-ENTEQPLNWNDRLNIALGSARGLAYLHHECC 432
L GY P LL YDY+ GSL DLLH + L+W RL IA+G+A+GLAYLHH+C
Sbjct: 675 LHGYALSPFGNLLFYDYMENGSLWDLLHGPGKKVKLDWETRLKIAVGAAQGLAYLHHDCT 734
Query: 433 PKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRA 492
P+I+HRDIKSSNILL+ N E +SDFG+AK + + +T V GT GY+ PEY ++ R
Sbjct: 735 PRIIHRDIKSSNILLDGNFEARLSDFGIAKSIPATKTYASTYVLGTIGYIDPEYARTSRL 794
Query: 493 TEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRR--CTDA 550
EKSD+YSFG++LLEL+TGK+ D N+ + + +N + + VD T
Sbjct: 795 NEKSDIYSFGIVLLELLTGKKAVD-----NEANLHQMILSKADDNTVMEAVDAEVSVTCM 849
Query: 551 DAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVMSPCP 592
D+G ++ +LA CT N +RP+M +V ++L V SP P
Sbjct: 850 DSGHIKKTFQLALLCTKRNPLERPTMQEVSRVLLSLVPSPPP 891
Score = 98.2 bits (243), Expect = 1e-20
Identities = 56/153 (36%), Positives = 83/153 (53%), Gaps = 3/153 (1%)
Query: 33 LEIKGALNDTKNVLSNWQEFDESP-CAWTGITCHPGDGEQRVRSINLPYSQLGGIISPSI 91
+ IK + ++ N+L +W + C+W G+ C + V S+NL LGG IS ++
Sbjct: 1 MAIKASFSNVANMLLDWDDVHNHDFCSWRGVFCD--NVSLNVVSLNLSNLNLGGEISSAL 58
Query: 92 GKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNILDLS 151
G L LQ + L N L G IP+EI NC L + N G IP I L L L+L
Sbjct: 59 GDLMNLQSIDLQGNKLGGQIPDEIGNCVSLAYVDFSTNLLFGDIPFSISKLKQLEFLNLK 118
Query: 152 SNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
+N G IP++L ++P+L+ L+L+ N +GEIP
Sbjct: 119 NNQLTGPIPATLTQIPNLKTLDLARNQLTGEIP 151
Score = 93.2 bits (230), Expect = 4e-19
Identities = 49/113 (43%), Positives = 69/113 (60%)
Query: 72 RVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYF 131
R+ + L ++L G I P +GKL +L L L N+L G+IP+ I++C L + N+
Sbjct: 302 RLSYLQLNDNELVGKIPPELGKLEQLFELNLANNNLVGLIPSNISSCAALNQFNVHGNFL 361
Query: 132 QGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
G +P + NL L L+LSSNSFKG IP+ LG + +L L+LS N FSG IP
Sbjct: 362 SGAVPLEFRNLGSLTYLNLSSNSFKGKIPAELGHIINLDTLDLSGNNFSGSIP 414
Score = 74.7 bits (182), Expect = 1e-13
Identities = 49/137 (35%), Positives = 72/137 (51%), Gaps = 6/137 (4%)
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
Q + ++L ++L G I P +G LS +L LH N L G IP E+ N + L L L N
Sbjct: 253 QALAVLDLSDNELTGPIPPILGNLSFTGKLYLHGNKLTGQIPPELGNMSRLSYLQLNDNE 312
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP----DI 186
G IP ++G L L L+L++N+ G IPS++ L N+ NF SG +P ++
Sbjct: 313 LVGKIPPELGKLEQLFELNLANNNLVGLIPSNISSCAALNQFNVHGNFLSGAVPLEFRNL 372
Query: 187 GVLS--TFQKNSFIGNL 201
G L+ NSF G +
Sbjct: 373 GSLTYLNLSSNSFKGKI 389
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/121 (37%), Positives = 62/121 (51%), Gaps = 1/121 (0%)
Query: 81 SQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIG 140
++L G I P +G +SRL L L+ N L G IP E+ +L L L N G IPS+I
Sbjct: 287 NKLTGQIPPELGNMSRLSYLQLNDNELVGKIPPELGKLEQLFELNLANNNLVGLIPSNIS 346
Query: 141 NLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP-DIGVLSTFQKNSFIG 199
+ LN ++ N GA+P L L LNLS+N F G+IP ++G + G
Sbjct: 347 SCAALNQFNVHGNFLSGAVPLEFRNLGSLTYLNLSSNSFKGKIPAELGHIINLDTLDLSG 406
Query: 200 N 200
N
Sbjct: 407 N 407
Score = 70.1 bits (170), Expect = 3e-12
Identities = 55/177 (31%), Positives = 85/177 (47%), Gaps = 8/177 (4%)
Query: 13 IFTTVFTPSSLALTQDGLTLLEIKG-----ALNDTKNVLSNWQEFDESPCAWTGITCHPG 67
+ T +P LT GL +++G + ++ ++++ D S TG+ +
Sbjct: 169 MLTGTLSPDMCQLT--GLWYFDVRGNNLTGTIPESIGNCTSFEILDVSYNQITGVIPY-N 225
Query: 68 DGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLR 127
G +V +++L ++L G I IG + L L L N L G IP + N + LYL
Sbjct: 226 IGFLQVATLSLQGNKLTGRIPEVIGLMQALAVLDLSDNELTGPIPPILGNLSFTGKLYLH 285
Query: 128 ANYFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
N G IP ++GN+ L+ L L+ N G IP LG+L L LNL+ N G IP
Sbjct: 286 GNKLTGQIPPELGNMSRLSYLQLNDNELVGKIPPELGKLEQLFELNLANNNLVGLIP 342
Score = 59.3 bits (142), Expect = 6e-09
Identities = 42/136 (30%), Positives = 68/136 (49%), Gaps = 5/136 (3%)
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
+++ +NL +QL G I ++ ++ L+ L L +N L G IP + L+ L LR N
Sbjct: 110 KQLEFLNLKNNQLTGPIPATLTQIPNLKTLDLARNQLTGEIPRLLYWNEVLQYLGLRGNM 169
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP-DIGVL 189
G + D+ L L D+ N+ G IP S+G ++L++S N +G IP +IG L
Sbjct: 170 LTGTLSPDMCQLTGLWYFDVRGNNLTGTIPESIGNCTSFEILDVSYNQITGVIPYNIGFL 229
Query: 190 S----TFQKNSFIGNL 201
+ Q N G +
Sbjct: 230 QVATLSLQGNKLTGRI 245
>At3g13380 brassinosteroid receptor kinase, putative
Length = 1164
Score = 305 bits (781), Expect = 5e-83
Identities = 183/513 (35%), Positives = 282/513 (54%), Gaps = 31/513 (6%)
Query: 100 LALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSFKGAI 159
L L N++ G IP L+ L L N G IP G L + +LDLS N +G +
Sbjct: 644 LDLSYNAVSGSIPLGYGAMGYLQVLNLGHNLLTGTIPDSFGGLKAIGVLDLSHNDLQGFL 703
Query: 160 PSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKPCRTSFGFP 219
P SLG L L L++S N +G IP G L+TF + N LCG + PC S G
Sbjct: 704 PGSLGGLSFLSDLDVSNNNLTGPIPFGGQLTTFPLTRYANNSGLCGVPLP-PC--SSGSR 760
Query: 220 VVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSFLWIRLLSKKERAVMRY 279
HA + ++ T S+ G + + ++++ ++ R + KKE+ +Y
Sbjct: 761 PTRSHAHPKKQSIATGMSA---------GIVFSFMCIVMLIMALYRARKVQKKEKQREKY 811
Query: 280 TDVKKQVDPEASTKLITFHGDLPY------------TSSEIIEKLESLEEEDIVGSGGFG 327
+ +S KL + H L T + ++E + ++GSGGFG
Sbjct: 812 IE-SLPTSGSSSWKLSSVHEPLSINVATFEKPLRKLTFAHLLEATNGFSADSMIGSGGFG 870
Query: 328 TVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLI 387
VY+ + D A+K++ + D+ F E+E +G IKH NLV L GYC++ RLL+
Sbjct: 871 DVYKAKLADGSVVAIKKLIQVTGQGDREFMAEMETIGKIKHRNLVPLLGYCKIGEERLLV 930
Query: 388 YDYLAIGSLDDLLHENTEQP---LNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSN 444
Y+Y+ GSL+ +LHE T++ L+W+ R IA+G+ARGLA+LHH C P I+HRD+KSSN
Sbjct: 931 YEYMKYGSLETVLHEKTKKGGIFLDWSARKKIAIGAARGLAFLHHSCIPHIIHRDMKSSN 990
Query: 445 ILLNENMEPHISDFGLAKLLVDEDAHVT-TVVAGTFGYLAPEYLQSGRATEKSDVYSFGV 503
+LL+++ +SDFG+A+L+ D H++ + +AGT GY+ PEY QS R T K DVYS+GV
Sbjct: 991 VLLDQDFVARVSDFGMARLVSALDTHLSVSTLAGTPGYVPPEYYQSFRCTAKGDVYSYGV 1050
Query: 504 LLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRCTDADAGTLEVI--LEL 561
+LLEL++GK+P DP N+VGW L +E R +++D +G +E++ L++
Sbjct: 1051 ILLELLSGKKPIDPEEFGEDNNLVGWAKQLYREKRGAEILDPELVTDKSGDVELLHYLKI 1110
Query: 562 AARCTDANADDRPSMNQVLQLLEQEVMSPCPSD 594
A++C D RP+M QV+ + ++ V +D
Sbjct: 1111 ASQCLDDRPFKRPTMIQVMTMFKELVQVDTEND 1143
Score = 73.6 bits (179), Expect = 3e-13
Identities = 41/116 (35%), Positives = 69/116 (59%), Gaps = 4/116 (3%)
Query: 73 VRSINLPYSQLGG-IISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYF 131
++S+NL ++L G +S + KLSR+ L L N++ G +P +TNC+ LR L L +N F
Sbjct: 328 LQSLNLGNNKLSGDFLSTVVSKLSRITNLYLPFNNISGSVPISLTNCSNLRVLDLSSNEF 387
Query: 132 QGGIPSDIGNLPFLNILD---LSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
G +PS +L ++L+ +++N G +P LG+ L+ ++LS N +G IP
Sbjct: 388 TGEVPSGFCSLQSSSVLEKLLIANNYLSGTVPVELGKCKSLKTIDLSFNALTGLIP 443
Score = 66.2 bits (160), Expect = 5e-11
Identities = 45/125 (36%), Positives = 67/125 (53%), Gaps = 6/125 (4%)
Query: 66 PGDGE----QRVRSINLPYSQLGGIISPSIGKLSR-LQRLALHQNSLHGIIPNEITNCTE 120
PGD Q +R ++L ++ G I P + L R L+ L L NSL G +P T+C
Sbjct: 268 PGDDYWGNFQNLRQLSLAHNLYSGEIPPELSLLCRTLEVLDLSGNSLTGQLPQSFTSCGS 327
Query: 121 LRALYLRANYFQGGIPSD-IGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFF 179
L++L L N G S + L + L L N+ G++P SL +L+VL+LS+N F
Sbjct: 328 LQSLNLGNNKLSGDFLSTVVSKLSRITNLYLPFNNISGSVPISLTNCSNLRVLDLSSNEF 387
Query: 180 SGEIP 184
+GE+P
Sbjct: 388 TGEVP 392
Score = 65.9 bits (159), Expect = 6e-11
Identities = 39/101 (38%), Positives = 53/101 (51%)
Query: 84 GGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLP 143
GGI L+ L L+ N L G +P I+ CT + + L +N G IP IG L
Sbjct: 464 GGIPESICVDGGNLETLILNNNLLTGSLPESISKCTNMLWISLSSNLLTGEIPVGIGKLE 523
Query: 144 FLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
L IL L +NS G IPS LG +L L+L++N +G +P
Sbjct: 524 KLAILQLGNNSLTGNIPSELGNCKNLIWLDLNSNNLTGNLP 564
Score = 64.7 bits (156), Expect = 1e-10
Identities = 45/129 (34%), Positives = 66/129 (50%), Gaps = 13/129 (10%)
Query: 83 LGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDI--- 139
L G + +GK L+ + L N+L G+IP EI +L L + AN GGIP I
Sbjct: 414 LSGTVPVELGKCKSLKTIDLSFNALTGLIPKEIWTLPKLSDLVMWANNLTGGIPESICVD 473
Query: 140 -GNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP-DIGVLSTFQ---- 193
GN L L L++N G++P S+ + ++ ++LS+N +GEIP IG L
Sbjct: 474 GGN---LETLILNNNLLTGSLPESISKCTNMLWISLSSNLLTGEIPVGIGKLEKLAILQL 530
Query: 194 -KNSFIGNL 201
NS GN+
Sbjct: 531 GNNSLTGNI 539
Score = 63.2 bits (152), Expect = 4e-10
Identities = 43/136 (31%), Positives = 65/136 (47%), Gaps = 7/136 (5%)
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEI-TNCTELRALYLRANYF 131
+++I+L ++ L G+I I L +L L + N+L G IP I + L L L N
Sbjct: 428 LKTIDLSFNALTGLIPKEIWTLPKLSDLVMWANNLTGGIPESICVDGGNLETLILNNNLL 487
Query: 132 QGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP-DIG--- 187
G +P I + + LSSN G IP +G+L L +L L N +G IP ++G
Sbjct: 488 TGSLPESISKCTNMLWISLSSNLLTGEIPVGIGKLEKLAILQLGNNSLTGNIPSELGNCK 547
Query: 188 --VLSTFQKNSFIGNL 201
+ N+ GNL
Sbjct: 548 NLIWLDLNSNNLTGNL 563
Score = 54.3 bits (129), Expect = 2e-07
Identities = 38/96 (39%), Positives = 48/96 (49%), Gaps = 3/96 (3%)
Query: 90 SIGKLSRLQRLALHQNSLHGIIPNEIT--NCTELRALYLRANYFQGGIPSDIGNL-PFLN 146
S+ L+ L L +NSL G IP + N LR L L N + G IP ++ L L
Sbjct: 246 SLSNCKLLETLNLSRNSLIGKIPGDDYWGNFQNLRQLSLAHNLYSGEIPPELSLLCRTLE 305
Query: 147 ILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGE 182
+LDLS NS G +P S LQ LNL N SG+
Sbjct: 306 VLDLSGNSLTGQLPQSFTSCGSLQSLNLGNNKLSGD 341
Score = 53.1 bits (126), Expect = 4e-07
Identities = 32/97 (32%), Positives = 52/97 (52%), Gaps = 1/97 (1%)
Query: 90 SIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNILD 149
S+ S L++L + N L G +P E+ C L+ + L N G IP +I LP L+ L
Sbjct: 397 SLQSSSVLEKLLIANNYLSGTVPVELGKCKSLKTIDLSFNALTGLIPKEIWTLPKLSDLV 456
Query: 150 LSSNSFKGAIPSSL-GRLPHLQVLNLSTNFFSGEIPD 185
+ +N+ G IP S+ +L+ L L+ N +G +P+
Sbjct: 457 MWANNLTGGIPESICVDGGNLETLILNNNLLTGSLPE 493
Score = 50.8 bits (120), Expect = 2e-06
Identities = 60/224 (26%), Positives = 97/224 (42%), Gaps = 18/224 (8%)
Query: 5 FPIWVFILIFTTVFTPSSLALTQDG-----LTLLEIKGALNDTKNVLSNWQEFD-ESPCA 58
F I +++F TV + L+ D LT + +D N L NW+ PC
Sbjct: 7 FLILCLLVLFLTVDSRGRRLLSDDVNDTALLTAFKQTSIKSDPTNFLGNWRYGSGRDPCT 66
Query: 59 WTGITCHPGDGEQRVRSINLPYSQLGGIIS-PSIGKLSRLQRLALHQNSLHGIIPNEITN 117
W G++C + RV ++L L G ++ ++ LS L+ L L N+ + +
Sbjct: 67 WRGVSC---SSDGRVIGLDLRNGGLTGTLNLNNLTALSNLRSLYLQGNNFSSGDSSSSSG 123
Query: 118 CTELRALYLRANYFQGGIPSDIGNLPFLNI--LDLSSNSFKGAIPSS-LGRLPHLQVLNL 174
C+ L L L +N D LN+ ++ S N G + SS + ++L
Sbjct: 124 CS-LEVLDLSSNSLTDSSIVDYVFSTCLNLVSVNFSHNKLAGKLKSSPSASNKRITTVDL 182
Query: 175 STNFFSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKP-CRTSFG 217
S N FS EIP+ ++ F + + +LDL G + R SFG
Sbjct: 183 SNNRFSDEIPET-FIADFPNS--LKHLDLSGNNVTGDFSRLSFG 223
Score = 50.1 bits (118), Expect = 4e-06
Identities = 31/81 (38%), Positives = 42/81 (51%)
Query: 83 LGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNL 142
L G + SI K + + ++L N L G IP I +L L L N G IPS++GN
Sbjct: 487 LTGSLPESISKCTNMLWISLSSNLLTGEIPVGIGKLEKLAILQLGNNSLTGNIPSELGNC 546
Query: 143 PFLNILDLSSNSFKGAIPSSL 163
L LDL+SN+ G +P L
Sbjct: 547 KNLIWLDLNSNNLTGNLPGEL 567
Score = 47.0 bits (110), Expect = 3e-05
Identities = 27/66 (40%), Positives = 37/66 (55%)
Query: 76 INLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGI 135
I+L + L G I IGKL +L L L NSL G IP+E+ NC L L L +N G +
Sbjct: 504 ISLSSNLLTGEIPVGIGKLEKLAILQLGNNSLTGNIPSELGNCKNLIWLDLNSNNLTGNL 563
Query: 136 PSDIGN 141
P ++ +
Sbjct: 564 PGELAS 569
Score = 37.0 bits (84), Expect = 0.032
Identities = 40/129 (31%), Positives = 51/129 (39%), Gaps = 15/129 (11%)
Query: 75 SINLPYSQLGGII--SPSIGKLSRLQRLALHQNSLHGIIPNEITNC--TELRALYLRANY 130
S+N +++L G + SPS R+ + L N IP L+ L L N
Sbjct: 154 SVNFSHNKLAGKLKSSPSASN-KRITTVDLSNNRFSDEIPETFIADFPNSLKHLDLSGNN 212
Query: 131 FQGGIPSDIGNLPF-----LNILDLSSNSFKG-AIPSSLGRLPHLQVLNLSTNFFSGEIP 184
G D L F L + LS NS G P SL L+ LNLS N G+IP
Sbjct: 213 VTG----DFSRLSFGLCENLTVFSLSQNSISGDRFPVSLSNCKLLETLNLSRNSLIGKIP 268
Query: 185 DIGVLSTFQ 193
FQ
Sbjct: 269 GDDYWGNFQ 277
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,669,101
Number of Sequences: 26719
Number of extensions: 605507
Number of successful extensions: 9877
Number of sequences better than 10.0: 1191
Number of HSP's better than 10.0 without gapping: 1015
Number of HSP's successfully gapped in prelim test: 178
Number of HSP's that attempted gapping in prelim test: 1534
Number of HSP's gapped (non-prelim): 3489
length of query: 603
length of database: 11,318,596
effective HSP length: 105
effective length of query: 498
effective length of database: 8,513,101
effective search space: 4239524298
effective search space used: 4239524298
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0170.5


