

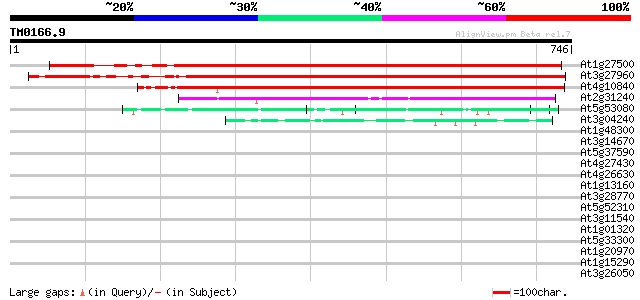
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.9
(746 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g27500 unknown protein 745 0.0
At3g27960 hypothetical protein 717 0.0
At4g10840 unknown protein 642 0.0
At2g31240 putative kinesin light chain protein (At2g31240) 172 5e-43
At5g53080 unknown protein 66 6e-11
At3g04240 O-linked GlcNAc transferase like protein 49 1e-05
At1g48300 unknown protein 42 0.001
At3g14670 hypothetical protein 39 0.008
At5g37590 kinesin -like protein 39 0.014
At4g27430 COP1-interacting protein 7 (CIP7) 38 0.018
At4g26630 unknown protein 38 0.018
At1g13160 unknown protein 37 0.031
At3g28770 hypothetical protein 37 0.053
At5g52310 low-temperature-induced protein 78 (sp|Q06738) 36 0.090
At3g11540 spindly (gibberellin signal transduction protein) 36 0.090
At1g01320 unknown protein 36 0.090
At5g33300 putative protein 35 0.12
At1g20970 hypothetical protein 35 0.12
At1g15290 unknown protein 35 0.12
At3g26050 unknown protein (At3g26050) 35 0.15
>At1g27500 unknown protein
Length = 650
Score = 745 bits (1923), Expect = 0.0
Identities = 395/684 (57%), Positives = 503/684 (72%), Gaps = 57/684 (8%)
Query: 53 DGVIEPSIEQLYENVCDMQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEE 112
D + + +IE+L +N+C++QSS+QSPSR+SFGS GDES+IDS+L+ L G MR+++I+E+E
Sbjct: 14 DQMFDTTIEELCKNLCELQSSNQSPSRQSFGSYGDESKIDSDLQHLALGEMRDIDILEDE 73
Query: 113 VEVEVEKERGGSSSGEISSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDA 172
+E ++ K E + SS N L+
Sbjct: 74 -------------------------GDEDEVAKPEEFDVKSNSS----------NLDLEV 98
Query: 173 SPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVEN-SAE 231
P+ +E++ K + K GV K K V +++N + E
Sbjct: 99 MPRD------------MEKQTGKKNVTKSNVGVGGMRK--------KKVGGTKLQNGNEE 138
Query: 232 SALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLE-KLGNGKPSLELVMCLHVT 290
+ + E A LL QAR+L+SSGD+ KALEL +A L E NGKP LE +MCLHVT
Sbjct: 139 PSSENVELARFLLNQARNLVSSGDSTHKALELTHRAAKLFEASAENGKPCLEWIMCLHVT 198
Query: 291 AAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCY 350
AA++C L +YNEAIP+L+RS+EIPV+ E ++HALAKFAG MQLGDTYAM+GQLE+SI CY
Sbjct: 199 AAVHCKLKEYNEAIPVLQRSVEIPVVEEGEEHALAKFAGLMQLGDTYAMVGQLESSISCY 258
Query: 351 SSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEE 410
+ G +Q++VLGE DPRVGETCRY+AEA VQAL+FDEA+++C+ AL IH+ + S+ E
Sbjct: 259 TEGLNIQKKVLGENDPRVGETCRYLAEALVQALRFDEAQQVCETALSIHRESGLPGSIAE 318
Query: 411 AADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEA 470
AADRRLMGLI +TKG+HE ALEHLVLASMAM ANGQE EVA VD SIGD+YLS+SR+DEA
Sbjct: 319 AADRRLMGLICETKGDHENALEHLVLASMAMAANGQESEVAFVDTSIGDSYLSLSRFDEA 378
Query: 471 VFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGV 530
+ AY+K+LT KT KGENHPAVGSV++RLADL+NRTGK++E+KSYC++ALRIYE+ +
Sbjct: 379 ICAYQKSLTALKTAKGENHPAVGSVYIRLADLYNRTGKVREAKSYCENALRIYESHNLEI 438
Query: 531 NPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYML 590
+PEEIASGLT++S I ESMNE+E+A+ LLQKAL IY DSPGQ+ IAGIEAQMGV+YYM+
Sbjct: 439 SPEEIASGLTDISVICESMNEVEQAITLLQKALKIYADSPGQKIMIAGIEAQMGVLYYMM 498
Query: 591 GNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILE 650
G Y ESYNT K+AI+KLRA G+K+S+F GIALNQMGLAC+QL A EA ELFEEAK ILE
Sbjct: 499 GKYMESYNTFKSAISKLRATGKKQSTFFGIALNQMGLACIQLDAIEEAVELFEEAKCILE 558
Query: 651 HEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLS 710
E GPYHPETLG+YSNLAG YDAIGRLD+AI++L +VV VREEKLGTANP +DEKRRL+
Sbjct: 559 QECGPYHPETLGLYSNLAGAYDAIGRLDDAIKLLGHVVGVREEKLGTANPVTEDEKRRLA 618
Query: 711 ELLKEAGRVRSRKVMSLENLLDAN 734
+LLKEAG V RK SL+ L+D++
Sbjct: 619 QLLKEAGNVTGRKAKSLKTLIDSD 642
>At3g27960 hypothetical protein
Length = 663
Score = 717 bits (1852), Expect = 0.0
Identities = 401/720 (55%), Positives = 503/720 (69%), Gaps = 78/720 (10%)
Query: 25 KETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPSIEQLYENVCDMQSSD-QSPSRKSFG 83
K+ A SPR LS + +DG + SIEQLY NVC+M+SSD QSPSR SF
Sbjct: 11 KDDSALQASPRSPLS-------SIDLAIDGAMNASIEQLYHNVCEMESSDDQSPSRASFI 63
Query: 84 SDGDESRIDSELRQLVGGRMREVEIMEEEVEVEVEKERGGSSSGEISSGVGGLSSNEKKL 143
S G ESRID ELR LVG E E + E+ +EK+ E S+G G LS +
Sbjct: 64 SYGAESRIDLELRHLVGDVGEEGE---SKKEIILEKK-------EESNGEGSLSQKKP-- 111
Query: 144 DKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDKPLLRKQTK 203
+S K V K+ P P
Sbjct: 112 -------------LSNGKKVA----------KTSPNNPKMPG------------------ 130
Query: 204 GVASGVKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARDLISSGDNPQKALEL 263
S + S KS LGK VS++ + PE +LLKQAR+L+SSG+N KAL+L
Sbjct: 131 ---SRISSRKSPDLGK-VSVDE---------ESPELGVVLLKQARELVSSGENLNKALDL 177
Query: 264 ALQAMNLLEKLGNGKPSL--ELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQ 321
AL+A+ + EK G G+ L LVM LH+ AAIY LG+YN+A+P+LERSIEIP+I + +
Sbjct: 178 ALRAVKVFEKCGEGEKQLGLNLVMSLHILAAIYAGLGRYNDAVPVLERSIEIPMIEDGED 237
Query: 322 HALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQ 381
HALAKFAG MQLGD Y ++GQ+ENSIM Y++G E+QRQVLGE+D RVGETCRY+AEA+VQ
Sbjct: 238 HALAKFAGCMQLGDMYGLMGQVENSIMLYTAGLEIQRQVLGESDARVGETCRYLAEAHVQ 297
Query: 382 ALQFDEAERLCQMALDIHKANSSAP--SLEEAADRRLMGLILDTKGNHEAALEHLVLASM 439
A+QF+EA RLCQMALDIHK N +A S+EEAADR+LMGLI D KG++E ALEH VLASM
Sbjct: 298 AMQFEEASRLCQMALDIHKENGAAATASIEEAADRKLMGLICDAKGDYEVALEHYVLASM 357
Query: 440 AMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRL 499
AM + +VA+VDCSIGD Y+S++R+DEA+FAY+KAL VFK GKGE H +V V+VRL
Sbjct: 358 AMSSQNHREDVAAVDCSIGDAYMSLARFDEAIFAYQKALAVFKQGKGETHSSVALVYVRL 417
Query: 500 ADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLL 559
ADL+N+ GK ++SKSYC++AL+IY P PG EE+A+G +SAIY+SMNEL++ALKLL
Sbjct: 418 ADLYNKIGKTRDSKSYCENALKIYLKPTPGTPMEEVATGFIEISAIYQSMNELDQALKLL 477
Query: 560 QKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIG 619
++AL IY ++PGQQ++IAGIEAQMGV+ YM+GNY+ESY+ K+AI+K R GEKK++ G
Sbjct: 478 RRALKIYANAPGQQNTIAGIEAQMGVVTYMMGNYSESYDIFKSAISKFRNSGEKKTALFG 537
Query: 620 IALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDE 679
IALNQMGLACVQ YA EA +LFEEAK+ILE E GPYHP+TL VYSNLAGTYDA+GRLD+
Sbjct: 538 IALNQMGLACVQRYAINEAADLFEEAKTILEKECGPYHPDTLAVYSNLAGTYDAMGRLDD 597
Query: 680 AIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANARIPN 739
AI+ILEYVV REEKLGTANP+V+DEK+RL+ LLKEAGR RS++ +L LLD N I N
Sbjct: 598 AIEILEYVVGTREEKLGTANPEVEDEKQRLAALLKEAGRGRSKRNRALLTLLDNNPEIAN 657
>At4g10840 unknown protein
Length = 609
Score = 642 bits (1656), Expect = 0.0
Identities = 334/579 (57%), Positives = 431/579 (73%), Gaps = 21/579 (3%)
Query: 171 DASPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVENSA 230
D P S P ++P K P + P K + ++G K SP S S V +
Sbjct: 25 DTQPLSNPP-RTPMKKTP----SSTPSRSKPSPNRSTGKKD---SPTVSS-STAAVIDVD 75
Query: 231 ESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKL-----------GNGKP 279
+ +LD P+ P LLK ARD I+SG+ P KAL+ A++A E+ +G P
Sbjct: 76 DPSLDNPDLGPFLLKLARDAIASGEGPNKALDYAIRATKSFERCCAAVAPPIPGGSDGGP 135
Query: 280 SLELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAM 339
L+L M LHV AAIYC+LG+++EA+P LER+I++P H+LA F+GHMQLGDT +M
Sbjct: 136 VLDLAMSLHVLAAIYCSLGRFDEAVPPLERAIQVPDPTRGPDHSLAAFSGHMQLGDTLSM 195
Query: 340 LGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIH 399
LGQ++ SI CY G ++Q Q LG+TDPRVGETCRY+AEA VQA+QF++AE LC+ L+IH
Sbjct: 196 LGQIDRSIACYEEGLKIQIQTLGDTDPRVGETCRYLAEAYVQAMQFNKAEELCKKTLEIH 255
Query: 400 KANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGD 459
+A+S SLEEAADRRLM +I + KG++E ALEHLVLASMAM+A+GQE EVAS+D SIG+
Sbjct: 256 RAHSEPASLEEAADRRLMAIICEAKGDYENALEHLVLASMAMIASGQESEVASIDVSIGN 315
Query: 460 TYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSA 519
Y+S+ R+DEAVF+Y+KALTVFK KGE HP V SVFVRLA+L++RTGK++ESKSYC++A
Sbjct: 316 IYMSLCRFDEAVFSYQKALTVFKASKGETHPTVASVFVRLAELYHRTGKLRESKSYCENA 375
Query: 520 LRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGI 579
LRIY P+PG EEIA GLT +SAIYES++E E+ALKLLQK++ + D PGQQS+IAG+
Sbjct: 376 LRIYNKPVPGTTVEEIAGGLTEISAIYESVDEPEEALKLLQKSMKLLEDKPGQQSAIAGL 435
Query: 580 EAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEAT 639
EA+MGVMYY +G Y ++ N ++A+TKLRA GE KS+F G+ LNQMGLACVQL+ EA
Sbjct: 436 EARMGVMYYTVGRYEDARNAFESAVTKLRAAGE-KSAFFGVVLNQMGLACVQLFKIDEAG 494
Query: 640 ELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTAN 699
ELFEEA+ ILE E GP +TLGVYSNLA TYDA+GR+++AI+ILE V+ +REEKLGTAN
Sbjct: 495 ELFEEARGILEQERGPCDQDTLGVYSNLAATYDAMGRIEDAIEILEQVLKLREEKLGTAN 554
Query: 700 PDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANARIP 738
PD +DEK+RL+ELLKEAGR R+ K SL+NL+D NAR P
Sbjct: 555 PDFEDEKKRLAELLKEAGRSRNYKAKSLQNLIDPNARPP 593
>At2g31240 putative kinesin light chain protein (At2g31240)
Length = 617
Score = 172 bits (437), Expect = 5e-43
Identities = 134/509 (26%), Positives = 238/509 (46%), Gaps = 13/509 (2%)
Query: 225 RVENSAESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLELV 284
R+ E + + E LK L G++P+K L A +A+ + GN KP+L +
Sbjct: 103 RLFKEMELSFEGNELGLSALKLGLHLDREGEDPEKVLSYADKALKSFDGDGN-KPNLLVA 161
Query: 285 MCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAK------FAGHMQLGDTYA 338
M + + L ++++++ L R+ I V E+ + + A ++L +
Sbjct: 162 MASQLMGSANYGLKRFSDSLGYLNRANRILVKLEADGDCVVEDVRPVLHAVQLELANVKN 221
Query: 339 MLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDI 398
+G+ E +I E++ E +G R +A+A V L F+EA AL+I
Sbjct: 222 AMGRREEAIENLKKSLEIKEMTFDEDSKEMGVANRSLADAYVAVLNFNEALPYALKALEI 281
Query: 399 HKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIG 458
HK S E A DRRL+G+I H+ ALE L+ + G ++E+ +
Sbjct: 282 HKKELGNNSAEVAQDRRLLGVIYSGLEQHDKALEQNRLSQRVLKNWGMKLELIRAEIDAA 341
Query: 459 DTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDS 518
+ +++ +Y+EA+ + V +T K A+ VF+ ++ K ESK +
Sbjct: 342 NMKVALGKYEEAIDILKSV--VQQTDKDSEMRAM--VFISMSKALVNQQKFAESKRCLEF 397
Query: 519 ALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAG 578
A I E + P E+A + V+ YESMNE E A+ LLQK L I P +Q S
Sbjct: 398 ACEILEKKETAL-PVEVAEAYSEVAMQYESMNEFETAISLLQKTLGILEKLPQEQHSEGS 456
Query: 579 IEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEA 638
+ A++G + G +++ L++A +L+ K +G N +G A ++L A
Sbjct: 457 VSARIGWLLLFSGRVSQAVPYLESAAERLKESFGAKHFGVGYVYNNLGAAYLELGRPQSA 516
Query: 639 TELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTA 698
++F AK I++ GP H +++ NL+ Y +G A++ + V++ + +A
Sbjct: 517 AQMFAVAKDIMDVSLGPNHVDSIDACQNLSKAYAGMGNYSLAVEFQQRVINAWDNHGDSA 576
Query: 699 NPDVDDEKRRLSEL-LKEAGRVRSRKVMS 726
++ + KR L +L LK G V + K+++
Sbjct: 577 KDEMKEAKRLLEDLRLKARGGVSTNKLLN 605
>At5g53080 unknown protein
Length = 564
Score = 66.2 bits (160), Expect = 6e-11
Identities = 120/553 (21%), Positives = 218/553 (38%), Gaps = 26/553 (4%)
Query: 150 QSATTSSVSTEKSV---KALNSQLDASPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVA 206
QS++ SS+S S L +Q K KPK PA+ L D T A
Sbjct: 20 QSSSQSSLSPRYSTWQCVCLRNQ-----KRKPKLYLIPARHFLSTPIDSVSSSSIT---A 71
Query: 207 SGVKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARDLISSGDNPQKALELALQ 266
S + S + +S S N V E ++ E L + + ++ G L
Sbjct: 72 SRYATSGVSEVQRSTSSNNVTEMEEFEMELQE----LFNEVKSMVKIGKESDAMDLLRAN 127
Query: 267 AMNLLEKLGNGKPSLELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAK 326
+ + E+L +G +E L + A Y +G +L+ +I + + L
Sbjct: 128 YVAVKEELDSGLKGIEQAAVLDIIALGYMAVGDLKPVPALLDMINKIVDNLKDSEPLLDS 187
Query: 327 FAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFD 386
H+ G Y+++G+ EN+I+ + + G+ + + +A++ +
Sbjct: 188 VLMHV--GSMYSVIGKFENAILVHQRAIRILENRYGKCNTLLVTPLLGMAKSFASDGKAT 245
Query: 387 EAERLCQMALDIHKANSSAPSLEEAADRRLMGLILDTKGNH-EAALEHLVLASMAMVANG 445
+A + + L I + N + S + +G +L +G EA + + ++ G
Sbjct: 246 KAIGVYERTLTILERNRGSESEDLVVPLFSLGKLLLKEGKAAEAEIPFTSIVNIYKKIYG 305
Query: 446 Q-EVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGE--NHPAVGSVFVRLADL 502
+ + V CS+ + S +EAV Y AL + K ++ + ++ + LA+L
Sbjct: 306 ERDGRVGMAMCSLANAKCSKGDANEAVDIYRNALRIIKDSNYMTIDNSILENMRIDLAEL 365
Query: 503 HNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKA 562
+ G+ E + + L I E G N +A+ L N++A Y +A +LL+
Sbjct: 366 LHFVGRGDEGRELLEECLLINER-FKGKNHPSMATHLINLAASYSRSKNYVEAERLLRTC 424
Query: 563 LLIYNDSPGQQS-SIAGIEAQMGVMYYMLGNYTESYN-TLKNAITKLRAIGEKKSSFIGI 620
L I S G + SI + V L E+ LK + +A GE S +G
Sbjct: 425 LNIMEVSVGSEGQSITFPMLNLAVTLSQLNRDEEAEQIALKVLRIREKAFGED-SLPVGE 483
Query: 621 ALNQMGLACVQLYAF-GEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDE 679
AL+ + +L GE L + I E E+GP E + + + + D+
Sbjct: 484 ALDCLVSIQARLGRDDGEILGLLKRVMMIQEKEFGPSAQELIVTLQKIIHFLEKLEMKDD 543
Query: 680 AIQILEYVVSVRE 692
+ + +RE
Sbjct: 544 KFKFRRRLALLRE 556
Score = 47.8 bits (112), Expect = 2e-05
Identities = 76/362 (20%), Positives = 145/362 (39%), Gaps = 40/362 (11%)
Query: 395 ALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAM------------- 441
A+D+ +AN A ++E D L G+ A L+ + L MA+
Sbjct: 120 AMDLLRANYVA--VKEELDSGLKGI------EQAAVLDIIALGYMAVGDLKPVPALLDMI 171
Query: 442 ---VANGQEVE--VASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVF 496
V N ++ E + SV +G Y + +++ A+ +++A+ + + G+ + + +
Sbjct: 172 NKIVDNLKDSEPLLDSVLMHVGSMYSVIGKFENAILVHQRAIRILENRYGKCNTLLVTPL 231
Query: 497 VRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKAL 556
+ +A GK ++ + L I E G E++ L ++ + + +A
Sbjct: 232 LGMAKSFASDGKATKAIGVYERTLTILERNR-GSESEDLVVPLFSLGKLLLKEGKAAEAE 290
Query: 557 KLLQKALLIYNDSPGQQSSIAGIE-AQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKS 615
+ IY G++ G+ + G+ E+ + +NA LR I K S
Sbjct: 291 IPFTSIVNIYKKIYGERDGRVGMAMCSLANAKCSKGDANEAVDIYRNA---LRII--KDS 345
Query: 616 SFIGI---ALNQMGLACVQLYAF----GEATELFEEAKSILEHEYGPYHPETLGVYSNLA 668
+++ I L M + +L F E EL EE I E G HP NLA
Sbjct: 346 NYMTIDNSILENMRIDLAELLHFVGRGDEGRELLEECLLINERFKGKNHPSMATHLINLA 405
Query: 669 GTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLE 728
+Y EA ++L +++ E +G+ + L+ L + R + ++L+
Sbjct: 406 ASYSRSKNYVEAERLLRTCLNIMEVSVGSEGQSITFPMLNLAVTLSQLNRDEEAEQIALK 465
Query: 729 NL 730
L
Sbjct: 466 VL 467
Score = 47.0 bits (110), Expect = 4e-05
Identities = 52/262 (19%), Positives = 101/262 (37%), Gaps = 4/262 (1%)
Query: 460 TYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSA 519
++ S + +A+ YE+ LT+ + +G + L L + GK E++ S
Sbjct: 237 SFASDGKATKAIGVYERTLTILERNRGSESEDLVVPLFSLGKLLLKEGKAAEAEIPFTSI 296
Query: 520 LRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQ---QSSI 576
+ IY+ + G + + +++ S + +A+ + + AL I DS S +
Sbjct: 297 VNIYKK-IYGERDGRVGMAMCSLANAKCSKGDANEAVDIYRNALRIIKDSNYMTIDNSIL 355
Query: 577 AGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFG 636
+ + + + +G E L+ + K + L + + + +
Sbjct: 356 ENMRIDLAELLHFVGRGDEGRELLEECLLINERFKGKNHPSMATHLINLAASYSRSKNYV 415
Query: 637 EATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLG 696
EA L +I+E G NLA T + R +EA QI V+ +RE+ G
Sbjct: 416 EAERLLRTCLNIMEVSVGSEGQSITFPMLNLAVTLSQLNRDEEAEQIALKVLRIREKAFG 475
Query: 697 TANPDVDDEKRRLSELLKEAGR 718
+ V + L + GR
Sbjct: 476 EDSLPVGEALDCLVSIQARLGR 497
>At3g04240 O-linked GlcNAc transferase like protein
Length = 977
Score = 48.9 bits (115), Expect = 1e-05
Identities = 90/455 (19%), Positives = 166/455 (35%), Gaps = 85/455 (18%)
Query: 287 LHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENS 346
L + AIY L +Y+ I E ++ I Q A+ G+M + + G + +
Sbjct: 90 LLLIGAIYYQLQEYDMCIARNEEALRI-------QPQFAECYGNM--ANAWKEKGDTDRA 140
Query: 347 IMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAP 406
I Y E++ P + +A A ++ + EA + CQ AL ++ P
Sbjct: 141 IRYYLIAIELR--------PNFADAWSNLASAYMRKGRLSEATQCCQQALSLN------P 186
Query: 407 SLEEAADRRLMGLILDTKGN-HEAALEHLVLASMAMVANGQEVEVASVDCSIGDTYLSMS 465
L +A +G ++ +G HEA +L + +A + GD ++
Sbjct: 187 LLVDAHSN--LGNLMKAQGLIHEAYSCYLEAVRIQPTFAIAWSNLAGLFMESGDLNRALQ 244
Query: 466 RYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYEN 525
Y EAV + PA ++ L +++ G+ E+ AL++ N
Sbjct: 245 YYKEAV---------------KLKPAFPDAYLNLGNVYKALGRPTEAIMCYQHALQMRPN 289
Query: 526 PMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALL-------IYNDSPGQQSSIAG 578
A N+++IY +L+ A++ ++AL YN+ I
Sbjct: 290 S---------AMAFGNIASIYYEQGQLDLAIRHYKQALSRDPRFLEAYNNLGNALKDIGR 340
Query: 579 IEAQMGVMYYMLG---NYTESYNTLKNAITKLRAIGEKKSSF---------IGIALNQMG 626
++ + L N+ ++ L N + +G S F + N +
Sbjct: 341 VDEAVRCYNQCLALQPNHPQAMANLGNIYMEWNMMGPASSLFKATLAVTTGLSAPFNNLA 400
Query: 627 LACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEY 686
+ Q + +A + E I P N TY IGR+ EAIQ +
Sbjct: 401 IIYKQQGNYSDAISCYNEVLRI--------DPLAADALVNRGNTYKEIGRVTEAIQDYMH 452
Query: 687 VVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRS 721
++ R P + + L+ K++G V +
Sbjct: 453 AINFR--------PTMAEAHANLASAYKDSGHVEA 479
Score = 40.8 bits (94), Expect = 0.003
Identities = 65/315 (20%), Positives = 118/315 (36%), Gaps = 55/315 (17%)
Query: 386 DEAERLCQMALDIHKANSSAPSL--------EEAADRRL-MGLILDTKGNHEAALEHLVL 436
D + + LD+ ++SS+ SL E D RL + L G+ + ALEH
Sbjct: 19 DRVDEVFSRKLDLSVSSSSSSSLLQQFNKTHEGDDDARLALAHQLYKGGDFKQALEH--- 75
Query: 437 ASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVF 496
++M N + + IG Y + YD + E+AL + P +
Sbjct: 76 SNMVYQRNPLRTDNLLL---IGAIYYQLQEYDMCIARNEEALRI--------QPQFAECY 124
Query: 497 VRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKAL 556
+A+ G + Y A+ + N A +N+++ Y L +A
Sbjct: 125 GNMANAWKEKGDTDRAIRYYLIAIELRPN---------FADAWSNLASAYMRKGRLSEAT 175
Query: 557 KLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSS 616
+ Q+AL + S++ + G+++ E+Y+ A+ S+
Sbjct: 176 QCCQQALSLNPLLVDAHSNLGNLMKAQGLIH-------EAYSCYLEAVRIQPTFAIAWSN 228
Query: 617 FIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGR 676
G+ + L A + ++EA + P P+ Y NL Y A+GR
Sbjct: 229 LAGLFMESGDL--------NRALQYYKEAVKLK-----PAFPDA---YLNLGNVYKALGR 272
Query: 677 LDEAIQILEYVVSVR 691
EAI ++ + +R
Sbjct: 273 PTEAIMCYQHALQMR 287
Score = 36.2 bits (82), Expect = 0.069
Identities = 55/304 (18%), Positives = 112/304 (36%), Gaps = 62/304 (20%)
Query: 291 AAIYCNLGQYNEAIPILERSIEI-PVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMC 349
A ++ G N A+ + ++++ P ++ ++ LG+ Y LG+ +IMC
Sbjct: 230 AGLFMESGDLNRALQYYKEAVKLKPAFPDA----------YLNLGNVYKALGRPTEAIMC 279
Query: 350 YSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMAL-----------DI 398
Y +++ P +A + Q D A R + AL ++
Sbjct: 280 YQHALQMR--------PNSAMAFGNIASIYYEQGQLDLAIRHYKQALSRDPRFLEAYNNL 331
Query: 399 HKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHL--VLASMAMVANGQEVEVASVDCS 456
A ++EA R L + NH A+ +L + M+ + A++ +
Sbjct: 332 GNALKDIGRVDEAV--RCYNQCLALQPNHPQAMANLGNIYMEWNMMGPASSLFKATLAVT 389
Query: 457 IGDT---------YLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTG 507
G + Y Y +A+ Y + L + P V + + G
Sbjct: 390 TGLSAPFNNLAIIYKQQGNYSDAISCYNEVLRI--------DPLAADALVNRGNTYKEIG 441
Query: 508 KIKESKSYCDSALRIYENPMPGVN-PEEIASGLTNVSAIYESMNELEKALKLLQKALLIY 566
++ E+ ++ M +N +A N+++ Y+ +E A+ ++ALL+
Sbjct: 442 RVTEA----------IQDYMHAINFRPTMAEAHANLASAYKDSGHVEAAITSYKQALLLR 491
Query: 567 NDSP 570
D P
Sbjct: 492 PDFP 495
Score = 30.8 bits (68), Expect = 2.9
Identities = 22/71 (30%), Positives = 29/71 (39%), Gaps = 8/71 (11%)
Query: 658 PETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAG 717
P +SNLA Y GRL EA Q + +S+ NP + D L L+K G
Sbjct: 152 PNFADAWSNLASAYMRKGRLSEATQCCQQALSL--------NPLLVDAHSNLGNLMKAQG 203
Query: 718 RVRSRKVMSLE 728
+ LE
Sbjct: 204 LIHEAYSCYLE 214
>At1g48300 unknown protein
Length = 285
Score = 42.4 bits (98), Expect = 0.001
Identities = 44/147 (29%), Positives = 68/147 (45%), Gaps = 9/147 (6%)
Query: 146 VHEIQSATTSSVSTEKSVKALNSQLDASPKS-KPKGKSPPAKAPLERKNDKPLLRKQTKG 204
V EIQ+ T S +TE VK L QL A K K + K AKA K K + ++
Sbjct: 39 VGEIQTKTISE-ATEILVKQLE-QLKAEEKILKKQRKEEKAKA----KAMKKMTEMDSES 92
Query: 205 VASGVKSLKSSPLGKSVSLNRVENSAESALD--KPERAPILLKQARDLISSGDNPQKALE 262
+S S GK V ++ + N A+ L+ +PE L + ++ S N +AL+
Sbjct: 93 SSSSESSDSDCDKGKVVDMSSLRNKAKPVLEPLQPEATVATLPRIQEDAISCKNTSEALQ 152
Query: 263 LALQAMNLLEKLGNGKPSLELVMCLHV 289
+ALQ + + N +L+ V + V
Sbjct: 153 IALQTSTIFPSMANPGQTLKTVEAVSV 179
>At3g14670 hypothetical protein
Length = 232
Score = 39.3 bits (90), Expect = 0.008
Identities = 44/183 (24%), Positives = 67/183 (36%), Gaps = 29/183 (15%)
Query: 27 TLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPSIEQLYENVCDMQSSDQSPSRKSFGSDG 86
T A + P + P + GV E I + V ++ ++S + + S+
Sbjct: 27 TKATTEPPMTTEEPSASKQNPVVIEGRGVEEEQIPTIITTV--VEEGEKSDNNEEENSEK 84
Query: 87 DESRIDSELRQLVGGRMREVEIMEEEVEVEVEKER-----GGSSSGEISSGVGGLSSNEK 141
DE E E E EEE + E EKE GG SS + S +G SS+++
Sbjct: 85 DEKEESEEEES-------EEEEKEEEEKEEEEKEEEGNVAGGGSSDDSSRTLGKESSSDE 137
Query: 142 KLDKVHEIQSATTSSVSTEKSVKALNSQLDASPKSK-------------PKGKSPPAKAP 188
+D E + + + + D PK K PK K P K P
Sbjct: 138 NMD--DETAVGKQVDIPAAMKINEMGQENDGDPKEKDGDLEKDGDQEKDPKEKDPKEKDP 195
Query: 189 LER 191
E+
Sbjct: 196 KEK 198
>At5g37590 kinesin -like protein
Length = 225
Score = 38.5 bits (88), Expect = 0.014
Identities = 31/121 (25%), Positives = 50/121 (40%), Gaps = 5/121 (4%)
Query: 334 GDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQ 393
G Y G+LE + + S + ++ GE DP V C +AE +FD+AE L
Sbjct: 104 GRDYFFQGKLEPAERLFGSAIQEAKEGFGEKDPHVASACNNLAELYRVKKEFDKAEPLYL 163
Query: 394 MALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAA-----LEHLVLASMAMVANGQEV 448
A+ I + + A +G + + E A + V A ++ V N E+
Sbjct: 164 EAVSILEEFYGPDDVRVGATLHNLGQLYLVQRKLEEARACYEIRRCVFAVISYVKNYDEL 223
Query: 449 E 449
E
Sbjct: 224 E 224
Score = 31.6 bits (70), Expect = 1.7
Identities = 25/94 (26%), Positives = 37/94 (38%)
Query: 587 YYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAK 646
Y+ G + +AI + + +K + A N + F +A L+ EA
Sbjct: 107 YFFQGKLEPAERLFGSAIQEAKEGFGEKDPHVASACNNLAELYRVKKEFDKAEPLYLEAV 166
Query: 647 SILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEA 680
SILE YGP NL Y +L+EA
Sbjct: 167 SILEEFYGPDDVRVGATLHNLGQLYLVQRKLEEA 200
>At4g27430 COP1-interacting protein 7 (CIP7)
Length = 1058
Score = 38.1 bits (87), Expect = 0.018
Identities = 42/165 (25%), Positives = 73/165 (43%), Gaps = 22/165 (13%)
Query: 79 RKSFGSDGDESRIDSELRQLVGGRMREVEIMEEEVEVEVEKERGGSSSGEISSGVGGLSS 138
R ++GS S+ SE+ + RM E+ I ++ E K GGS S ++S
Sbjct: 745 RPAWGSRAAVSKSKSEMEEERKKRMEELLIQRQKRIAE--KSSGGSVSSSLAS------- 795
Query: 139 NEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDKPLL 198
K + + SS+ EK+ +A S+ +S + A+ + KP++
Sbjct: 796 -----KKTPTVTKSVKSSIKNEKTPEAAQSKAKPVLRSSTIERLAVARTAPKEPQQKPVI 850
Query: 199 RKQTKGVASGVKS-----LKSSPLGKSVSLNRVENSAESALDKPE 238
++ +K SG K+ KSS +G+S VE S + +L+ E
Sbjct: 851 KRTSK--PSGYKTEKAQEKKSSKIGQS-DAKSVELSRDPSLEIKE 892
>At4g26630 unknown protein
Length = 763
Score = 38.1 bits (87), Expect = 0.018
Identities = 39/167 (23%), Positives = 67/167 (39%), Gaps = 6/167 (3%)
Query: 11 DGAVNEMNGNHSPSKETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPSIEQLYENVC-- 68
D A +GN KE V +G+ ++ +NG N V+ V E EN
Sbjct: 168 DKANGTKDGNTGDIKEEGTLVDEDKGTDMDEKVENGDENKQVENVEGKEKEDKEENKTKE 227
Query: 69 -DMQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEEVEVEVEKERGGSSSG 127
+ ++ S+ +G E D+E + + E E ++ E E E+ +G G
Sbjct: 228 VEAAKAEVDESKVEDEKEGSEDENDNEKVESKDAKEDEKEETNDDKEDEKEESKGSKKRG 287
Query: 128 EISSGVGGLSSNEKKLDKVHEIQSATTSS---VSTEKSVKALNSQLD 171
+ +S G + K + + + T S V KSV+ L + +D
Sbjct: 288 KGTSSGGKVREKNKTEEVKKDAEPRTPFSDRPVRERKSVERLVALID 334
>At1g13160 unknown protein
Length = 804
Score = 37.4 bits (85), Expect = 0.031
Identities = 53/239 (22%), Positives = 96/239 (39%), Gaps = 22/239 (9%)
Query: 23 PSKETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPSIEQLYENVCDMQSSDQSPSRKSF 82
P+ E L + GS Q + G P+ +E ++L C S D++ +
Sbjct: 515 PNVELLQESDNESGSDGDQ--DDDGVELPIGDDVE---QELIPGDCG--SEDKAEEDSND 567
Query: 83 GSDGDESRIDSELRQLVGG-------RMREVEIMEEEVEVEVEKERGGSSSGEISSGVGG 135
G D + + DS++ +GG E + E E+E E+E G +S S V
Sbjct: 568 GDDMNNTEDDSDIDTSIGGDEDEEVNDSDEADTDSENEEIESEEEDGEAS----DSSVED 623
Query: 136 LSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDK 195
+ EK K +I + +S + S++AL +A + + + + + K
Sbjct: 624 SGNKEKAKGKKRKIVDFDANLLSADTSLRALKRFAEAKNEKPSFDEGDGILSNEDFRKIK 683
Query: 196 PLLRKQTKGVASGVKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARDLISSG 254
L K+ +A K K P +S RV+ + L+ R + +Q +L+ +G
Sbjct: 684 TLQAKKEAKIALARKGFK-VPNSDQLSKKRVD---PAKLEAHIRHKLTKEQRLELVKAG 738
>At3g28770 hypothetical protein
Length = 2081
Score = 36.6 bits (83), Expect = 0.053
Identities = 52/229 (22%), Positives = 85/229 (36%), Gaps = 14/229 (6%)
Query: 57 EPSIEQLYENVCDMQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEE---V 113
E S+E+ E + D S + K + G + + E VG + + +E
Sbjct: 624 ETSLEEKREQT--QKGHDNSINSKIVDNKGGNADSNKEKEVHVGDSTNDNNMESKEDTKS 681
Query: 114 EVEVEKERGGSSSGEISSGVGGLSSNEKKLD-KVHEIQSATTSSVSTEKSVKALNSQLDA 172
EVEV+K G S GE S +KKL+ K + S SV ++ +
Sbjct: 682 EVEVKKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDSKDDKSVDDKQEEAQIYGGESK 741
Query: 173 SPKS-KPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLG---KSVSLNRVEN 228
KS + KGK +K + K ++ +R + + V K + G +S VE
Sbjct: 742 DDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAKSVET 801
Query: 229 SAESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLGNG 277
L E +A++ + K Q++ EK NG
Sbjct: 802 KDNKKLSSTENR----DEAKERSGEDNKEDKEESKDYQSVEAKEKNENG 846
Score = 29.3 bits (64), Expect = 8.4
Identities = 44/215 (20%), Positives = 86/215 (39%), Gaps = 35/215 (16%)
Query: 53 DGVIEPSIEQLYENVCDMQSS--DQSPSRKSFGS---DGDESRIDSELRQLVGGRMREVE 107
D IE ++E + +++++ D S ++ G + S ID+E G E
Sbjct: 286 DSAIEKNLESKEDVKSEVEAAKNDGSSMTENLGEAQGNNGVSTIDNEKEVEGQGESIEDS 345
Query: 108 IMEEEVE--------VEVEKERGGSSSGEI-----SSGVG------------GLSSNEKK 142
+E+ +E VE K G S +G++ ++GV G S+N+K
Sbjct: 346 DIEKNLESKEDVKSEVEAAKNAGSSMTGKLEEAQRNNGVSTNETMNSENKGSGESTNDKM 405
Query: 143 L-----DKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDKPL 197
+ D+ H+ ++ + + +SVK N + A + KG++ K E
Sbjct: 406 VNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEESMKGENLENKVGNEELKGNAS 465
Query: 198 LRKQTKGVASGVKSLKSSPLGKSVSLNRVENSAES 232
+ +T +S + + S V +N+ E+
Sbjct: 466 VEAKTNNESSKEEKREESQRSNEVYMNKETTKGEN 500
>At5g52310 low-temperature-induced protein 78 (sp|Q06738)
Length = 710
Score = 35.8 bits (81), Expect = 0.090
Identities = 37/143 (25%), Positives = 59/143 (40%), Gaps = 8/143 (5%)
Query: 14 VNEMNGNHSPSKETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPSIEQLYENVCDMQSS 73
VNE + + T P+ + QRG++ V G + E+L D S
Sbjct: 539 VNEKDQETESAVTTKLPISGGGSGVEEQRGEDKS----VSGR-DYVAEKLTTEEEDKAFS 593
Query: 74 DQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEEVEVEVEKERGGSSSGEISSGV 133
D + G G+E + ++ ++ V E EE VE E + GG G I
Sbjct: 594 DMVAEKLQIG--GEEEKKETTTKE-VEKISTEKAASEEGEAVEEEVKGGGGMVGRIKGWF 650
Query: 134 GGLSSNEKKLDKVHEIQSATTSS 156
GG +++E K + H ++ A SS
Sbjct: 651 GGGATDEVKPESPHSVEEAPKSS 673
>At3g11540 spindly (gibberellin signal transduction protein)
Length = 914
Score = 35.8 bits (81), Expect = 0.090
Identities = 64/333 (19%), Positives = 118/333 (35%), Gaps = 67/333 (20%)
Query: 408 LEEAADRRLMGLILDTKGNHEAALEHLVLA----SMAMVANGQE-----VEVASVD---- 454
L EAA+ L+ D A +VL S+ + N QE E +D
Sbjct: 127 LVEAAESYQKALMADASYKPAAECLAIVLTDLGTSLKLAGNTQEGIQKYYEALKIDPHYA 186
Query: 455 ---CSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKE 511
++G Y M +YD A+ YEKA P + + ++ G ++
Sbjct: 187 PAYYNLGVVYSEMMQYDNALSCYEKAAL--------ERPMYAEAYCNMGVIYKNRGDLEM 238
Query: 512 SKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLI---YND 568
+ + + L + +P + +A LT++ + ++ + + +KAL Y D
Sbjct: 239 AITCYERCLAV--SPNFEIAKNNMAIALTDLGTKVKLEGDVTQGVAYYKKALYYNWHYAD 296
Query: 569 SPGQQSSIAG--IEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMG 626
+ G ++ M +++Y L + + A N +G
Sbjct: 297 AMYNLGVAYGEMLKFDMAIVFYELAFHFNPH--------------------CAEACNNLG 336
Query: 627 LACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEY 686
+ +A E ++ A SI P +NL Y G++D A ++E
Sbjct: 337 VLYKDRDNLDKAVECYQMALSI--------KPNFAQSLNNLGVVYTVQGKMDAAASMIEK 388
Query: 687 VVSVREEKLGTANPDVDDEKRRLSELLKEAGRV 719
+ ANP + L L ++AG +
Sbjct: 389 AI--------LANPTYAEAFNNLGVLYRDAGNI 413
>At1g01320 unknown protein
Length = 1047
Score = 35.8 bits (81), Expect = 0.090
Identities = 34/146 (23%), Positives = 60/146 (40%), Gaps = 2/146 (1%)
Query: 460 TYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSA 519
T L + ++AV KAL G H + LA + TG ++ Y A
Sbjct: 141 TALDKGKLEDAVTYGTKALAKLVAVCGPYHRMTAGAYSLLAVVLYHTGDFNQATIYQQKA 200
Query: 520 LRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPG-QQSSIAG 578
L I E + G++ + +++ Y + E ALK +++AL + + + G + A
Sbjct: 201 LDINEREL-GLDHPDTMKSYGDLAVFYYRLQHTELALKYVKRALYLLHLTCGPSHPNTAA 259
Query: 579 IEAQMGVMYYMLGNYTESYNTLKNAI 604
+ +M LGN + L A+
Sbjct: 260 TYINVAMMEEGLGNVHVALRYLHKAL 285
Score = 33.5 bits (75), Expect = 0.45
Identities = 17/48 (35%), Positives = 24/48 (49%)
Query: 654 GPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPD 701
GPYH T G YS LA G ++A + + + E +LG +PD
Sbjct: 167 GPYHRMTAGAYSLLAVVLYHTGDFNQATIYQQKALDINERELGLDHPD 214
Score = 32.3 bits (72), Expect = 0.99
Identities = 23/99 (23%), Positives = 45/99 (45%)
Query: 603 AITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLG 662
A+ KL A+ A + + + F +AT ++A I E E G HP+T+
Sbjct: 158 ALAKLVAVCGPYHRMTAGAYSLLAVVLYHTGDFNQATIYQQKALDINERELGLDHPDTMK 217
Query: 663 VYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPD 701
Y +LA Y + + A++ ++ + + G ++P+
Sbjct: 218 SYGDLAVFYYRLQHTELALKYVKRALYLLHLTCGPSHPN 256
Score = 31.2 bits (69), Expect = 2.2
Identities = 16/59 (27%), Positives = 27/59 (45%)
Query: 638 ATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLG 696
A + + A +L GP HP T Y N+A + +G + A++ L + + LG
Sbjct: 235 ALKYVKRALYLLHLTCGPSHPNTAATYINVAMMEEGLGNVHVALRYLHKALKCNQRLLG 293
>At5g33300 putative protein
Length = 439
Score = 35.4 bits (80), Expect = 0.12
Identities = 44/189 (23%), Positives = 82/189 (43%), Gaps = 19/189 (10%)
Query: 88 ESRIDSELRQLVGGRMREVEIMEEEVEVEVEKERGGSSSGEISSGVGGLSS----NEKKL 143
+S + + +L ++ ++E + E ++ +VE+ R + ISS VG L S ++K +
Sbjct: 11 DSSLGDDAEKLYEKKILDLESVNEALKSDVEELRSKLADVSISSSVGTLQSSRKFSQKSI 70
Query: 144 DKVHEIQSATTSSVSTEKSVKALNS--QLDASPKSKPKGKSPPAKAPLERKNDKPLLRKQ 201
E S+ + S+ + K K +S Q D + K K+ K + K D R
Sbjct: 71 ANKEEGMSSRSKSMCSTKKYKTESSVKQFDGEVQ---KLKAQKVKLHCKIKLDSMQFRLL 127
Query: 202 TKGVASGVKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARDLISSGDNPQKAL 261
+ V LK K + + E SAL+ ++ + LK + L + K L
Sbjct: 128 KASLEKEVLQLK-----KELRKSEFEKHVLSALNNRQKLILQLKNTQALTA-----LKRL 177
Query: 262 ELALQAMNL 270
++ LQ+ +
Sbjct: 178 KMLLQSKKI 186
>At1g20970 hypothetical protein
Length = 1498
Score = 35.4 bits (80), Expect = 0.12
Identities = 44/208 (21%), Positives = 84/208 (40%), Gaps = 36/208 (17%)
Query: 5 VTNGVCDGAVNEMNGNHSPSKETL-------------APVKSPRGSLSPQRGQNGGPNFP 51
V NG+ G N NGN + + P + ++ G N
Sbjct: 37 VVNGISHGGSNNGNGNDTDGSYDFITENDTVGDDFVESDYVKPVDDANVEKDLKEGENVK 96
Query: 52 VDGVIEPSIEQLYENVCDMQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEE 111
VD PSI ++V + Q+ + SEL G VEI +
Sbjct: 97 VDA---PSIAD--DDVLGVSQDSQTLEK-------------SELESTDDGPEEVVEIPKS 138
Query: 112 EVEVEVEK--ERGGSSSGEISSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQ 169
EVE +EK ++ +G + SG+ G ++++++++H+ S S T+ +V+ +
Sbjct: 139 EVEDSLEKSVDQQHPGNGHLESGLEGKVESKEEVEQLHD--SEVGSKDLTKNNVEEPEVE 196
Query: 170 LDASPKSKPKGKSPPAKAPLERKNDKPL 197
+++ ++ +G K + K+D+ L
Sbjct: 197 IESDSETDVEGHQGD-KIEAQEKSDRDL 223
>At1g15290 unknown protein
Length = 1558
Score = 35.4 bits (80), Expect = 0.12
Identities = 32/132 (24%), Positives = 54/132 (40%), Gaps = 2/132 (1%)
Query: 462 LSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALR 521
L + D+AV KAL G H + LA + TG ++ Y AL
Sbjct: 809 LDKGKLDDAVSYGTKALVKMIAVCGPYHRNTACAYSLLAVVLYHTGDFNQATIYQQKALD 868
Query: 522 IYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPG-QQSSIAGIE 580
I E + G++ + ++S Y + E ALK + +AL + + + G + A
Sbjct: 869 INEREL-GLDHPDTMKSYGDLSVFYYRLQHFELALKYVNRALFLLHFTCGLSHPNTAATY 927
Query: 581 AQMGVMYYMLGN 592
+ +M +GN
Sbjct: 928 INVAMMEKEVGN 939
Score = 33.5 bits (75), Expect = 0.45
Identities = 27/122 (22%), Positives = 53/122 (43%), Gaps = 2/122 (1%)
Query: 596 SYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGP 655
SY T A+ K+ A+ A + + + F +AT ++A I E E G
Sbjct: 819 SYGT--KALVKMIAVCGPYHRNTACAYSLLAVVLYHTGDFNQATIYQQKALDINERELGL 876
Query: 656 YHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKE 715
HP+T+ Y +L+ Y + + A++ + + + G ++P+ ++ + KE
Sbjct: 877 DHPDTMKSYGDLSVFYYRLQHFELALKYVNRALFLLHFTCGLSHPNTAATYINVAMMEKE 936
Query: 716 AG 717
G
Sbjct: 937 VG 938
>At3g26050 unknown protein (At3g26050)
Length = 533
Score = 35.0 bits (79), Expect = 0.15
Identities = 44/164 (26%), Positives = 71/164 (42%), Gaps = 20/164 (12%)
Query: 103 MREVEIMEEEVEVEVEKERGGSSSGEISSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKS 162
++ V + +E + KER SSSG S SS +LD H +++ T S+ S
Sbjct: 154 LKSVVVPDEGNSTSLSKERPPSSSGSKVSSSKLESSVVIELD--HSLKN--TKKESSSSS 209
Query: 163 VKALNSQLDASPKSKPKGKSPPAKAPL------ERKNDKPLLRKQTKGVASGVKSLKSSP 216
++L++ SK + +SPP + +K ++ + G S VK+ K
Sbjct: 210 TRSLSA-------SKNRSRSPPEPIHMSISCVSSSNTEKTIVGRPQNGSRSAVKADKKKR 262
Query: 217 LGKSVSLNRVENSAESALDKPERAPILLKQARDLISSGDNPQKA 260
G S S++ N A S L + AP L AR+ + G A
Sbjct: 263 SGPS-SVHMSLNFASSTLRTTKEAPKTL--ARNSTTQGTTSTNA 303
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,883,435
Number of Sequences: 26719
Number of extensions: 693103
Number of successful extensions: 2664
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 103
Number of HSP's that attempted gapping in prelim test: 2503
Number of HSP's gapped (non-prelim): 192
length of query: 746
length of database: 11,318,596
effective HSP length: 107
effective length of query: 639
effective length of database: 8,459,663
effective search space: 5405724657
effective search space used: 5405724657
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0166.9


