

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.7
(1602 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
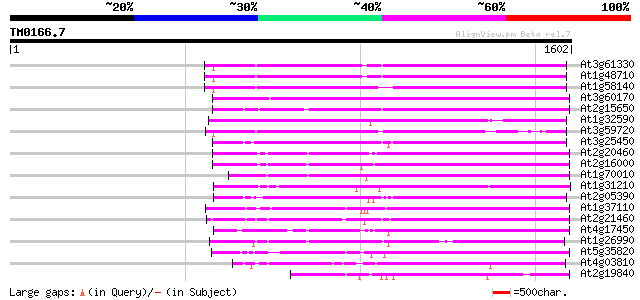
Score E
Sequences producing significant alignments: (bits) Value
At3g61330 copia-type polyprotein 766 0.0
At1g48710 hypothetical protein 762 0.0
At1g58140 hypothetical protein 760 0.0
At3g60170 putative protein 754 0.0
At2g15650 putative retroelement pol polyprotein 718 0.0
At1g32590 hypothetical protein, 5' partial 662 0.0
At3g59720 copia-type reverse transcriptase-like protein 655 0.0
At3g25450 hypothetical protein 637 0.0
At2g20460 putative retroelement pol polyprotein 634 0.0
At2g16000 putative retroelement pol polyprotein 628 e-180
At1g70010 hypothetical protein 592 e-169
At1g31210 putative reverse transcriptase 588 e-167
At2g05390 putative retroelement pol polyprotein 586 e-167
At1g37110 557 e-158
At2g21460 putative retroelement pol polyprotein 553 e-157
At4g17450 retrotransposon like protein 540 e-153
At1g26990 polyprotein, putative 538 e-152
At5g35820 copia-like retrotransposable element 531 e-150
At4g03810 putative retrotransposon protein 503 e-142
At2g19840 copia-like retroelement pol polyprotein 478 e-134
>At3g61330 copia-type polyprotein
Length = 1352
Score = 766 bits (1977), Expect = 0.0
Identities = 419/1049 (39%), Positives = 595/1049 (55%), Gaps = 32/1049 (3%)
Query: 555 KEDPESGYLRTRLSMLQISLIAPLK------HQSWYLDSGCSRHMTGEKRMFRELKLKPG 608
K + ++ Y+ ++ + L+A K + WYLDSG S HM G K MF EL
Sbjct: 301 KFEEKAHYVEEKIQEEDMLLMASYKKDEQKENHKWYLDSGASNHMCGRKSMFAELDESVR 360
Query: 609 GEVGFGGNEKGKIIGTGTICV----DSSPCIDNVLLVDGLTHNLLSISQLADKGYDVIFN 664
G V G K ++ G G I + I NV + + N+LS+ QL +KGYD+
Sbjct: 361 GNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLK 420
Query: 665 QKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMR 724
+ Q + KN ++ + + AQ +K + EE W+WH R GH +
Sbjct: 421 DNNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLK--MCYKEESWLWHLRFGHLNFG 478
Query: 725 KISQLSKLNLVRGLPNLKFASDALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGP 784
+ LS+ +VRGLP + + +CE C GK K+ F ++ +PLEL+H D+ GP
Sbjct: 479 GLELLSRKEMVRGLPCINHPNQ-VCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGP 537
Query: 785 VKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDH 844
+K +S+G Y ++ +DD+SR TWV FL K E +F F A V+ E I +RSD
Sbjct: 538 IKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDR 597
Query: 845 GGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWA 904
GGEF + +F + GI + PR+PQQNGVVERKNRT+ EMAR+ML+ + K WA
Sbjct: 598 GGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVVERKNRTILEMARSMLKSKRLPKELWA 657
Query: 905 EAVNTACYIQNRISVRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKS 964
EAV A Y+ NR + + KTP E W KP +S+ FG + + ++ K D KS
Sbjct: 658 EAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKS 717
Query: 965 SKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADLSINVSDKGK 1024
K + +GY + SKG++ YN D K S ++ FD++ + D + N D
Sbjct: 718 EKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNS----------NEEDYNF 767
Query: 1025 APEEVEPEEDEPEEEAGPSNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETL- 1083
P E +E EP E PS T + T++ +E + + R RS E T
Sbjct: 768 FPH-FEEDEPEPTREEPPSEEPTTPPTSPTSSQIEE---SSSERTPRFRSIQELYEVTEN 823
Query: 1084 ---LSLKGLVSLIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKW 1140
L+L L + EP +A++ K W AM EE+ KND W L P IG KW
Sbjct: 824 QENLTLFCLFAECEPMDFQKAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKW 883
Query: 1141 VFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILH 1200
V++ K N KG+V R KARLVA+GYSQ+ GIDY E FAPVARLE +RL+IS + + +H
Sbjct: 884 VYKAKKNSKGEVERYKARLVAKGYSQRVGIDYDEVFAPVARLETVRLIISLAAQNKWKIH 943
Query: 1201 QMDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLL 1260
QMDVKSAFLNG + EEVY+ QP G+ + + D V +LKK LYGLKQAPRAW R+ +
Sbjct: 944 QMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKVLYGLKQAPRAWNTRIDKYFK 1003
Query: 1261 ENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGE 1320
E +F++ + L+ K K+DILI +YVDD+IF N S+ +EF + M EFEM+ +G
Sbjct: 1004 EKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPSIFEEFKKEMTKEFEMTDIGL 1063
Query: 1321 LKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQ 1380
+ Y+LGI+V Q G +I Q Y KE+LKKF + +S TPM L K+++ V
Sbjct: 1064 MSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKIDDSNPVCTPMECGIKLSKKEEGEGVDP 1123
Query: 1381 KLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYK 1440
++ ++GSL YLT +RPDIL++V + +R+ P TH A KRILRY+KGT N GL Y
Sbjct: 1124 TTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYS 1183
Query: 1441 KTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAA 1500
TS+YKL GY D+D+ GD +RKSTSG ++G +W SK+Q + LST EAEY++A
Sbjct: 1184 TTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAAT 1243
Query: 1501 ICSTQMLWMKHQLEDYQI-FESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYV 1559
C +W+++ L++ + E I+ DN +AI+L+KNP+ H R+KHI+ +YH+IR+ V
Sbjct: 1244 SCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECV 1303
Query: 1560 QKGVLLLKFVDTDHQWADIFTKPLAEDRF 1588
K + L++V T Q AD FTKPL + F
Sbjct: 1304 SKKDVQLEYVKTHDQVADFFTKPLKRENF 1332
>At1g48710 hypothetical protein
Length = 1352
Score = 762 bits (1967), Expect = 0.0
Identities = 418/1049 (39%), Positives = 593/1049 (55%), Gaps = 32/1049 (3%)
Query: 555 KEDPESGYLRTRLSMLQISLIAPLK------HQSWYLDSGCSRHMTGEKRMFRELKLKPG 608
K + ++ Y+ ++ + L+A K + WYLDSG S HM G K MF EL
Sbjct: 301 KFEEKANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVR 360
Query: 609 GEVGFGGNEKGKIIGTGTICV----DSSPCIDNVLLVDGLTHNLLSISQLADKGYDVIFN 664
G V G K ++ G G I + I NV + + N+LS+ QL +KGYD+
Sbjct: 361 GNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLK 420
Query: 665 QKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMR 724
+ Q + KN ++ + + AQ +K + EE W+WH R GH +
Sbjct: 421 DNNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLK--MCYKEESWLWHLRFGHLNFG 478
Query: 725 KISQLSKLNLVRGLPNLKFASDALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGP 784
+ LS+ +VRGLP + + +CE C GK K+ F ++ + LEL+H D+ GP
Sbjct: 479 GLELLSRKEMVRGLPCINHPNQ-VCEGCLLGKQFKMSFPKESSSRAQKSLELIHTDVCGP 537
Query: 785 VKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDH 844
+K +S+G Y ++ +DD+SR TWV FL K E +F F A V+ E I +RSD
Sbjct: 538 IKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDR 597
Query: 845 GGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWA 904
GGEF + +F + GI + PR+PQQNGV ERKNRT+ EMAR+ML+ + K WA
Sbjct: 598 GGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWA 657
Query: 905 EAVNTACYIQNRISVRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKS 964
EAV A Y+ NR + + KTP E W K +S+ FG + + ++ K D KS
Sbjct: 658 EAVACAVYLLNRSPTKSVSGKTPQEAWSGRKSGVSHLRVFGSIAHAHVPDEKRSKLDDKS 717
Query: 965 SKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADLSINVSDKGK 1024
K + +GY + SKG++ YN D K S ++ FD++ + D + N D
Sbjct: 718 EKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNS----------NEEDYNF 767
Query: 1025 APEEVEPEEDEPEEEAGPSNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETL- 1083
P E +E EP E PS T + T++ +E + + R RS E T
Sbjct: 768 FPH-FEEDEPEPTREEPPSEEPTTPPTSPTSSQIEE---SSSERTPRFRSIQELYEVTEN 823
Query: 1084 ---LSLKGLVSLIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKW 1140
L+L L + EP EA++ K W AM EE+ KND W L P IG KW
Sbjct: 824 QENLTLFCLFAECEPMDFQEAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKTIGVKW 883
Query: 1141 VFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILH 1200
V++ K N KG+V R KARLVA+GY Q+ GIDY E FAPVARLE +RL+IS + + +H
Sbjct: 884 VYKAKKNSKGEVERYKARLVAKGYIQRAGIDYDEVFAPVARLETVRLIISLAAQNKWKIH 943
Query: 1201 QMDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLL 1260
QMDVKSAFLNG + EEVY+ QP G+ + + D V +LKK+LYGLKQAPRAW R+ +
Sbjct: 944 QMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFK 1003
Query: 1261 ENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGE 1320
E +F++ + L+ K K+DILI +YVDD+IF N S+ +EF + M EFEM+ +G
Sbjct: 1004 EKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGL 1063
Query: 1321 LKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQ 1380
+ Y+LGI+V Q G +I Q Y KE+LKKF M +S TPM L K+++ V
Sbjct: 1064 MSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDP 1123
Query: 1381 KLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYK 1440
++ ++GSL YLT +RPDIL++V + +R+ P TH A KRILRY+KGT N GL Y
Sbjct: 1124 TTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYS 1183
Query: 1441 KTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAA 1500
TS+YKL GY D+D+ GD +RKSTSG ++G +W SK+Q + LST EAEY++A
Sbjct: 1184 TTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVVLSTCEAEYVAAT 1243
Query: 1501 ICSTQMLWMKHQLEDYQI-FESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYV 1559
C +W+++ L++ + E I+ DN +AI+L+KNP+ H R+KHI+ +YH+IR+ V
Sbjct: 1244 SCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECV 1303
Query: 1560 QKGVLLLKFVDTDHQWADIFTKPLAEDRF 1588
K + L++V T Q ADIFTKPL + F
Sbjct: 1304 SKKDVQLEYVKTHDQVADIFTKPLKREDF 1332
>At1g58140 hypothetical protein
Length = 1320
Score = 760 bits (1962), Expect = 0.0
Identities = 410/1045 (39%), Positives = 582/1045 (55%), Gaps = 56/1045 (5%)
Query: 555 KEDPESGYLRTRLSMLQISLIAPLK------HQSWYLDSGCSRHMTGEKRMFRELKLKPG 608
K + ++ Y+ ++ + L+A K + WYLDSG S HM G K MF EL
Sbjct: 301 KFEEKANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVR 360
Query: 609 GEVGFGGNEKGKIIGTGTICV----DSSPCIDNVLLVDGLTHNLLSISQLADKGYDVIFN 664
G V G K ++ G G I + I NV + + N+LS+ QL +KGYD+
Sbjct: 361 GNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLK 420
Query: 665 QKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMR 724
+ Q + KN ++ + + AQ +K + EE W+WH R GH +
Sbjct: 421 DNNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLK--MCYKEESWLWHLRFGHLNFG 478
Query: 725 KISQLSKLNLVRGLPNLKFASDALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGP 784
+ LS+ +VRGLP + + +CE C GK K+ F ++ +PLEL+H D+ GP
Sbjct: 479 GLELLSRKEMVRGLPCINHPNQ-VCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGP 537
Query: 785 VKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDH 844
+K +S+G Y ++ +DD+SR TWV FL K E +F F A V+ E I +RSD
Sbjct: 538 IKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDR 597
Query: 845 GGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWA 904
GGEF + +F + GI + PR+PQQNGV ERKNRT+ EMAR+ML+ + K WA
Sbjct: 598 GGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWA 657
Query: 905 EAVNTACYIQNRISVRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKS 964
EAV A Y+ NR + + KTP E W KP +S+ FG + + ++ K D KS
Sbjct: 658 EAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKS 717
Query: 965 SKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADLSINVSDKGK 1024
K + +GY + SKG++ YN D K S ++ FD++ + D + E + DK +
Sbjct: 718 EKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDKPE 777
Query: 1025 APEEVEPEEDEPEEEAGPSNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLL 1084
E P E+ P++SQ +K
Sbjct: 778 PTREEPPSEEPTTPPTSPTSSQIEEKC--------------------------------- 804
Query: 1085 SLKGLVSLIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRN 1144
EP EA++ K W AM EE+ KND W L P IG KWV++
Sbjct: 805 ---------EPMDFQEAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKA 855
Query: 1145 KLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDV 1204
K N KG+V R KARLVA+GYSQ+ GIDY E FAPVARLE +RL+IS + + +HQMDV
Sbjct: 856 KKNSKGEVERYKARLVAKGYSQRAGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDV 915
Query: 1205 KSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEF 1264
KSAFLNG + EEVY+ QP G+ + + D V +LKK+LYGLKQAPRAW R+ + E +F
Sbjct: 916 KSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKDF 975
Query: 1265 VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYF 1324
++ + L+ K K+DILI +YVDD+IF N S+ +EF + M EFEM+ +G + Y+
Sbjct: 976 IKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYY 1035
Query: 1325 LGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYR 1384
LGI+V Q G +I Q Y KE+LKKF M +S TPM L K+++ V ++
Sbjct: 1036 LGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFK 1095
Query: 1385 GMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSE 1444
++GSL YLT +RPDIL++V + +R+ P TH A KRILRY+KGT N GL Y TS+
Sbjct: 1096 SLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSD 1155
Query: 1445 YKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICST 1504
YKL GY D+D+ GD +RKSTSG ++G +W SK+Q + LST EAEY++A C
Sbjct: 1156 YKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVC 1215
Query: 1505 QMLWMKHQLEDYQI-FESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGV 1563
+W+++ L++ + E I+ DN +AI+L+KNP+ H R+KHI+ +YH+IR+ V K
Sbjct: 1216 HAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKD 1275
Query: 1564 LLLKFVDTDHQWADIFTKPLAEDRF 1588
+ L++V T Q ADIFTKPL + F
Sbjct: 1276 VQLEYVKTHDQVADIFTKPLKREDF 1300
>At3g60170 putative protein
Length = 1339
Score = 754 bits (1947), Expect = 0.0
Identities = 419/1039 (40%), Positives = 606/1039 (57%), Gaps = 22/1039 (2%)
Query: 579 KHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICVDSS---PCI 635
+ + W+LDSGCS HMTG K F EL+ V G + + ++G G++ V + I
Sbjct: 296 RDEVWFLDSGCSNHMTGSKEWFSELEEGFNRTVKLGNDTRMSVVGKGSVKVKVNGVTQVI 355
Query: 636 DNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSE 695
V V L +NLLS+ QL ++G ++ +C+ G+++ + N ++ + S+
Sbjct: 356 PEVYYVPELRNNLLSLGQLQERGLAILIRDGTCKVYHPSKGAIMETNMSGNRMFFLLASK 415
Query: 696 LEAQNVKCLLS---VNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCETC 752
+ +N CL + +++E +WH R GH + + L+ +V GLP LK A+ +C C
Sbjct: 416 PQ-KNSLCLQTEEVMDKENHLWHCRFGHLNQEGLKLLAHKKMVIGLPILK-ATKEICAIC 473
Query: 753 QKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFL 812
GK + K +S L+L+H D+ GP+ S GKRY + +DD++R TWV FL
Sbjct: 474 LTGKQHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKTWVYFL 533
Query: 813 TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTP 872
K E+ A F F A V+ E + +R+D GGEF +++F S+GI+ + TP
Sbjct: 534 HEKSEAFATFKIFKASVEKEIGAFLTCLRTDRGGEFTSNEFGEFCRSHGISRQLTAAFTP 593
Query: 873 QQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWK 932
QQNGV ERKNRT+ R+ML E + K FW+EA + +IQNR + TP E W
Sbjct: 594 QQNGVAERKNRTIMNAVRSMLSERQVPKMFWSEATKWSVHIQNRSPTAAVEGMTPEEAWS 653
Query: 933 NIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYNTDAKTIEES 992
KP + YF FGC+ YV + K D KS KC+ LG S+ SK +R Y+ K I S
Sbjct: 654 GRKPVVEYFRVFGCIGYVHIPDQKRSKLDDKSKKCVFLGVSEESKAWRLYDPVMKKIVIS 713
Query: 993 IHVRFDD--KLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSN---SQT 1047
V FD+ D DQ+ + K L D K E VEP G N S
Sbjct: 714 KDVVFDEDKSWDWDQADVEAKEVTLECGDEDDEKNSEVVEPIAVASPNHVGSDNNVSSSP 773
Query: 1048 LKKSRITAAHPKELILGNKDEPVRTRSAFRPSE----ETLLSLKGLVSLIE--PKSIDEA 1101
+ A P + + P + + E E LS+ L+ + E P D+A
Sbjct: 774 ILAPSSPAPSPVAAKVTRERRPPGWMADYETGEGEEIEENLSVMLLMMMTEADPIQFDDA 833
Query: 1102 LQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGDVVRNKARLVA 1161
++DK W AM+ E+ KN+ W L P+ IG KWV++ KLNE G+V + KARLVA
Sbjct: 834 VKDKIWREAMEHEIESIVKNNTWELTTLPKGFTPIGVKWVYKTKLNEDGEVDKYKARLVA 893
Query: 1162 QGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQ 1221
+GY+Q GIDYTE FAPVARL+ +R +++ S N + Q+DVKSAFL+G + EEVYV Q
Sbjct: 894 KGYAQCYGIDYTEVFAPVARLDTVRTILAISSQFNWEIFQLDVKSAFLHGELKEEVYVRQ 953
Query: 1222 PPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDD 1281
P GF E + + V+KL+K+LYGLKQAPRAWY R+ ++ L+ EF R + TLF KT +
Sbjct: 954 PEGFIREGEEEKVYKLRKALYGLKQAPRAWYSRIEAYFLKEEFERCPSEHTLFTKTRVGN 1013
Query: 1282 ILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 1341
ILIV +YVDD+IF +++++C EF + M EFEMS +G++K+FLGI+V Q+ G +I Q
Sbjct: 1014 ILIVSLYVDDLIFTGSDKAMCDEFKKSMMLEFEMSDLGKMKHFLGIEVKQSDGGIFICQR 1073
Query: 1342 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDIL 1401
+Y +E+L +F M ES K P+ P L K++ KV + +++ ++GSL+YLT +RPD++
Sbjct: 1074 RYAREVLARFGMDESNAVKNPIVPGTKLTKDENGEKVDETMFKQLVGSLMYLTVTRPDLM 1133
Query: 1402 FSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMY--KKTSEYKLSGYCDADYAGDR 1459
+ V L +RF S+PR +H A KRILRYLKGT LG+ Y +K KL + D+DYAGD
Sbjct: 1134 YGVCLISRFMSNPRMSHWLAAKRILRYLKGTVELGIFYRRRKNRSLKLMAFTDSDYAGDL 1193
Query: 1460 TERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQIF 1519
+R+STSG + S + WASK+Q +ALST EAEYI+AA C+ Q +W++ LE
Sbjct: 1194 NDRRSTSGFVFLMASGAICWASKKQPVVALSTTEAEYIAAAFCACQCVWLRKVLEKLGAE 1253
Query: 1520 E-SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADI 1578
E S I CDN++ I LSK+P+LH ++KHIEV++H++RD V V+ L++ T+ Q ADI
Sbjct: 1254 EKSATVINCDNSSTIQLSKHPVLHGKSKHIEVRFHYLRDLVNGDVVKLEYCPTEDQVADI 1313
Query: 1579 FTKPLAEDRFNFILKNLNM 1597
FTKPL ++F + L M
Sbjct: 1314 FTKPLKLEQFEKLRALLGM 1332
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 718 bits (1853), Expect = 0.0
Identities = 400/1041 (38%), Positives = 593/1041 (56%), Gaps = 40/1041 (3%)
Query: 578 LKHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICV---DSSPC 634
L+ W +DSGC+ HMT E+R F + + + G G I V
Sbjct: 321 LREDVWLVDSGCTNHMTKEERYFSNINKSIKVPIRVRNGDIVMTAGKGDITVMTRHGKRI 380
Query: 635 IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLS 694
I NV LV GL NLLS+ Q+ GY V F K C + +G + N + + +KI+LS
Sbjct: 381 IKNVFLVPGLEKNLLSVPQIISSGYWVRFQDKRC-IIQDANGKEIMNIEMTDKSFKIKLS 439
Query: 695 ELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCETCQK 754
+E + + + E WH+RLGH S +++ Q+ LV GLP K + C+ C
Sbjct: 440 SVEEEAMTANVQTEE---TWHKRLGHVSNKRLQQMQDKELVNGLPRFKVTKET-CKACNL 495
Query: 755 GKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTR 814
GK ++ F ++ T LE++H D+ GP++ +SI G RY ++ +DDY+ WV FL +
Sbjct: 496 GKQSRKSFPKESQTKTREKLEIVHTDVCGPMQHQSIDGSRYYVLFLDDYTHMCWVYFLKQ 555
Query: 815 KDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQ 874
K E+ A F F A V+ + C I +R E + GI + P +PQQ
Sbjct: 556 KSETFATFKKFKALVEKQSNCSIKTLRP----------MEVFCEDEGINRQVTLPYSPQQ 605
Query: 875 NGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNK-TPYELWKN 933
NG ERKNR+L EMAR+ML E + WAEAV T+ Y+QNR+ + I + TP E W
Sbjct: 606 NGAAERKNRSLVEMARSMLVEQDLPLKLWAEAVYTSAYLQNRLPSKAIEDDVTPMEKWCG 665
Query: 934 IKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYNTDAKTIEESI 993
KPN+S+ FG +CYV + K D K+ +L+GYS+++KG+R + + + +E S
Sbjct: 666 HKPNVSHLRIFGSICYVHIPDQKRRKLDAKAKCGILIGYSNQTKGYRVFLLEDEKVEVSR 725
Query: 994 HVRF--DDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEE-----DEPEEEAGPSNSQ 1046
V F D K D D+ + V+K +SIN + + +E + D G ++S
Sbjct: 726 DVVFQEDKKWDWDKQEEVKKTFVMSINDIQESRDQQETSSHDLSQIDDHANNGEGETSSH 785
Query: 1047 TL-----KKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKG-LVSLIEPKSIDE 1100
L ++ R T+ PK+ + + ++ ++E ++ LV+ EP++ DE
Sbjct: 786 VLSQVNDQEERETSESPKKY---KSMKEILEKAPRMENDEAAQGIEACLVANEEPQTYDE 842
Query: 1101 ALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGDVVRNKARLV 1160
A DK+W AM EE+ KN W LV KPE +VI KW+++ K + G+ V++KARLV
Sbjct: 843 ARGDKEWEEAMNEEIKVIEKNRTWKLVDKPEKKNVISVKWIYKIKTDASGNHVKHKARLV 902
Query: 1161 AQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVH 1220
A+G+SQ+ GIDY ETFAPV+R + IR L++++ L+QMDVKSAFLNG + EEVYV
Sbjct: 903 ARGFSQEYGIDYLETFAPVSRYDTIRALLAYAAQMKWRLYQMDVKSAFLNGELEEEVYVT 962
Query: 1221 QPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKD 1280
QPPGF E K + V +L K+LYGLKQAPRAWYER+ S+ ++N F R D L+ K +
Sbjct: 963 QPPGFVIEGKEEKVLRLYKALYGLKQAPRAWYERIDSYFIQNGFARSMNDAALYSKKKGE 1022
Query: 1281 DILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 1340
D+LIV +YVDD+I N L F + M+ EFEM+ +G L YFLG++V+Q G ++ Q
Sbjct: 1023 DVLIVSLYVDDLIITGNNTHLINTFKKNMKDEFEMTDLGLLNYFLGMEVNQDDSGIFLSQ 1082
Query: 1341 SKYTKELLKKFNMLESTVAKTPMHPTCI---LEKEDKSGKVCQKLYRGMIGSLLYLTASR 1397
KY +L+ KF M ES TP+ P +E +DK K YR ++G LLYL ASR
Sbjct: 1083 EKYANKLIDKFGMKESKSVSTPLTPQGKRKGVEGDDKEFADPTK-YRRIVGGLLYLCASR 1141
Query: 1398 PDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAG 1457
PD++++ +R+ S P H KR+LRY+KGT+N G+++ +L GY D+D+ G
Sbjct: 1142 PDVMYASSYLSRYMSSPSIQHYQEAKRVLRYVKGTSNFGVLFTSKETPRLVGYSDSDWGG 1201
Query: 1458 DRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQ 1517
++KST+G LG + W S +Q T+A STAEAEYI+ + Q +W++ ED+
Sbjct: 1202 SLEDKKSTTGYVFTLGLAMFCWQSCKQQTVAQSTAEAEYIAVCAATNQAIWLQRLFEDFG 1261
Query: 1518 I-FESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWA 1576
+ F+ IPI CDN +AI++ +NP+ H R KHIE+KYHF+R+ KG++ L++ + Q A
Sbjct: 1262 LKFKEGIPILCDNKSAIAIGRNPVQHRRTKHIEIKYHFVREAEHKGLIQLEYCKGEDQLA 1321
Query: 1577 DIFTKPLAEDRFNFILKNLNM 1597
D+ TK L+ RF + + L +
Sbjct: 1322 DVLTKALSVSRFEGLRRKLGV 1342
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 662 bits (1708), Expect = 0.0
Identities = 370/1040 (35%), Positives = 562/1040 (53%), Gaps = 50/1040 (4%)
Query: 567 LSMLQISLIAPLKHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGT 626
L M + I + Q W+LDSGCS HM G + F EL V G + + + G G
Sbjct: 242 LLMAHVEQIGDEEKQIWFLDSGCSNHMCGTREWFLELDSGFKQNVRLGDDRRMAVEGKGK 301
Query: 627 ICVDSS---PCIDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSK 683
+ ++ I +V V GL +NL S+ QL KG I C + + ++ +S
Sbjct: 302 LRLEVDGRIQVISDVYFVPGLKNNLFSVGQLQQKGLRFIIEGDVCEVWHKTEKRMVMHST 361
Query: 684 RKNN----IYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLP 739
N ++ E + +CL + + +WH+R GH + + + L++ +V+GLP
Sbjct: 362 MTKNRMFVVFAAVKKSKETEETRCLQVIGKANNMWHKRFGHLNHQGLRSLAEKEMVKGLP 421
Query: 740 NLKFASD-ALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMV 798
+ A+C+ C KGK + ++ +++ L+L+H D+ GP+ S GKRY +
Sbjct: 422 KFDLGEEEAVCDICLKGKQIRESIPKESAWKSTQVLQLVHTDICGPINPASTSGKRYILN 481
Query: 799 IVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFD 858
+DD+SR W L+ K E+ F F A+V+ E ++V +RSD GGE+ + +F+
Sbjct: 482 FIDDFSRKCWTYLLSEKSETFQFFKEFKAEVERESGKKLVCLRSDRGGEYNSREFDEYCK 541
Query: 859 SYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRIS 918
+GI + TPQQNGV ERKNR++ M R ML E + + FW EAV A YI NR
Sbjct: 542 EFGIKRQLTAAYTPQQNGVAERKNRSVMNMTRCMLMEMSVPRKFWPEAVQYAVYILNRSP 601
Query: 919 VRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKG 978
+ + + TP E W + KP++ + FG + Y L + K D KS KC++ G S SK
Sbjct: 602 SKALNDITPEEKWSSWKPSVEHLRIFGSLAYALVPYQKRIKLDEKSIKCVMFGVSKESKA 661
Query: 979 FRFYNTDAKTIEESIHVRFDDKLD---SDQSKLVEKFADLSINVSDKGKAPE------EV 1029
+R Y+ I S V+FD++ D+S E D S + + PE +
Sbjct: 662 YRLYDPATGKILISRDVQFDEERGWEWEDKSLEEELVWDNSDHEPAGEEGPEINHNGQQD 721
Query: 1030 EPEEDEPEEEAGPSNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGL 1089
+ E +E EE + Q L ++ + KD V +E L
Sbjct: 722 QEETEEEEETVAETVHQNLPAVGTGGVRQRQQPVWMKDYVVGNARVLITQDEEDEVLALF 781
Query: 1090 VSLIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEK 1149
+ +P +EA Q + W AM+ E+ +N+ W LV+ PE VIG KW+F+ K NEK
Sbjct: 782 IGPDDPVCFEEAAQLEVWRKAMEAEITSIEENNTWELVELPEEAKVIGLKWIFKTKFNEK 841
Query: 1150 GDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFL 1209
G+V + KARLVA+GY Q+ G+D+ E FAPVA+ + IRL++ + + Q+DVKSAFL
Sbjct: 842 GEVDKFKARLVAKGYHQRYGVDFYEVFAPVAKWDTIRLILGLAAEKGWSVFQLDVKSAFL 901
Query: 1210 NGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKV 1269
+G + E+V+V QP GFE E++ V+KLKK+LYGLKQAPRAWY R+ F + F +
Sbjct: 902 HGDLKEDVFVEQPKGFEVEEESSKVYKLKKALYGLKQAPRAWYSRIEEFFGKEGFEKCYC 961
Query: 1270 DTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQV 1329
+ TLF K + D L+V +YVDD+I+ ++ + + F M EF M+ +G++KYFLG++V
Sbjct: 962 EHTLFVKKERSDFLVVSVYVDDLIYTGSSMEMIEGFKNSMMEEFAMTDLGKMKYFLGVEV 1021
Query: 1330 DQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGS 1389
Q G +I+Q KY E++KK+ M K P+ P +K K+G V
Sbjct: 1022 IQDERGIFINQRKYAAEIIKKYGMEGCNSVKNPIVPG---QKLTKAGAV----------- 1067
Query: 1390 LLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSG 1449
+R+ P E HL AVKRILRY++GT +LG+ Y++ +L G
Sbjct: 1068 ------------------SRYMESPNEQHLLAVKRILRYVQGTLDLGIQYERGGATELVG 1109
Query: 1450 YCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWM 1509
+ D+DYAGD +RKSTSG LG ++WASK+Q + LST EAE++SA+ + Q +W+
Sbjct: 1110 FVDSDYAGDVDDRKSTSGYVFMLGGGAIAWASKKQPIVTLSTTEAEFVSASYGACQAVWL 1169
Query: 1510 KHQLEDYQI-FESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKF 1568
++ LE+ E ++CDN++ I LSKNP+LH R+KHI V+YHF+R+ V++G + L +
Sbjct: 1170 RNVLEEIGCRQEGGTLVFCDNSSTIKLSKNPVLHGRSKHIHVRYHFLRELVKEGTIRLDY 1229
Query: 1569 VDTDHQWADIFTKPLAEDRF 1588
T Q ADI TK + + F
Sbjct: 1230 CTTTDQVADIMTKAVKREVF 1249
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 655 bits (1689), Expect = 0.0
Identities = 386/1044 (36%), Positives = 542/1044 (50%), Gaps = 110/1044 (10%)
Query: 559 ESGYLRTRLSMLQISLIAPLK------HQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVG 612
++ Y+ ++ + L+A K + WYLDSG S HM G K MF EL G V
Sbjct: 305 KANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVA 364
Query: 613 FGGNEKGKIIGTGTICV----DSSPCIDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSC 668
G K ++ G G I + I NV + + N+LS+ QL +KGYD+ +
Sbjct: 365 LGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNL 424
Query: 669 RAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQ 728
+ + KN ++ + + AQ +K + EE W+WH R GH + +
Sbjct: 425 SIRDKESNLITKVPMSKNRMFVLNIRNDIAQCLK--MCYKEESWLWHLRFGHLNFGGLEL 482
Query: 729 LSKLNLVRGLPNLKFASDALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTE 788
LS+ +VRGLP + + +CE C G K+ F ++ +PLEL+H D+ GP+K +
Sbjct: 483 LSRKEMVRGLPCINHPNQ-VCEGCLLGNQFKMSFPKESSSRAQKPLELIHTDVCGPIKPK 541
Query: 789 SIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEF 848
S+G Y ++ +DD+SR TWV FL K E +F F A V+ E I +RSD GGEF
Sbjct: 542 SLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDSGGEF 601
Query: 849 ENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVN 908
+ +F + GI + PR+PQQNGV ERKNRT+ EMAR+ML+ + K WAEAV
Sbjct: 602 TSKEFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVA 661
Query: 909 TACYIQNRISVRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCL 968
A Y+ NR + + KTP E W KP +S+ FG + + ++ +K D KS K +
Sbjct: 662 CAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRNKLDDKSEKYI 721
Query: 969 LLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEE 1028
+GY + SKG++ YN D K S ++ FD++ + D + E + DK + E
Sbjct: 722 FIGYDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDKPEPTRE 781
Query: 1029 VEPEEDEPEEEAGPSNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETL----L 1084
P E+ P++SQ + S + R RS E T L
Sbjct: 782 EPPSEEPTTPPTSPTSSQIEESS--------------SERTPRFRSIQELYEVTENQENL 827
Query: 1085 SLKGLVSLIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRN 1144
+L L + EP EA++ K W AM EE+ KND W L P IG KWV++
Sbjct: 828 TLFCLFAECEPMDFQEAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKA 887
Query: 1145 KLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDV 1204
K N KG+V R KARLVA+GYSQ+ GIDY E FAPVARLE +RL+IS + + +HQMDV
Sbjct: 888 KKNSKGEVERYKARLVAKGYSQRAGIDYDEIFAPVARLETVRLIISLAAQNKWKIHQMDV 947
Query: 1205 KSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEF 1264
KSAFLNG + EEVY+ QP G+ + + D V +LKK LYGLKQAPRAW R+ + E +F
Sbjct: 948 KSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKVLYGLKQAPRAWNTRIDKYFKEKDF 1007
Query: 1265 VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYF 1324
++ + L+ K K+DILI +YVDD+IF N S+ +EF + M EFEM+ +G + Y+
Sbjct: 1008 IKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYY 1067
Query: 1325 LGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYR 1384
LGI+V Q G +I Q Y KE+LKKF M +S +
Sbjct: 1068 LGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPS------------------------- 1102
Query: 1385 GMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSE 1444
++GSL YLT +RPDIL++V + +R+ P TH A KRILRY+KGT N GL Y TS+
Sbjct: 1103 -LVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSD 1161
Query: 1445 YKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICST 1504
YKL C A W ++L
Sbjct: 1162 YKLV-VCHA------------------------IWLRNLLKELSLP-------------- 1182
Query: 1505 QMLWMKHQLEDYQIFESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVL 1564
Q E +IF N +AI+L+KNP+ H R+KHI+ +YH+IR+ V K +
Sbjct: 1183 -------QEEPTKIFVDN-------KSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDV 1228
Query: 1565 LLKFVDTDHQWADIFTKPLAEDRF 1588
L++V T Q ADIFTKPL + F
Sbjct: 1229 QLEYVKTHDQVADIFTKPLKREDF 1252
>At3g25450 hypothetical protein
Length = 1343
Score = 637 bits (1642), Expect = 0.0
Identities = 369/1038 (35%), Positives = 568/1038 (54%), Gaps = 41/1038 (3%)
Query: 580 HQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICVDSS----PCI 635
+ +WYLD+G S HMTG + F +L G+V FG + I G G+I S +
Sbjct: 289 NNAWYLDNGASNHMTGNRAWFCKLDEMITGKVRFGDDSCINIKGKGSIPFISKGGERKIL 348
Query: 636 DNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKR-KNNIYKIRLS 694
+V + L N+LS+ Q + G D+ + + +G++L ++R +N +YK+
Sbjct: 349 FDVYYIPDLKSNILSLGQATESGCDIRMREDYL-TLHDREGNLLIKAQRSRNRLYKV--- 404
Query: 695 ELEAQNVKCL-LSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCETCQ 753
LE +N KCL L+ E +WH RLGH S I + K LV G+ + C +C
Sbjct: 405 SLEVENSKCLQLTTTNESTIWHARLGHISFETIKAMIKKELVIGISSSVPQEKETCGSCL 464
Query: 754 KGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLT 813
GK + F ++ LEL+H DL GP+ + KRY V++DD+SR+ W L
Sbjct: 465 FGKQARHSFPKATSYRAAQVLELIHGDLCGPISPSTAAKKRYVFVLIDDHSRYMWSILLK 524
Query: 814 RKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQ 873
K E+ F F A V+ E I R+D GGEF + +F+ GI + P TPQ
Sbjct: 525 EKSEAFGKFKEFKALVEQECGAIIKTFRTDRGGEFLSHEFQEFCAKEGINRHLTAPYTPQ 584
Query: 874 QNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKN 933
QNGVVER+NRTL M R++L+ M + W EAV + Y+ NR+ R + N+TPYE++K+
Sbjct: 585 QNGVVERRNRTLLGMTRSILKHMNMPNYLWGEAVRHSTYLINRVGTRSLSNQTPYEVFKH 644
Query: 934 IKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYNTDAKTIEESI 993
KPN+ + FGCV Y L K D +S + LG SK +R + + I S
Sbjct: 645 KKPNVEHLRVFGCVSYAKVEVPNLKKLDDRSRMLVYLGTEPGSKAYRLLDPTKRRIFVSR 704
Query: 994 HVRFDDKLD----SDQSKLVEKFADLSINVSD---KGKAPEEVEPEEDEPEEEAGPSNSQ 1046
V FD+ S+ ++ +I +S+ G ++ E +E EE +
Sbjct: 705 DVVFDENRSWMWQESSSETDKESGTFTITLSEFGNNGVTENDISTEPEETEEAEINGEDE 764
Query: 1047 TLKKSRITAAHPKELILGNKDEPVRT--RSAFRPS-------------EETLLSLKGLVS 1091
+ + T H + + +PVR R RP+ E LL++
Sbjct: 765 NIIEEAETEEHDQSQ---EEPQPVRRSQRQVIRPNYLKDYVLCAEIEAEHLLLAVND--- 818
Query: 1092 LIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGD 1151
EP EA + K+W A KEE+ KN WSLV P IG KWVF+ K N G
Sbjct: 819 --EPWDFKEANKSKEWRDACKEEIQSIEKNRTWSLVDLPVGSKAIGVKWVFKLKHNSDGS 876
Query: 1152 VVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNG 1211
+ + KARLVA+GY Q+ G+D+ E FAPVAR+E +RL+I+ + ++ +H +DVK+AFL+G
Sbjct: 877 INKYKARLVAKGYVQRHGVDFEEVFAPVARIETVRLIIALAASNGWEIHHLDVKTAFLHG 936
Query: 1212 YISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDT 1271
+ E+VYV QP GF +++ + V+KL K+LYGL+QAPRAW +L+ L E +F + +
Sbjct: 937 ELREDVYVSQPEGFTNKESKEKVYKLHKALYGLRQAPRAWNTKLNEILKELKFEKCHKEP 996
Query: 1272 TLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQ 1331
+L+ K ++IL+V +YVDD++ +N + F + M +FEMS +G+L Y+LGI+V Q
Sbjct: 997 SLYRKQEGENILVVAVYVDDLLVTGSNLDIILNFKKGMVGKFEMSDLGKLTYYLGIEVLQ 1056
Query: 1332 TPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLL 1391
+ +G + Q +Y K++L++ M + TPM + L K ++ + YR IG L
Sbjct: 1057 SKDGITLKQERYAKKILEEAGMSKCNTVNTPMIASLELSKAQDEKRIDETDYRRNIGCLR 1116
Query: 1392 YLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYC 1451
YL +RPD+ ++V + +R+ +PRE+H A+K+ILRYL+GTT+ GL +KK L GY
Sbjct: 1117 YLLHTRPDLSYNVGILSRYLQEPRESHGAALKQILRYLQGTTSHGLYFKKGENAGLIGYS 1176
Query: 1452 DADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH 1511
D+ + D + KST G+ +L ++W S++Q + LS+ EAE+++A + Q +W++
Sbjct: 1177 DSSHNVDLDDGKSTGGHIFYLNDCPITWCSQKQQVVTLSSCEAEFMAATEAAKQAIWLQE 1236
Query: 1512 QLEDYQIFE-SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVD 1570
L + E + I DN +AI+L+KNP+ H R+KHI +YHFIR+ V+ G + ++ V
Sbjct: 1237 LLAEVIGTECEKVTIRVDNKSAIALTKNPVFHGRSKHIHRRYHFIRECVENGQIEVEHVP 1296
Query: 1571 TDHQWADIFTKPLAEDRF 1588
Q ADI TK L + +F
Sbjct: 1297 GVRQKADILTKALGKIKF 1314
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 634 bits (1635), Expect = 0.0
Identities = 365/1037 (35%), Positives = 567/1037 (54%), Gaps = 37/1037 (3%)
Query: 578 LKHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICVDSSPCIDN 637
L +W +DSG + H++ ++++F+ L V +I G GT+ ++ + N
Sbjct: 437 LSSDTWVIDSGATHHVSHDRKLFQTLDTSIVSFVNLPTGPNVRISGVGTVLINKDIILQN 496
Query: 638 VLLVDGLTHNLLSISQLA-DKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSEL 696
VL + NL+SIS L D G VIF+ C+ G L KR N+Y + ++
Sbjct: 497 VLFIPEFRLNLISISSLTTDLGTRVIFDPSCCQIQDLTKGLTLGEGKRIGNLYVLD-TQS 555
Query: 697 EAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCETCQKGK 756
A +V ++ V+ VWH+RLGH S ++ LS+ V G K A C C K
Sbjct: 556 PAISVNAVVDVS----VWHKRLGHPSFSRLDSLSE---VLGTTRHKNKKSAYCHVCHLAK 608
Query: 757 FTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKD 816
K+ F + N + S ELLHID++GP E++ G +Y + IVDD+SR TW+ L K
Sbjct: 609 QKKLSFPSANNICNST-FELLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWIYLLKSKS 667
Query: 817 ESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNG 876
+ VF FI V+N+ R+ VRSD+ E F + + GI SCP TP+QN
Sbjct: 668 DVLTVFPAFIDLVENQYDTRVKSVRSDNAKELA---FTEFYKAKGIVSFHSCPETPEQNS 724
Query: 877 VVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKP 936
VVERK++ + +AR ++ ++ M+ +W + V TA ++ NR + NKTP+E+ P
Sbjct: 725 VVERKHQHILNVARALMFQSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGKLP 784
Query: 937 NISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVR 996
+ S FGC+CY + + HKF P+S C+ LGY KG++ + ++ + S +V
Sbjct: 785 DYSQLKTFGCLCYSSTSSKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVE 844
Query: 997 FDDKL----DSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSR 1052
F ++L S QS ++ G + P + PS + K R
Sbjct: 845 FHEELFPLASSQQSATTASDVFTPMDPLSSGNSITS-----HLPSPQISPSTQ--ISKRR 897
Query: 1053 ITA--AHPKEL--ILGNKDE--PVRTRSAFRP-SEETLLSLKGLVSLIEPKSIDEALQDK 1105
IT AH ++ NKD+ P+ + ++ S +L + + + P+S EA K
Sbjct: 898 ITKFPAHLQDYHCYFVNKDDSHPISSSLSYSQISPSHMLYINNISKIPIPQSYHEAKDSK 957
Query: 1106 DWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYS 1165
+W A+ +E+ + D W + P +G KWVF K + G + R KAR+VA+GY+
Sbjct: 958 EWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWVFTVKFHADGSLERFKARIVAKGYT 1017
Query: 1166 QQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGF 1225
Q+EG+DYTETF+PVA++ ++LL+ S + L+Q+D+ +AFLNG + E +Y+ P G+
Sbjct: 1018 QKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQLDISNAFLNGDLEETIYMKLPDGY 1077
Query: 1226 EDEK----KPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDD 1281
D K P+ V +LKKS+YGLKQA R W+ + S+ LL F + D TLF + +
Sbjct: 1078 ADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSNSLLALGFEKQHGDHTLFVRCIGSE 1137
Query: 1282 ILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 1341
+++ +YVDDI+ S + + +E ++A F++ +G LKYFLG++V +T EG + Q
Sbjct: 1138 FIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRELGPLKYFLGLEVARTSEGISLSQR 1197
Query: 1342 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDIL 1401
KY ELL +ML+ + PM P L K D +++YR ++G L+YLT +RPDI
Sbjct: 1198 KYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLLEDKEMYRRLVGKLMYLTITRPDIT 1257
Query: 1402 FSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTE 1461
F+V+ +F S PR HL AV ++L+Y+KGT GL Y + L GY DAD+
Sbjct: 1258 FAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGLFYSAEDDLTLKGYTDADWGTCPDS 1317
Query: 1462 RKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQIFES 1521
R+ST+G F+GS+L+SW SK+Q T++ S+AEAEY + A+ S +M W+ L ++ S
Sbjct: 1318 RRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYRALALASCEMAWLSTLLLALRV-HS 1376
Query: 1522 NIPI-YCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFT 1580
+PI Y D+TAA+ ++ NP+ H R KHIE+ H +R+ + G L L V T Q ADI T
Sbjct: 1377 GVPILYSDSTAAVYIATNPVFHERTKHIEIDCHTVREKLDNGQLKLLHVKTKDQVADILT 1436
Query: 1581 KPLAEDRFNFILKNLNM 1597
KPL +F +L +++
Sbjct: 1437 KPLFPYQFAHLLSKMSI 1453
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 628 bits (1619), Expect = e-180
Identities = 359/1037 (34%), Positives = 565/1037 (53%), Gaps = 33/1037 (3%)
Query: 578 LKHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICVDSSPCIDN 637
L +W +DSG + H++ ++ +F L V KI G GT+ ++ + N
Sbjct: 426 LSSATWVIDSGATHHVSHDRSLFSSLDTSVLSAVNLPTGPTVKISGVGTLKLNDDILLKN 485
Query: 638 VLLVDGLTHNLLSISQLADK-GYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSEL 696
VL + NL+SIS L D G VIF++ SC I G +L +R N+Y + + +
Sbjct: 486 VLFIPEFRLNLISISSLTDDIGSRVIFDKNSCEIQDLIKGRMLGQGRRVANLYLLDVGD- 544
Query: 697 EAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCETCQKGK 756
++ +V ++ ++ +WHRRLGHAS++++ +S G K C C K
Sbjct: 545 QSISVNAVVDIS----MWHRRLGHASLQRLDAISDS---LGTTRHKNKGSDFCHVCHLAK 597
Query: 757 FTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKD 816
K+ F N V +LLHID++GP E++ G +Y + IVDD+SR TW+ L K
Sbjct: 598 QRKLSFPTSNKVC-KEIFDLLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWMYLLKTKS 656
Query: 817 ESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNG 876
E VF FI QV+N+ ++ VRSD+ E KF S + GI SCP TP+QN
Sbjct: 657 EVLTVFPAFIQQVENQYKVKVKAVRSDNAPEL---KFTSFYAEKGIVSFHSCPETPEQNS 713
Query: 877 VVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKP 936
VVERK++ + +AR ++ ++ + W + V TA ++ NR + ++NKTPYE+ P
Sbjct: 714 VVERKHQHILNVARALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEILTGTAP 773
Query: 937 NISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVR 996
FGC+CY + + HKF P+S CL LGY KG++ + ++ T+ S +V+
Sbjct: 774 VYEQLRTFGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQ 833
Query: 997 FDDKL--------DSDQSKLVEKFADLSINV-SDKGKAPEEVEPE-EDEPEEEAGPSNSQ 1046
F +++ KL +S + SD +P + + D P + +SQ
Sbjct: 834 FHEEVFPLAKNPGSESSLKLFTPMVPVSSGIISDTTHSPSSLPSQISDLPPQ----ISSQ 889
Query: 1047 TLKK--SRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQD 1104
++K + + H + +K T S + S + + + + P + EA
Sbjct: 890 RVRKPPAHLNDYHCNTMQSDHKYPISSTISYSKISPSHMCYINNITKIPIPTNYAEAQDT 949
Query: 1105 KDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGY 1164
K+W A+ E+ K + W + P+ +G KWVF K G++ R KARLVA+GY
Sbjct: 950 KEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGY 1009
Query: 1165 SQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPG 1224
+Q+EG+DYT+TF+PVA++ I+LL+ S + L Q+DV +AFLNG + EE+++ P G
Sbjct: 1010 TQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEG 1069
Query: 1225 FEDEK----KPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKD 1280
+ + K + V +LK+S+YGLKQA R W+++ SS LL F + D TLF K Y
Sbjct: 1070 YAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDG 1129
Query: 1281 DILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 1340
+ +IV +YVDDI+ S +++ + +E + F++ +G+LKYFLG++V +T G I Q
Sbjct: 1130 EFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVARTTAGISICQ 1189
Query: 1341 SKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDI 1400
KY ELL+ ML PM P + K+D + YR ++G L+YLT +RPDI
Sbjct: 1190 RKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDI 1249
Query: 1401 LFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRT 1460
F+V+ +F S PR THLTA R+L+Y+KGT GL Y +S+ L G+ D+D+A +
Sbjct: 1250 TFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQD 1309
Query: 1461 ERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQIFE 1520
R+ST+ F+G +L+SW SK+Q T++ S+AEAEY + A+ + +M+W+ L Q
Sbjct: 1310 SRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQASP 1369
Query: 1521 SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFT 1580
+Y D+TAAI ++ NP+ H R KHI++ H +R+ + G L L V T+ Q ADI T
Sbjct: 1370 PVPILYSDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGELKLLHVRTEDQVADILT 1429
Query: 1581 KPLAEDRFNFILKNLNM 1597
KPL +F + +++
Sbjct: 1430 KPLFPYQFEHLKSKMSI 1446
>At1g70010 hypothetical protein
Length = 1315
Score = 592 bits (1527), Expect = e-169
Identities = 347/1002 (34%), Positives = 534/1002 (52%), Gaps = 35/1002 (3%)
Query: 624 TGTICVDSSPCIDNVLLVDGLTHNLLSISQLADK-GYDVIFNQKSCRAVSQIDGSVLFNS 682
+G++ + +++VL + NLLS+S L G + F++ SC ++
Sbjct: 314 SGSVHLGRHLILNDVLFIPQFKFNLLSVSSLTKSMGCRIWFDETSCVLQDATRELMVGMG 373
Query: 683 KRKNNIYKIRLSELEAQNVKCLLSVNE--EQWVWHRRLGHASMRKISQLSKLNLVRGLPN 740
K+ N+Y + L L ++V +WH+RLGH S++K+ +S L P
Sbjct: 374 KQVANLYIVDLDSLSHPGTDSSITVASVTSHDLWHKRLGHPSVQKLQPMSSL---LSFPK 430
Query: 741 LKFASDALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIV 800
K +D C C K +PF + N S SRP +L+HID +GP ++ G RY + IV
Sbjct: 431 QKNNTDFHCRVCHISKQKHLPFVSHNNKS-SRPFDLIHIDTWGPFSVQTHDGYRYFLTIV 489
Query: 801 DDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSY 860
DDYSR TWV L K + V TF+ V+N+ I VRSD+ E F + S
Sbjct: 490 DDYSRATWVYLLRNKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPELN---FTQFYHSK 546
Query: 861 GIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVR 920
GI SCP TPQQN VVERK++ + +AR++ ++ + +W + + TA Y+ NR+
Sbjct: 547 GIVPYHSCPETPQQNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINRLPAP 606
Query: 921 PILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFR 980
+ +K P+E+ P + FGC+CY + HKF P++ C +GY KG++
Sbjct: 607 ILEDKCPFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYK 666
Query: 981 FYNTDAKTIEESIHVRFDDKL----DSDQSKLVEKF-ADLS------------INVSDKG 1023
+ + +I S HV F ++L SD S+ + F DL+ +N SD
Sbjct: 667 LLDLETHSIIVSRHVVFHEELFPFLGSDLSQEEQNFFPDLNPTPPMQRQSSDHVNPSDSS 726
Query: 1024 KAPE---EVEPEEDEPEEEAGPSNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRPSE 1080
+ E P + PE S+ + K + + + ++ E + S R ++
Sbjct: 727 SSVEILPSANPTNNVPEPSVQTSHRKAKKPAYLQDYYCHSVVSSTPHEIRKFLSYDRIND 786
Query: 1081 ETLLSLKGLVSLIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKW 1140
L L L EP + EA + + W AM E + W + P + IG +W
Sbjct: 787 PYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRW 846
Query: 1141 VFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILH 1200
+F+ K N G V R KARLVAQGY+Q+EGIDY ETF+PVA+L +++LL+ + + L
Sbjct: 847 IFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLT 906
Query: 1201 QMDVKSAFLNGYISEEVYVHQPPGFE----DEKKPDHVFKLKKSLYGLKQAPRAWYERLS 1256
Q+D+ +AFLNG + EE+Y+ P G+ D P+ V +LKKSLYGLKQA R WY + S
Sbjct: 907 QLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFS 966
Query: 1257 SFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMS 1316
S LL F++ D T F K L V +Y+DDII S N + M++ F++
Sbjct: 967 STLLGLGFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLR 1026
Query: 1317 MMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSG 1376
+GELKYFLG+++ ++ +G +I Q KY +LL + L + PM P+ + +
Sbjct: 1027 DLGELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGD 1086
Query: 1377 KVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLG 1436
V YR +IG L+YL +RPDI F+V+ A+F PR+ HL AV +IL+Y+KGT G
Sbjct: 1087 FVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQG 1146
Query: 1437 LMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEY 1496
L Y TSE +L Y +ADY R R+STSG C FLG +L+ W S++Q ++ S+AEAEY
Sbjct: 1147 LFYSATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEY 1206
Query: 1497 ISAAICSTQMLWMKHQLEDYQI-FESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFI 1555
S ++ + +++W+ + L++ Q+ ++CDN AAI ++ N + H R KHIE H +
Sbjct: 1207 RSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSV 1266
Query: 1556 RDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1597
R+ + KG+ L ++T+ Q AD FTKPL F+ ++ + +
Sbjct: 1267 RERLLKGLFELYHINTELQIADPFTKPLYPSHFHRLISKMGL 1308
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 588 bits (1515), Expect = e-167
Identities = 355/1058 (33%), Positives = 561/1058 (52%), Gaps = 49/1058 (4%)
Query: 581 QSWYLDSGCSRHMTGEKRMFRELKLKPGGE-VGFGGNEKGKIIGTGTICVDSSPC---ID 636
+ W+ DS + H+T + G + V G I TG+ + SS ++
Sbjct: 320 KEWHPDSAATAHVTSSTNGLQSATEYEGDDAVLVGDGTYLPITHTGSTTIKSSNGKIPLN 379
Query: 637 NVLLVDGLTHNLLSISQLADK-GYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSE 695
VL+V + +LLS+S+L D V F+ + V+ R+N +Y + E
Sbjct: 380 EVLVVPNIQKSLLSVSKLCDDYPCGVYFDANKVCIIDLQTQKVVTTGPRRNGLYVLENQE 439
Query: 696 LEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCETCQKG 755
A + EE VWH RLGHA+ + + L ++ K + +CE CQ G
Sbjct: 440 FVALYSNRQCAATEE--VWHHRLGHANSKALQHLQNSKAIQ---INKSRTSPVCEPCQMG 494
Query: 756 KFTKVPFKAKNVVSTSR---PLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFL 812
K +++PF ++S SR PL+ +H DL+GP S G +Y + VDDYSR++W L
Sbjct: 495 KSSRLPF----LISDSRVLHPLDRIHCDLWGPSPVVSNQGLKYYAIFVDDYSRYSWFYPL 550
Query: 813 TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTP 872
K E +VF +F V+N+ +I +SD GGEF ++K ++ +GI H SCP TP
Sbjct: 551 HNKSEFLSVFISFQKLVENQLNTKIKVFQSDGGGEFVSNKLKTHLSEHGIHHRISCPYTP 610
Query: 873 QQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWK 932
QQNG+ ERK+R L E+ +ML + + FW E+ TA YI NR+ + N +PYE
Sbjct: 611 QQNGLAERKHRHLVELGLSMLFHSHTPQKFWVESFFTANYIINRLPSSVLKNLSPYEALF 670
Query: 933 NIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFR-FYNTDAK---- 987
KP+ S FG CY +KFDP+S +C+ LGY+ + KG+R FY K
Sbjct: 671 GEKPDYSSLRVFGSACYPCLRPLAQNKFDPRSLQCVFLGYNSQYKGYRCFYPPTGKVYIS 730
Query: 988 --TIEESIHVRFDDKLDS----DQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAG 1041
I + F +K S + L++ + I+ AP ++ + + AG
Sbjct: 731 RNVIFNESELPFKEKYQSLVPQYSTPLLQAWQHNKISEISVPAAPVQLFSKPIDLNTYAG 790
Query: 1042 PSNSQTLKKSRITA----------------AHPKELILGNKDEPVRTRSAFRPSEETLLS 1085
++ L T+ A +E ++ + R+++ +
Sbjct: 791 SQVTEQLTDPEPTSNNEGSDEEVNPVAEEIAANQEQVINSHAMTTRSKAGIQKPNTRYAL 850
Query: 1086 LKGLVSLIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNK 1145
+ ++ EPK++ A++ W A+ EE+N+ WSLV ++++++ +KWVF+ K
Sbjct: 851 ITSRMNTAEPKTLASAMKHPGWNEAVHEEINRVHMLHTWSLVPPTDDMNILSSKWVFKTK 910
Query: 1146 LNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVK 1205
L+ G + + KARLVA+G+ Q+EG+DY ETF+PV R IRL++ S + + Q+DV
Sbjct: 911 LHPDGSIDKLKARLVAKGFDQEEGVDYLETFSPVVRTATIRLVLDVSTSKGWPIKQLDVS 970
Query: 1206 SAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFV 1265
+AFL+G + E V+++QP GF D +KP HV +L K++YGLKQAPRAW++ S+FLL+ FV
Sbjct: 971 NAFLHGELQEPVFMYQPSGFIDPQKPTHVCRLTKAIYGLKQAPRAWFDTFSNFLLDYGFV 1030
Query: 1266 RGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFL 1325
K D +LF IL + +YVDDI+ ++QSL ++ + ++ F M +G +YFL
Sbjct: 1031 CSKSDPSLFVCHQDGKILYLLLYVDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFL 1090
Query: 1326 GIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRG 1385
GIQ++ G ++HQ+ Y ++L++ M + TP+ L+ + +R
Sbjct: 1091 GIQIEDYANGLFLHQTAYATDILQQAGMSDCNPMPTPLPQQ--LDNLNSELFAEPTYFRS 1148
Query: 1386 MIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEY 1445
+ G L YLT +RPDI F+V+ + P + +KRILRY+KGT +GL K+ S
Sbjct: 1149 LAGKLQYLTITRPDIQFAVNFICQRMHSPTTSDFGLLKRILRYIKGTIGMGLPIKRNSTL 1208
Query: 1446 KLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQ 1505
LS Y D+D+AG + R+ST+G C LGSNL+SW++KRQ T++ S+ EAEY + + +
Sbjct: 1209 TLSAYSDSDHAGCKNTRRSTTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTYAARE 1268
Query: 1506 MLWMKHQLEDYQIFESNIP--IYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGV 1563
+ W+ L D I +P +YCDN +A+ LS NP LH+R+KH + YH+IR+ V G+
Sbjct: 1269 ITWISFLLRDLGI-PQYLPTQVYCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVALGL 1327
Query: 1564 LLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNMDFCP 1601
+ + + Q AD+FTK L F + L + P
Sbjct: 1328 IETQHISATFQLADVFTKSLPRRAFVDLRSKLGVSGSP 1365
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 586 bits (1511), Expect = e-167
Identities = 358/1032 (34%), Positives = 549/1032 (52%), Gaps = 60/1032 (5%)
Query: 582 SWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICVDSS----PCIDN 637
SWYLD+G S HMTG + F +L G+V FG + + I G G+I + + + +
Sbjct: 279 SWYLDNGASNHMTGNLQWFSKLNEMITGKVRFGDDSRIDIKGKGSIVLITKGGIRKTLTD 338
Query: 638 VLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKR-KNNIYKIRLSEL 696
V + L N++S+ Q + G DV + +G +L + R +N +YK+ +L
Sbjct: 339 VYFIPDLKSNIISLGQATEAGCDVRMKDDQL-TLHDREGCLLLRATRSRNRLYKV---DL 394
Query: 697 EAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCETCQKGK 756
+NVKCL + + + + LV G+ N+ + C +C GK
Sbjct: 395 NVENVKCL------------------QLEAATMVRKELVIGISNIPKEKET-CGSCLLGK 435
Query: 757 FTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKD 816
+ PF S+ LEL+H DL GP+ + KRY +V++DD++R+ W L K
Sbjct: 436 QARQPFPKATTYRASQVLELVHGDLCGPITQSTTAKKRYILVLIDDHTRYMWSMLLKEKS 495
Query: 817 ESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNG 876
E+ F F +V+ E +I R+D GGEF + +F+ GI + P TPQQNG
Sbjct: 496 EAFEKFRDFKTKVEQESGVKIKTFRTDKGGEFVSQEFQDFCAKEGINRHLTAPYTPQQNG 555
Query: 877 VVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKP 936
VVER+NRTL M R++L+ M + W EAV + YI NR+ R + N+TPYE++K KP
Sbjct: 556 VVERRNRTLLGMTRSILKHMKMPNYLWGEAVRHSTYIINRVGTRSLQNQTPYEVFKQRKP 615
Query: 937 NISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYN-TDAKTIEESIHV 995
N+ + FGC+ Y L K D +S + LG SK +R + T+ K I+ +
Sbjct: 616 NVEHLRVFGCIGYAKIEGPHLRKLDDRSKMLVYLGTEPGSKAYRLLDPTNRKIIKWNNSD 675
Query: 996 RFDDKLDSDQSKLVEKFADLSINVSD------KGKAPEEVEPEEDEPE---------EEA 1040
+ S + +F + I SD G+ E EE E E EE
Sbjct: 676 SETRDISGTFSLTLGEFGNNGIQESDDIETEKNGEESENSHEEEGENEHNEQEQIDAEET 735
Query: 1041 GPSNSQ---TLKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKS 1097
PS++ TL++S P L D+ V E+ LL++ EP
Sbjct: 736 QPSHATPLPTLRRSTRQVGKPNYL-----DDYVLMAEI--EGEQVLLAIND-----EPWD 783
Query: 1098 IDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGDVVRNKA 1157
EA + K+W A KEE+ KN WSL+ P VIG KWVF+ K N G + + KA
Sbjct: 784 FKEANKLKEWRDACKEEILSIEKNKTWSLIDLPVRRKVIGLKWVFKIKRNSDGSINKYKA 843
Query: 1158 RLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEV 1217
RLVA+GY Q+ GIDY E FA VAR+E IR++I+ + ++ +H +DVK+AFL+G + E+V
Sbjct: 844 RLVAKGYVQRHGIDYDEVFAHVARIETIRVIIALAASNGWEVHHLDVKTAFLHGELREDV 903
Query: 1218 YVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKT 1277
YV QP GF ++ V+KL K+LYGLKQAPRAW +L+ L E FV+ + +++ +
Sbjct: 904 YVTQPEGFTNKDNEGKVYKLHKALYGLKQAPRAWNTKLNKILQELNFVKCSKEPSVYRRQ 963
Query: 1278 YKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTY 1337
+ +LIV IYVDD++ ++ L F + M +FEMS +G+L Y+LGI+V G
Sbjct: 964 EEKKLLIVAIYVDDLLVTGSSLDLILCFKKDMAGKFEMSDLGQLTYYLGIEVLHRKNGII 1023
Query: 1338 IHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASR 1397
+ Q +Y +++++ M PM L K + + ++ YR MIG L Y+ +R
Sbjct: 1024 LRQERYAMKIIEEAGMSNCNPVLIPMAAGLELCKAQEEKCITERDYRRMIGCLRYIVHTR 1083
Query: 1398 PDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAG 1457
PD+ + V + +R+ PRE+H A+K++LRYLKGT + GL K+ + L GY D+ ++
Sbjct: 1084 PDLSYCVGVLSRYLQQPRESHGNALKQVLRYLKGTMSHGLYLKRGFKSGLVGYSDSSHSA 1143
Query: 1458 DRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKHQL-EDY 1516
D + KST+G+ +L ++W S++Q +ALS+ EAE+++A + Q +W++ E
Sbjct: 1144 DLDDGKSTAGHIFYLHQCPITWCSQKQQVVALSSCEAEFMAATEAAKQAIWLQDLFAEVC 1203
Query: 1517 QIFESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWA 1576
+ I DN +AI+L+KN + H R+KHI +YHFIR+ V+ ++ + V Q A
Sbjct: 1204 GTTSEKVMIRVDNKSAIALTKNLVFHGRSKHIHRRYHFIRECVENNLVEVDHVPGVEQRA 1263
Query: 1577 DIFTKPLAEDRF 1588
DI TKPL +F
Sbjct: 1264 DILTKPLGRIKF 1275
>At1g37110
Length = 1356
Score = 557 bits (1436), Expect = e-158
Identities = 356/1093 (32%), Positives = 566/1093 (51%), Gaps = 77/1093 (7%)
Query: 559 ESGYLRTRLSMLQ-ISLIAPLKHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNE 617
E+G + +L + +S+ + W LDSGC+ HMT + F + K + G +
Sbjct: 283 EAGVITEKLVFSEALSVNEQMVKDLWILDSGCTSHMTSRRDWFISFQEKGNTTILLGDDH 342
Query: 618 KGKIIGTGTICVDSS----PCIDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQ 673
+ G GTI +D+ ++NV V L NL+S L GY + R
Sbjct: 343 SVESQGQGTIRIDTHGGTIKILENVKYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFK- 401
Query: 674 IDGSVLFNSKRKNNIYKIR----LSEL---EAQNVKCLLSVNEEQWVWHRRLGHASMRKI 726
+ N +Y + +SEL E VK L WH RLGH SM +
Sbjct: 402 -NNKTALRGSLSNGLYVLDGSTVMSELCNAETDKVKTAL--------WHSRLGHMSMNNL 452
Query: 727 SQLSKLNLV--RGLPNLKFASDALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFG- 783
L+ L+ + + L+F CE C GK KV F S L +H DL+G
Sbjct: 453 KVLAGKGLIDRKEINELEF-----CEHCVMGKSKKVSFNVGKHTSEDA-LSYVHADLWGS 506
Query: 784 PVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSD 843
P T SI GK+Y + I+DD +R W+ FL KDE+ F + + V+N+ ++ +R+D
Sbjct: 507 PNVTPSISGKQYFLSIIDDKTRKVWLYFLKSKDETFDKFCEWKSLVENQVNKKVKCLRTD 566
Query: 844 HGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFW 903
+G EF N +F+S +GI +C TPQQNGV ER NRT+ E R +L ++G+ + FW
Sbjct: 567 NGLEFCNSRFDSYCKEHGIERHRTCTYTPQQNGVAERMNRTIMEKVRCLLNKSGVEEVFW 626
Query: 904 AEAVNTACYIQNRISVRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPK 963
AEA TA Y+ NR I + P E+W N KP + FG + YV + +L P+
Sbjct: 627 AEAAATAAYLINRSPASAINHNVPEEMWLNRKPGYKHLRKFGSIAYVHQDQGKLK---PR 683
Query: 964 SSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKL----------DSDQSKLVE--- 1010
+ K LGY +KG++ + + + S +V F + + D+D E
Sbjct: 684 ALKGFFLGYPAGTKGYKVWLLEEEKCVISRNVVFQESVVYRDLKVKEDDTDNLNQKETTS 743
Query: 1011 ------KFADLSIN------VSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSRITAAH- 1057
KFA+ S + SD E + + E E E +T K++ +T
Sbjct: 744 SEVEQNKFAEASGSGGVIQLQSDSEPITEGEQSSDSEEEVEYSEKTQETPKRTGLTTYKL 803
Query: 1058 PKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLI--EPKSIDEALQDKD---WILAMK 1112
++ + N + P R F +L + + I EP+S EA++ +D W +A
Sbjct: 804 ARDRVRRNINPPTR----FTEESSVTFALVVVENCIVQEPQSYQEAMESQDCEKWDMATH 859
Query: 1113 EELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKG-DVVRNKARLVAQGYSQQEGID 1171
+E++ KN W LV KP++ +IG +W+F+ K G + R KARLVA+GY+Q+EG+D
Sbjct: 860 DEMDSLMKNGTWDLVDKPKDRKIIGCRWLFKLKSGIPGVEPTRFKARLVAKGYTQREGVD 919
Query: 1172 YTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFEDEKKP 1231
Y E FAPV + +IR+L+S V+ ++ L QMDVK+ FL+G + EE+Y+ QP GF +
Sbjct: 920 YQEIFAPVVKHVSIRILMSLVVDKDLELEQMDVKTTFLHGDLEEELYMEQPEGFVSDSSE 979
Query: 1232 DHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKD-DILIVQIYVD 1290
+ V +LKKSLYGLKQ+PR W +R F+ +F+R + D ++ K + D + + +YVD
Sbjct: 980 NKVCRLKKSLYGLKQSPRQWNKRFDRFMSSQQFIRSEHDACVYVKHVSEHDFIYLLLYVD 1039
Query: 1291 DIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSK--YTKELL 1348
D++ A+++ E + EFEM MG LGI + + +G + S+ Y +++L
Sbjct: 1040 DMLIAGASKAEINRVKEQLSTEFEMKDMGGASRILGIDIYRDRKGGVLKLSQEIYIRKVL 1099
Query: 1349 KKFNMLESTVAKTPM---HPTCILEKEDKSGKVCQKLYRGMIGSLLY-LTASRPDILFSV 1404
+FNM + + P+ + +ED+ Y +GS++Y + +RPD+ +++
Sbjct: 1100 DRFNMSGAKMTNAPVGAHFKLAAVREEDECVDTDVVPYSSAVGSIMYAMLGTRPDLAYAI 1159
Query: 1405 HLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKS 1464
L +R+ S P H AVK ++RYLKG +L L++ K ++ ++GYCD++YA D R+S
Sbjct: 1160 CLISRYMSKPGSMHWEAVKWVMRYLKGAQDLNLVFTKEKDFTVTGYCDSNYAADLDRRRS 1219
Query: 1465 TSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQIFESNIP 1524
SG +G N VSW + Q +A+ST EAEYI+ A + + +W+K L+D + + +
Sbjct: 1220 ISGYVFTIGGNTVSWKASLQPVVAMSTTEAEYIALAEAAKEAMWIKGLLQDMGMQQDKVK 1279
Query: 1525 IYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLA 1584
I+CD+ +AI LSKN + H R KHI+V++++IRD V+ G + + + T D TK +
Sbjct: 1280 IWCDSQSAICLSKNSVYHERTKHIDVRFNYIRDVVESGDVDVLKIHTSRNPVDALTKCIP 1339
Query: 1585 EDRFNFILKNLNM 1597
++F L L +
Sbjct: 1340 VNKFKSALGVLKL 1352
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 553 bits (1424), Expect = e-157
Identities = 356/1079 (32%), Positives = 567/1079 (51%), Gaps = 62/1079 (5%)
Query: 562 YLRTRLSMLQISLIAPLKHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKI 621
Y+ LS I L W +D+GCS HMT ++ F +L GG V G K+
Sbjct: 271 YVSEALSSTDIHL-----EDEWVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKV 325
Query: 622 IGTGTICVDSSPC----IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGS 677
G GTI V + + NV + + NLLS+ GY + ++ S
Sbjct: 326 RGIGTIRVKNEAGMVVRLTNVRYIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIA--GDS 383
Query: 678 VLFNSKRKNNIYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRG 737
VL +R +Y ++ + +++ ++ ++ +WHRRLGH S + + L K L
Sbjct: 384 VLLTVRRCYTLYLLQWRPVTEESLS-VVKRQDDTILWHRRLGHMSQKNMDLLLKKGL--- 439
Query: 738 LPNLKFASDALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFG-PVKTESIGGKRYG 796
L K + CE C GK ++ F T LE +H DL+G P S+G +Y
Sbjct: 440 LDKKKVSKLETCEDCIYGKAKRIGFNLAQH-DTREKLEYVHSDLWGAPSVPFSLGKCQYF 498
Query: 797 MVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESL 856
+ +DDY+R + FL KDE+ F + V+N+ RI +R+D+G EF N F+
Sbjct: 499 ISFIDDYTRKVRIYFLKTKDEAFDKFVEWANLVENQTDKRIKTLRTDNGLEFCNRSFDEF 558
Query: 857 FDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNR 916
GI +C TPQQNGV ER NRTL E R+ML ++G+ K FWAEA +T + N+
Sbjct: 559 CSQKGILWHRTCAYTPQQNGVAERMNRTLMEKVRSMLSDSGLPKKFWAEATHTTAILINK 618
Query: 917 ISVRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRS 976
+ + P + W P SY FGC+ +V +T D K +P++ K +L+GY
Sbjct: 619 TPSSALNYEVPDKRWSGKSPIYSYLRRFGCIAFV-HTDDG--KLNPRAKKGILVGYPIGV 675
Query: 977 KGFRFYNTDAKTIEESIHVRFDDKL---DSDQSKLVEK---------FADLSIN----VS 1020
KG++ + + K S +V F + D QSK EK + DL ++ ++
Sbjct: 676 KGYKIWLLEEKKCVVSRNVIFQENASYKDMMQSKDAEKDENEAPPSSYLDLDLDHEEVIT 735
Query: 1021 DKGKAP--EEVEPEEDEPEEEAGPSNSQTLKKSRITAAHPKELILGNKDEPVRT-RSAFR 1077
G P E P P S + I + +L+ +D RT R+ R
Sbjct: 736 SGGDDPIVEAQSPFNPSPATTQTYSEGVNSETDIIQSPLSYQLV---RDRDRRTIRAPVR 792
Query: 1078 PSEETLLSLKGLVSL-----IEPKSIDEALQDKDWI---LAMKEELNQFSKNDVWSLVKK 1129
+E L+ + L + IEP EA + +W LAM EE+ KN W++VK+
Sbjct: 793 FDDEDYLA-EALYTTEDSGEIEPADYSEAKRSMNWNKWKLAMNEEMESQIKNHTWTVVKR 851
Query: 1130 PENVHVIGTKWVFRNKLNEKG-DVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLL 1188
P++ VIG++W+++ KL G + R KARLVA+GY+Q++GIDY E FAPV + +IR+L
Sbjct: 852 PQHQKVIGSRWIYKFKLGIPGVEEGRFKARLVAKGYAQRKGIDYHEIFAPVVKHVSIRIL 911
Query: 1189 ISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAP 1248
+S ++ L Q+DVK+AFL+G + E++Y+ P G+E+ K D V L KSLYGLKQAP
Sbjct: 912 MSIVAQEDLELEQLDVKTAFLHGELKEKIYMVPPEGYEEMFKEDEVCLLNKSLYGLKQAP 971
Query: 1249 RAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILI-VQIYVDDIIFGSANQSLCKEFSE 1307
+ W E+ ++++ E F+R D+ + K D + + +YVDD++ + N+ + E
Sbjct: 972 KQWNEKFNAYMSEIGFIRSLYDSCAYIKELSDGSRVYLLLYVDDMLVAAKNKEDISQLKE 1031
Query: 1308 MMQAEFEMSMMGELKYFLGIQVDQTPEGT--YIHQSKYTKELLKKFNMLESTVAKTPMHP 1365
+ F+M +G K LG+++ + E ++ Q+ Y ++L+ +NM ES TP+
Sbjct: 1032 ELSQRFDMKDLGAAKRILGMEIIRNREENTLWLSQNGYLNKILETYNMAESKHVVTPLGA 1091
Query: 1366 -----TCILEKEDKSGKVCQKL-YRGMIGSLLY-LTASRPDILFSVHLCARFQSDPRETH 1418
+EK+++ + + Y +GS++Y + +RPD+ + V + +R+ S P H
Sbjct: 1092 HLKMRAATVEKQEQDEDYMKSIPYSSAVGSIMYAMIGTRPDLAYPVGIISRYMSQPAREH 1151
Query: 1419 LTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVS 1478
VK +LRY+KG+ L YK++S++K+ GYCDAD+A + R+S +G LG + +S
Sbjct: 1152 WLGVKWVLRYIKGSLGTKLQYKRSSDFKVVGYCDADHAACKDRRRSITGLVFTLGGSTIS 1211
Query: 1479 WASKRQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQIFESNIPIYCDNTAAISLSKN 1538
W S +Q +ALST EAEY+S + +WMK L+++ + ++ I+CD+ +AI+LSKN
Sbjct: 1212 WKSGQQRVVALSTTEAEYMSLTEAVKEAVWMKGLLKEFGYEQKSVEIFCDSQSAIALSKN 1271
Query: 1539 PILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1597
+ H R KHI+V+Y +IRD + G + +DT+ ADIFTK + ++F L L +
Sbjct: 1272 NVHHERTKHIDVRYQYIRDIIANGDGDVVKIDTEKNPADIFTKIVPVNKFQAALTLLQV 1330
>At4g17450 retrotransposon like protein
Length = 1433
Score = 540 bits (1392), Expect = e-153
Identities = 329/1040 (31%), Positives = 525/1040 (49%), Gaps = 68/1040 (6%)
Query: 581 QSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICVDSSPCIDNVLL 640
+ W +DSG + H+T + ++ + V + KI G G I + + + NVL
Sbjct: 430 RGWVIDSGATHHVTHNRDLYLNFRSLENTFVRLPNDCTVKIAGIGFIQLSDAISLHNVLY 489
Query: 641 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 700
+ NL+S + + SQ+ + + N+ ++ +
Sbjct: 490 IPEFKFNLIS----------ELTKELMIGRGSQVGNLYVLDFNENNHTVSLKGTTSMCPE 539
Query: 701 VKCLLSVNEEQWVWHRRLGHASMRKISQLSK-LNL-VRGLPNLKFASDALCETCQKGKFT 758
SV + WH+RLGH + KI LS LNL V+ + +C C K
Sbjct: 540 FSVCSSVVVDSVTWHKRLGHPAYSKIDLLSDVLNLKVKKINKEHSPVCHVCHVCHLSKQK 599
Query: 759 KVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDES 818
+ F+++ + S +L+HID +GP + TW+ L K +
Sbjct: 600 HLSFQSRQNMC-SAAFDLVHIDTWGPFSVPTNDA--------------TWIYLLKNKSDV 644
Query: 819 HAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVV 878
VF FI V + ++ VRSD+ E KF LF ++GI SCP TP+QN VV
Sbjct: 645 LHVFPAFINMVHTQYQTKLKSVRSDNAHEL---KFTDLFAAHGIVAYHSCPETPEQNSVV 701
Query: 879 ERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYELWKNIKPNI 938
ERK++ + +AR +L ++ + FW + V TA ++ NR+ + NK+PYE KNI P
Sbjct: 702 ERKHQHILNVARALLFQSNIPLEFWGDCVLTAVFLINRLPTPVLNNKSPYEKLKNIPPAY 761
Query: 939 SYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFD 998
FGC+CY + + HKF+P++ C+ LGY KG++ + + + S HV F
Sbjct: 762 ESLKTFGCLCYSSTSPKQRHKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISRHVIFH 821
Query: 999 DKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEEDEPEEEAGPSNSQTLKKSRITAAHP 1058
+ + F +S + D K + P P ++ L+++ I HP
Sbjct: 822 EDI----------FPFISSTIKDDIK---DFFPLLQFPAR----TDDLPLEQTSIIDTHP 864
Query: 1059 KELILGNKD----EPVRTRSAFRP------------SEETLLSLKGLVSLIEPKSIDEAL 1102
+ + +K +P+ R P +E + + + + P+ EA
Sbjct: 865 HQDVSSSKALVPFDPLSKRQKKPPKHLQDFHCYNNTTEPFHAFINNITNAVIPQRYSEAK 924
Query: 1103 QDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGDVVRNKARLVAQ 1162
K W AMKEE+ + + WS+V P N IG KWVF K N G + R KARLVA+
Sbjct: 925 DFKAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCKWVFTIKHNADGSIERYKARLVAK 984
Query: 1163 GYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQP 1222
GY+Q+EG+DY ETF+PVA+L ++R+++ + +HQ+D+ +AFLNG + EE+Y+ P
Sbjct: 985 GYTQEEGLDYEETFSPVAKLTSVRMMLLLAAKMKWSVHQLDISNAFLNGDLDEEIYMKIP 1044
Query: 1223 PGFED---EKKPDH-VFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTY 1278
PG+ D E P H + +L KS+YGLKQA R WY +LS+ L F + D TLF K
Sbjct: 1045 PGYADLVGEALPPHAICRLHKSIYGLKQASRQWYLKLSNTLKGMGFQKSNADHTLFIKYA 1104
Query: 1279 KDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYI 1338
++ V +YVDDI+ S + +F+ +++ F++ +G KYFLGI++ ++ +G I
Sbjct: 1105 NGVLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKLRDLGAAKYFLGIEIARSEKGISI 1164
Query: 1339 HQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRP 1398
Q KY ELL L S + P+ P+ L KED YR ++G L+YL +RP
Sbjct: 1165 CQRKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGVPLTDSTSYRKLVGKLMYLQITRP 1224
Query: 1399 DILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGD 1458
DI ++V+ +F P HL+AV ++LRYLKGT GL Y ++ L GY D+D+
Sbjct: 1225 DIAYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQGLFYSADDKFDLRGYTDSDFGSC 1284
Query: 1459 RTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQI 1518
R+ + C F+G LVSW SK+Q T+++STAEAE+ + + + +M+W+ +D+++
Sbjct: 1285 TDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAEFRAMSQGTKEMIWLSRLFDDFKV 1344
Query: 1519 -FESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWAD 1577
F +YCDNTAA+ + N + H R K +E+ + R+ V+ G L FV+T Q AD
Sbjct: 1345 PFIPPAYLYCDNTAALHIVNNSVFHERTKFVELDCYKTREAVESGFLKTMFVETGEQVAD 1404
Query: 1578 IFTKPLAEDRFNFILKNLNM 1597
TK + +F+ ++ + +
Sbjct: 1405 PLTKAIHPAQFHKLIGKMGV 1424
>At1g26990 polyprotein, putative
Length = 1436
Score = 538 bits (1386), Expect = e-152
Identities = 335/1044 (32%), Positives = 534/1044 (51%), Gaps = 71/1044 (6%)
Query: 572 ISLIAPLKHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICVDS 631
I + L ++W +DSG S H+T E+ ++ K V KI GTG I +
Sbjct: 410 IPIETELSLRAWVIDSGASHHVTHERNLYHTYKALDRTFVRLPNGHTVKIEGTGFIQLTD 469
Query: 632 SPCIDNVLLVDGLTHNLLSISQLADKGYD-VIFNQKSCRAVSQIDGSVLFNSKRKNNIYK 690
+ + NVL + NLLS+S L V F C + +L + N+Y
Sbjct: 470 ALSLHNVLFIPEFKFNLLSVSVLTKTLQSKVSFTSDECMIQALTKELMLGKGSQVGNLYI 529
Query: 691 IRL-------SELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKF 743
+ L S ++V SV E +WH+RLGH S KI LS + + LP K
Sbjct: 530 LNLDKSLVDVSSFPGKSV--CSSVKNESEMWHKRLGHPSFAKIDTLSDVLM---LPKQKI 584
Query: 744 ASDAL-CETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDD 802
D+ C C K +PFK+ N + + EL+HID +GP ++ RY + IVDD
Sbjct: 585 NKDSSHCHVCHLSKQKHLPFKSVNHIR-EKAFELVHIDTWGPFSVPTVDSYRYFLTIVDD 643
Query: 803 YSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGI 862
+SR TW+ L +K + VF +F+ V+ + ++ VRSD+ E KF LF GI
Sbjct: 644 FSRATWIYLLKQKSDVLTVFPSFLKMVETQYHTKVCSVRSDNAHEL---KFNELFAKEGI 700
Query: 863 AHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPI 922
D CP TP+QN VVERK++ L +AR ++ ++G+ +W + V TA ++ NR+ I
Sbjct: 701 KADHPCPETPEQNFVVERKHQHLLNVARALMFQSGIPLEYWGDCVLTAVFLINRLLSPVI 760
Query: 923 LNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFY 982
N+TPYE KP+ S FGC+CY + KFDP++ C+ LGY KG++
Sbjct: 761 NNETPYERLTKGKPDYSSLKAFGCLCYCSTSPKSRTKFDPRAKACIFLGYPMGYKGYKLL 820
Query: 983 NTDAKTIEESIHVRF-DDKLDSDQSKLVEKFADLSINVSDKGKAPEE----VEPEEDEPE 1037
+ + ++ S HV F +D S + + D ++ +E V+ D P
Sbjct: 821 DIETYSVSISRHVIFYEDIFPFASSNITDAAKDFFPHIYLPAPNNDEHLPLVQSSSDAPH 880
Query: 1038 --EEAG-----PSNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRP----------SE 1080
+E+ PS ++ ++ ++ + H ++ N + P T+++ P SE
Sbjct: 881 NHDESSSMIFVPSEPKSTRQRKLPS-HLQDFHCYN-NTPTTTKTSPYPLTNYISYSYLSE 938
Query: 1081 ETLLSLKGLVSLIEPKSIDEALQDKDWILAMKEELNQFSKNDVWSLVKKPENVHVIGTKW 1140
+ + + P+ EA DK W AM +E++ F + WS+ P +G KW
Sbjct: 939 PFGAFINIITATKLPQKYSEARLDKVWNDAMGKEISAFVRTGTWSICDLPAGKVAVGCKW 998
Query: 1141 VFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILH 1200
+ K G + R+KARLVA+GY+QQEGID+ TF+PVA++ +++L+S + LH
Sbjct: 999 IITIKFLADGSIERHKARLVAKGYTQQEGIDFFNTFSPVAKMVTVKVLLSLAPKMKWYLH 1058
Query: 1201 QMDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLL 1260
Q+D+ +A LNG + EE+Y+ PPG+ + + G + +P A
Sbjct: 1059 QLDISNALLNGDLEEEIYMKLPPGYSE-------------IQGQEVSPNA---------- 1095
Query: 1261 ENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGE 1320
+ D TLF K L+V +YVDDI+ S ++ E + + + F++ +GE
Sbjct: 1096 -----KCHGDHTLFVKAQDGFFLVVLVYVDDILIASTTEAASAELTSQLSSFFQLRDLGE 1150
Query: 1321 LKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQ 1380
K+FLGI++ + +G + Q KY +LL + + + PM P L K+ +
Sbjct: 1151 PKFFLGIEIARNADGISLCQRKYVLDLLASSDFSDCKPSSIPMEPNQKLSKDTGTLLEDG 1210
Query: 1381 KLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYK 1440
K YR ++G L YL +RPDI F+V A++ S P + HL A+ +ILRYLKGT GL Y
Sbjct: 1211 KQYRRILGKLQYLCLTRPDINFAVSKLAQYSSAPTDIHLQALHKILRYLKGTIGQGLFYG 1270
Query: 1441 KTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAA 1500
+ + L G+ D+D+ R+ +G F+G++LVSW SK+Q +++S+AEAEY + +
Sbjct: 1271 ADTNFDLRGFSDSDWQTCPDTRRCVTGFAIFVGNSLVSWRSKKQDVVSMSSAEAEYRAMS 1330
Query: 1501 ICSTQMLWMKHQLEDYQI-FESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYV 1559
+ + +++W+ + L ++I F +YCDN AA+ ++ N + H R KHIE H +R+ +
Sbjct: 1331 VATKELIWLGYILTAFKIPFTHPAYLYCDNEAALHIANNSVFHERTKHIENDCHKVRECI 1390
Query: 1560 QKGVLLLKFVDTDHQWADIFTKPL 1583
+ G+L FV TD+Q AD TKPL
Sbjct: 1391 EAGILKTIFVRTDNQLADTLTKPL 1414
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 531 bits (1368), Expect = e-150
Identities = 341/1071 (31%), Positives = 564/1071 (51%), Gaps = 85/1071 (7%)
Query: 576 APLKHQSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTICVD----S 631
A + +W LD+GCS HMT K + K G+V G + ++ G G + + S
Sbjct: 305 AEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKNEDGS 364
Query: 632 SPCIDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI 691
+ + +V + ++ NL+S+ L DKG ++K + + D +VL K+++ +Y +
Sbjct: 365 TILLTDVRYIPEMSKNLISLGTLEDKGC-WFESKKGILTIFKNDLTVL-TGKKESTLYFL 422
Query: 692 RLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQL-SKLNLVRGLPNLKFASDALCE 750
+ + L A + +E +WH RLGH + + L SK +L + + + F +
Sbjct: 423 QGTTL-AGEANVIDKEKDETSLWHSRLGHIGAKGLQVLVSKGHLDKNIM-ISFGA----- 475
Query: 751 TCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTE-SIGGKRYGMVIVDDYSRWTWV 809
AK+V T L+ +H DL+G SIG +Y + +DD++R TW+
Sbjct: 476 -------------AKHV--TKDKLDYVHSDLWGSTNVPFSIGKCQYFITFIDDFTRRTWI 520
Query: 810 KFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCP 869
F+ KDE+ + F + Q++N++ ++ + +D+G EF N +F+S G+ +C
Sbjct: 521 YFIRTKDEAFSKFVEWKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKEGVIRHRTCA 580
Query: 870 RTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILNKTPYE 929
TPQQNGV ER NRT+ R ML E+G+ K FWAEA +TA ++ N+ I P E
Sbjct: 581 YTPQQNGVAERMNRTIMNKVRCMLSESGLGKQFWAEAASTAVFLINKSPSSSIEFDIPEE 640
Query: 930 LWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYNTDAKTI 989
W P+ FG V Y+ + + +L+ P++ K + LGY D K F+ + + +
Sbjct: 641 KWTGHPPDYKILKKFGSVAYIHSDQGKLN---PRAKKGIFLGYPDGVKRFKVWLLEDRKC 697
Query: 990 EESIHVRFDDKLDSDQSKLVEKFADLSIN-VSDKGKAPEEVE---------PEEDEPEEE 1039
S + F + + + +L N +S++ K EVE +DE + E
Sbjct: 698 VVSRDIVFQEN---------QMYKELQKNDMSEEDKQLTEVERTLIELKNLSADDENQSE 748
Query: 1040 AGPSNSQTLKKSRITAAHPKELILGNKDEP---------------VRTRSAFRPSEETLL 1084
G +++Q + +A+ K++ + D+ +R F +++L+
Sbjct: 749 GGDNSNQEQASTTRSASKDKQVEETDSDDDCLENYLLARDRIRRQIRAPQRFVEEDDSLV 808
Query: 1085 SLKGLVS----LIEPKSIDEALQDKD---WILAMKEELNQFSKNDVWSLVKKPENVHVIG 1137
++ + EP++ +EA++ + W A EE++ KND W ++ KPE VIG
Sbjct: 809 GFALTMTEDGEVYEPETYEEAMRSPECEKWKQATIEEMDSMKKNDTWDVIDKPEGKRVIG 868
Query: 1138 TKWVFRNKLNEKG-DVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHN 1196
KW+F+ K G + R KARLVA+G+SQ+EGIDY E F+PV + +IR L+S V +
Sbjct: 869 CKWIFKRKAGIPGVEPPRYKARLVAKGFSQREGIDYQEIFSPVVKHVSIRYLLSIVVQFD 928
Query: 1197 IILHQMDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLS 1256
+ L Q+DVK+AFL+G + E + + QP G+EDE + V LKKSLYGLKQ+PR W +R
Sbjct: 929 MELEQLDVKTAFLHGNLDEYILMSQPEGYEDEDSTEKVCLLKKSLYGLKQSPRQWNQRFD 988
Query: 1257 SFLLENEFVRGKVDTTLFCKTYKDDILI-VQIYVDDIIFGSANQSLCKEFSEMMQAEFEM 1315
SF++ + + R K + ++ + D I + +YVDD++ S N+ ++ E + EFEM
Sbjct: 989 SFMINSGYQRSKYNPCVYTQQLNDGSYIYLLLYVDDMLIASQNKDQIQKLKESLNREFEM 1048
Query: 1316 SMMGELKYFLGIQVDQTPEGTYIH--QSKYTKELLKKFNMLESTVAKTPMHPTCIL---- 1369
+G + LG+++ + E + QS+Y +L+ F M +S V++TP+ L
Sbjct: 1049 KDLGPARKILGMEITRNREQGILDLSQSEYVAGVLRAFGMDQSKVSQTPLGAHFKLRAAN 1108
Query: 1370 EKEDKSGKVCQKL--YRGMIGSLLY-LTASRPDILFSVHLCARFQSDPRETHLTAVKRIL 1426
EK KL Y IGS++Y + SRPD+ + V + +RF S P + H AVK ++
Sbjct: 1109 EKTLARDAEYMKLVPYPNAIGSIMYSMIGSRPDLAYPVGVVSRFMSKPSKEHWQAVKWVM 1168
Query: 1427 RYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQST 1486
RY+KGT + L +KK ++++ GYCD+DYA D R+S +G G N +SW S Q
Sbjct: 1169 RYMKGTQDTCLRFKKDDKFEIRGYCDSDYATDLDRRRSITGFVFTAGGNTISWKSGLQRV 1228
Query: 1487 IALSTAEAEYISAAICSTQMLWMKHQLEDYQIFESNIPIYCDNTAAISLSKNPILHSRAK 1546
+ALST EAEY++ A + +W++ + + + + CD+ +AI+LSKN + H R K
Sbjct: 1229 VALSTTEAEYMALAEAVKEAIWLRGLAAEMGFEQDAVEVMCDSQSAIALSKNSVHHERTK 1288
Query: 1547 HIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1597
HI+V+YHFIR+ + G + + + T ADIFTK + + LK L +
Sbjct: 1289 HIDVRYHFIREKIADGEIQVVKISTTWNPADIFTKTVPVSKLQEALKLLRV 1339
>At4g03810 putative retrotransposon protein
Length = 964
Score = 503 bits (1295), Expect = e-142
Identities = 324/984 (32%), Positives = 509/984 (50%), Gaps = 70/984 (7%)
Query: 635 IDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNN------- 687
+ N V + N++S+S L +G+ K C S + + S +N
Sbjct: 5 LKNCYYVPAINKNIISVSCLDMEGFHFSIKNKCC---SFDRDDMFYGSAPLDNGLHVLNQ 61
Query: 688 ---IYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFA 744
IY IR + ++ ++ ++WH RLGH + + I +L L L + +
Sbjct: 62 SMPIYNIRTKKFKSNDLN-------PTFLWHCRLGHINEKHIQKLHSDGL---LNSFDYE 111
Query: 745 SDALCETCQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYS 804
S CE+C GK TK PF + S L L+H D+ GP+ T + G +Y + DD+S
Sbjct: 112 SYETCESCLLGKMTKAPFTGHSE-RASDLLGLIHTDVCGPMSTSARGNYQYFITFTDDFS 170
Query: 805 RWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAH 864
R+ +V + K +S F F +VQN+ I +RSD GGE+ + F GI
Sbjct: 171 RYGYVYLMKHKSKSFENFKEFQNEVQNQFGKSIKALRSDRGGEYLSQVFSDHLRECGIVS 230
Query: 865 DFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVRPILN 924
+ P TPQ NGV ER+NRTL +M R+M+ T + FW A+ T+ ++ NR + +
Sbjct: 231 QLTPPGTPQWNGVSERRNRTLLDMVRSMMSHTDLPSPFWGYALETSAFMLNRCPSKSV-E 289
Query: 925 KTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFRFYN- 983
KTPYE+W PN+S+ +GC Y + K PKS KC +GY +KG+ FY+
Sbjct: 290 KTPYEIWTGKVPNLSFLKIWGCESYA--KRLITDKLGPKSDKCYFVGYPKETKGYYFYHP 347
Query: 984 TDAKTIEESIHVRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEE-DEPEEEAGP 1042
TD K +V A L KG + +V EE EP+ +
Sbjct: 348 TDNKVF------------------VVRNGAFLEREFLSKGTSGSKVLLEEVREPQGDVPT 389
Query: 1043 SNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRPS--EETLLSLKGL--VSLIEPKSI 1098
S ++ ++ E IL + RS P + ++ L + EP S
Sbjct: 390 SQ----EEHQLDLRRVVEPILVEPEVRRSERSRHEPDRFRDWVMDDHALFMIESDEPTSY 445
Query: 1099 DEALQDKD---WILAMKEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGDVVRN 1155
+EAL D W+ A K E+ S+N VW+LV P+ V I KW+F+ K++ G++
Sbjct: 446 EEALMGPDSDKWLEAAKSEMESMSQNKVWTLVDLPDGVKPIECKWIFKKKIDMDGNIQIY 505
Query: 1156 KARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSAFLNGYISE 1215
KA LVA+GY Q GIDY ET++PVA L++IR+L++ + +++ + QMDVK+AFLNG + E
Sbjct: 506 KAGLVAKGYKQVHGIDYDETYSPVAMLKSIRILLATAAHYDYEIWQMDVKTAFLNGNLEE 565
Query: 1216 EVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFC 1275
VY+ QP GF + V KL +S+YGLKQA R+W R + + E +F+R + + ++
Sbjct: 566 HVYMTQPEGFTVPEAARKVCKLHRSIYGLKQASRSWNLRFNEAIKEFDFIRNEEEPCVYK 625
Query: 1276 KTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQV--DQTP 1333
KT + + +YVDDI+ + L + + + F M MGE Y LGI++ D+
Sbjct: 626 KTSGSAVAFLVLYVDDILLLGNDIPLLQSVKTWLGSCFSMKDMGEAAYILGIRIYRDRLN 685
Query: 1334 EGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEK------EDKSGKVCQKLYRGMI 1387
+ + Q Y ++L +FNM +S PM L K D+ ++ + Y I
Sbjct: 686 KIIGLSQDTYIDKVLHRFNMHDSKKGFIPMSHGITLSKTQCPSTHDERERMSKIPYASAI 745
Query: 1388 GSLLY-LTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYK 1446
GS++Y + +RPD+ ++ + +R+QSDP E+H V+ I +YL+ T + L+Y + E
Sbjct: 746 GSIMYAMLYTRPDVACALSMTSRYQSDPGESHWIVVRNIFKYLRRTKDKFLVYGGSEELV 805
Query: 1447 LSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQM 1506
+SGY DA + D+ + +S SG L VSW S +QST+A ST EAEYI+A+ + ++
Sbjct: 806 VSGYTDASFQTDKDDFRSQSGFFFCLNGGAVSWKSTKQSTVADSTTEAEYIAASEAAKEV 865
Query: 1507 LWMKHQLEDYQIFES---NIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGV 1563
+W++ + + + S I +YCDN AI+ +K P H ++KHI+ +YH IR+ + +G
Sbjct: 866 VWIRKFITELGVVPSISGPIDLYCDNNGAIAQAKEPKSHQKSKHIQRRYHLIREIIDRGD 925
Query: 1564 LLLKFVDTDHQWADIFTKPLAEDR 1587
+ + V TD AD FTKPL + +
Sbjct: 926 VKISRVSTDANVADHFTKPLPQPK 949
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 478 bits (1230), Expect = e-134
Identities = 297/838 (35%), Positives = 448/838 (53%), Gaps = 63/838 (7%)
Query: 801 DDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSY 860
+D+SR WV FL KDE+ A F+ + V+ + ++ +R+D+G EF N KF+ +
Sbjct: 319 NDWSRKVWVYFLKTKDEAFASFTEWKKMVETQSERKLKHLRTDNGLEFCNHKFDEVCKKE 378
Query: 861 GIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIQNRISVR 920
GI +C TPQQNGV ER NRT+ R+ML E+G+ K FWA+A +TA Y+ NR
Sbjct: 379 GIVRHRTCTYTPQQNGVAERLNRTIMNKVRSMLSESGLDKKFWAKAASTAVYLINRSPSS 438
Query: 921 PILNKTPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDPKSSKCLLLGYSDRSKGFR 980
I NK P ELW + PN S FGCV YV + + +L DP++ K + +GY + KGFR
Sbjct: 439 SIENKIPEELWTSAVPNFSGLKRFGCVVYVYSQEGKL---DPRAKKGVFVGYPNGVKGFR 495
Query: 981 FYNTDAKTIEESIHVRF------DDKLDSDQSKLVEKFADLSINVSDKGKAPEEVEPEED 1034
+ + + S +V F D L+ S + F + + A +ED
Sbjct: 496 VWMIEEERCSISRNVVFREDVMYKDILNQSTSGMSFDFPLATNRIPSFECAGNR---KED 552
Query: 1035 EPEEEAGPSNSQTLKKSRITAAHP--------KELILGNKDEPVRTRSA------FRPSE 1080
E + G S+ T + S + + +D+P R + +E
Sbjct: 553 EISVQGGVSDDDTKQSSEESPISTGSSGQNSGQRTYQIARDKPKRQTKIPDKLRDYELNE 612
Query: 1081 ETLLSLKGLVSLI-------EPKSIDEALQDKD---WILAMKEELNQFSKNDVWSLVKKP 1130
E L + G +I EP +ALQD D W+ A+ EE+ KN+ W LV +
Sbjct: 613 EVLDEIAGYAYMITEDGGNPEPNDYQKALQDSDYKMWLKAVDEEIESLLKNNTWVLVNRD 672
Query: 1131 ENVHVIGTKWVFRNKLNEKG-DVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLI 1189
+ IG KWVF+ K G + R KARLV +GYSQ+EGIDY E F+PV + +IRLL+
Sbjct: 673 QFQKPIGCKWVFKRKSGIVGVEKPRFKARLVVKGYSQKEGIDYQEIFSPVVKHVSIRLLL 732
Query: 1190 SFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFEDEKKPDHVFKLKKSLYGLKQAPR 1249
S + ++ L QMDVK+AFL+GY+ E +Y+ QP G+ ++ PD V LK+SLYGL+Q+PR
Sbjct: 733 SMVTHCDMELQQMDVKTAFLHGYLDETIYIEQPEGYVHKRYPDKVCLLKRSLYGLRQSPR 792
Query: 1250 AWYERLSSFLLENEFVRGKVDTTLFCKTYKD-DILIVQIYVDDIIFGSANQSLCKEFSEM 1308
W R + F+ + + R K D+ ++ K + + + + +YVDDI+ S ++ + +
Sbjct: 793 QWNNRFNEFMQKIGYERSKYDSCVYFKELQSGEYIYLLLYVDDILIASRDKRTVCDLKAL 852
Query: 1309 MQAEFEMSMMGELKYFLGIQV--DQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPM--- 1363
+ +EFEM +G+ K LG+++ D+ I Q Y ++L F M ++ TPM
Sbjct: 853 LNSEFEMKDLGDAKKILGMEIVRDRKAGTMSISQEGYLLKVLGNFGMDQAKPVFTPMGAH 912
Query: 1364 ---HPTCILEKEDKSGKVCQKLYRGMIGSLLY-LTASRPDILFSVHLCARFQSDPRETHL 1419
P E +S + Y+ +GSL+Y + +RPD+ SV L RF S P + H
Sbjct: 913 FKLKPATDEEVMRQSEVMRAVPYQSAVGSLMYSMIGTRPDLAHSVGLVCRFMSKPLKEHW 972
Query: 1420 TAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSW 1479
AVK ILRY++G+ + L YK E L GYCD+DYA D+ R+STSG
Sbjct: 973 QAVKWILRYIRGSIDRKLCYKNEGELILEGYCDSDYAADKEGRRSTSG------------ 1020
Query: 1480 ASKRQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQIFESNIPIYCDNTAAISLSKNP 1539
+ALS+ EAEY++ + + +W+K + + + + I+CD+ +AI+L+KN
Sbjct: 1021 ----VKVVALSSTEAEYMALTDGAKEAIWLKGHVSELGFVQKTVNIHCDSQSAIALAKNA 1076
Query: 1540 ILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNLNM 1597
+ H R KHI+VKYHFIRD V G + + +DT+ ADIFTK L +F L+ L +
Sbjct: 1077 VYHERTKHIDVKYHFIRDLVNNGEVQVLKIDTEDNPADIFTKVLPVSKFQDALELLRV 1134
Score = 34.3 bits (77), Expect = 0.60
Identities = 14/47 (29%), Positives = 22/47 (46%)
Query: 581 QSWYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIIGTGTI 627
+ W +D+GCS HMT K + G+V N ++ G G +
Sbjct: 264 EEWIMDTGCSFHMTPRKEYLMDFVEAKSGKVRMANNSFSEVKGIGKV 310
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.332 0.142 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,041,099
Number of Sequences: 26719
Number of extensions: 1375378
Number of successful extensions: 5596
Number of sequences better than 10.0: 155
Number of HSP's better than 10.0 without gapping: 115
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 4833
Number of HSP's gapped (non-prelim): 307
length of query: 1602
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1489
effective length of database: 8,299,349
effective search space: 12357730661
effective search space used: 12357730661
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0166.7


