

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.14
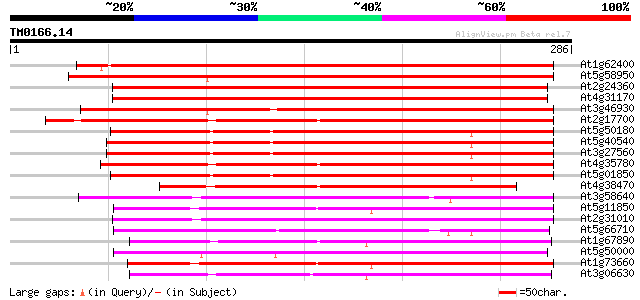
(286 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g62400 hypothetical protein 322 2e-88
At5g58950 protein kinase 6 - like 270 7e-73
At2g24360 putative protein kinase (T28I24.9/At2g24360) 258 3e-69
At4g31170 protein kinase - like protein 249 1e-66
At3g46930 protein kinase 6-like protein 215 3e-56
At2g17700 putative protein kinase 205 2e-53
At5g50180 protein kinase ATN1-like protein 203 1e-52
At5g40540 protein kinase - like protein 202 2e-52
At3g27560 putative protein kinase, ATN1 200 8e-52
At4g35780 unknown protein 199 1e-51
At5g01850 protein kinase ATN1-like protein 194 5e-50
At4g38470 protein kinase like protein 186 1e-47
At3g58640 unknown protein 171 3e-43
At5g11850 MAP3K like protein kinase 166 2e-41
At2g31010 putative protein kinase 165 2e-41
At5g66710 protein kinase ATN1-like protein 163 1e-40
At1g67890 putative protein kinase (At1g67890) 160 7e-40
At5g50000 protein kinase 160 1e-39
At1g73660 putative protein kinase 159 1e-39
At3g06630 protein kinase, putative 158 3e-39
>At1g62400 hypothetical protein
Length = 345
Score = 322 bits (824), Expect = 2e-88
Identities = 161/247 (65%), Positives = 193/247 (77%), Gaps = 5/247 (2%)
Query: 35 SWSKYLVSPGA----AIKGEGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKL 90
SWS L S A KGE EEW+AD+SQL IG KFASG HSRIY+G+YK+R VA+K+
Sbjct: 11 SWSMILESENVETWEASKGE-REEWTADLSQLFIGNKFASGAHSRIYRGIYKQRAVAVKM 69
Query: 91 VSQPEEDEDLACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRK 150
V P E+ LE+QF SEV+LL L HPNI+ F+AACKKPPV+CIITEY++ G+LR
Sbjct: 70 VRIPTHKEETRAKLEQQFKSEVALLSRLFHPNIVQFIAACKKPPVYCIITEYMSQGNLRM 129
Query: 151 YLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGIS 210
YL+ +EP+S+ + VL+LALDI+RGM+YLHSQG+IHRDLKS NLLL ++M VKV DFG S
Sbjct: 130 YLNKKEPYSLSIETVLRLALDISRGMEYLHSQGVIHRDLKSNNLLLNDEMRVKVADFGTS 189
Query: 211 CLESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQA 270
CLE+QC AKG GTYRWMAPEMIKEK +T+KVDVYSFGIVLWEL T L PF MTP QA
Sbjct: 190 CLETQCREAKGNMGTYRWMAPEMIKEKPYTRKVDVYSFGIVLWELTTALLPFQGMTPVQA 249
Query: 271 AYAVSYK 277
A+AV+ K
Sbjct: 250 AFAVAEK 256
>At5g58950 protein kinase 6 - like
Length = 525
Score = 270 bits (690), Expect = 7e-73
Identities = 129/250 (51%), Positives = 177/250 (70%), Gaps = 3/250 (1%)
Query: 31 KRAVSWSKYLVSPGAAIKG-EGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIK 89
K+ WSK + G + E EE+ DMS+L G+KFA G +SR+Y G Y+++ VA+K
Sbjct: 175 KKDTGWSKLFDNTGRRVSAVEASEEFRVDMSKLFFGLKFAHGLYSRLYHGKYEDKAVAVK 234
Query: 90 LVSQPEEDED--LACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGS 147
L++ P++D++ L LEKQFT EV+LL L HPN+I FV A K PPV+C++T+YL GS
Sbjct: 235 LITVPDDDDNGCLGARLEKQFTKEVTLLSRLTHPNVIKFVGAYKDPPVYCVLTQYLPEGS 294
Query: 148 LRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDF 207
LR +LH E S+PL +++ A+DIARGM+Y+HS+ IIHRDLK EN+L+ E+ +K+ DF
Sbjct: 295 LRSFLHKPENRSLPLKKLIEFAIDIARGMEYIHSRRIIHRDLKPENVLIDEEFHLKIADF 354
Query: 208 GISCLESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTP 267
GI+C E C GTYRWMAPEMIK K H +K DVYSFG+VLWE++ G P+++M P
Sbjct: 355 GIACEEEYCDMLADDPGTYRWMAPEMIKRKPHGRKADVYSFGLVLWEMVAGAIPYEDMNP 414
Query: 268 EQAAYAVSYK 277
QAA+AV +K
Sbjct: 415 IQAAFAVVHK 424
>At2g24360 putative protein kinase (T28I24.9/At2g24360)
Length = 411
Score = 258 bits (659), Expect = 3e-69
Identities = 119/222 (53%), Positives = 162/222 (72%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
+EW+ D+ +L +G FA G ++YKG Y DVAIK++ +PE + A F+E+QF EV
Sbjct: 121 DEWTIDLRKLNMGPAFAQGAFGKLYKGTYNGEDVAIKILERPENSPEKAQFMEQQFQQEV 180
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
S+L L+HPNI+ F+ AC+KP V+CI+TEY GGS+R++L ++ +VPL L +K ALD+
Sbjct: 181 SMLANLKHPNIVRFIGACRKPMVWCIVTEYAKGGSVRQFLTRRQNRAVPLKLAVKQALDV 240
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H + IHRDLKS+NLL+ D +K+ DFG++ +E Q TGTYRWMAPE
Sbjct: 241 ARGMAYVHGRNFIHRDLKSDNLLISADKSIKIADFGVARIEVQTEGMTPETGTYRWMAPE 300
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAV 274
MI+ + + +KVDVYSFGIVLWEL+TGL PF NMT QAA+AV
Sbjct: 301 MIQHRAYNQKVDVYSFGIVLWELITGLLPFQNMTAVQAAFAV 342
>At4g31170 protein kinase - like protein
Length = 412
Score = 249 bits (636), Expect = 1e-66
Identities = 118/222 (53%), Positives = 160/222 (71%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW+ D+ +L +G FA G ++Y+G Y DVAIKL+ + + + + A LE+QF EV
Sbjct: 122 EEWTIDLRKLHMGPAFAQGAFGKLYRGTYNGEDVAIKLLERSDSNPEKAQALEQQFQQEV 181
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
S+L L+HPNI+ F+ AC KP V+CI+TEY GGS+R++L ++ +VPL L + ALD+
Sbjct: 182 SMLAFLKHPNIVRFIGACIKPMVWCIVTEYAKGGSVRQFLTKRQNRAVPLKLAVMQALDV 241
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H + IHRDLKS+NLL+ D +K+ DFG++ +E Q TGTYRWMAPE
Sbjct: 242 ARGMAYVHERNFIHRDLKSDNLLISADRSIKIADFGVARIEVQTEGMTPETGTYRWMAPE 301
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAV 274
MI+ + +T+KVDVYSFGIVLWEL+TGL PF NMT QAA+AV
Sbjct: 302 MIQHRPYTQKVDVYSFGIVLWELITGLLPFQNMTAVQAAFAV 343
>At3g46930 protein kinase 6-like protein
Length = 475
Score = 215 bits (547), Expect = 3e-56
Identities = 113/245 (46%), Positives = 158/245 (64%), Gaps = 7/245 (2%)
Query: 37 SKYLVSPGAAIKGEGE-EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPE 95
SK + G+ + G EE D+S+L G +FA G++S+IY G Y+ + VA+K+++ PE
Sbjct: 135 SKSVDYRGSKVSSAGVLEECLIDVSKLSYGDRFAHGKYSQIYHGEYEGKAVALKIITAPE 194
Query: 96 EDED--LACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLH 153
+ +D L LEK+F E +LL L HPN++ FV CIITEY+ GSLR YLH
Sbjct: 195 DSDDIFLGARLEKEFIVEATLLSRLSHPNVVKFVGVNTGN---CIITEYVPRGSLRSYLH 251
Query: 154 HQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLE 213
E S+PL ++ LDIA+GM+Y+HS+ I+H+DLK EN+L+ D +K+ DFGI+C E
Sbjct: 252 KLEQKSLPLEQLIDFGLDIAKGMEYIHSREIVHQDLKPENVLIDNDFHLKIADFGIACEE 311
Query: 214 SQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMT-PEQAAY 272
C GTYRWMAPE++K H +K DVYSFG++LWE++ G P++ M EQ AY
Sbjct: 312 EYCDVLGDNIGTYRWMAPEVLKRIPHGRKCDVYSFGLLLWEMVAGALPYEEMKFAEQIAY 371
Query: 273 AVSYK 277
AV YK
Sbjct: 372 AVIYK 376
>At2g17700 putative protein kinase
Length = 546
Score = 205 bits (522), Expect = 2e-53
Identities = 106/259 (40%), Positives = 161/259 (61%), Gaps = 8/259 (3%)
Query: 19 GKAGRRLSLGEYKRAVSWSKYLVSPGAAIKGEGEEEWSADMSQLLIGMKFASGRHSRIYK 78
G + +S E+ ++ S L+ I +G +EW D++QL I K ASG + +++
Sbjct: 246 GSKQKSISFFEHDKS---SNELIPACIEIPTDGTDEWEIDVTQLKIEKKVASGSYGDLHR 302
Query: 79 GVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCI 138
G Y ++VAIK + + ++ ++F+ EV ++ +RH N++ F+ AC + P CI
Sbjct: 303 GTYCSQEVAIKFLKPDRVNNEML----REFSQEVFIMRKVRHKNVVQFLGACTRSPTLCI 358
Query: 139 ITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGE 198
+TE++A GS+ +LH Q+ + L +LK+ALD+A+GM YLH IIHRDLK+ NLL+ E
Sbjct: 359 VTEFMARGSIYDFLHKQKC-AFKLQTLLKVALDVAKGMSYLHQNNIIHRDLKTANLLMDE 417
Query: 199 DMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTG 258
VKV DFG++ ++ + G TGTYRWMAPE+I+ K + K DV+S+ IVLWELLTG
Sbjct: 418 HGLVKVADFGVARVQIESGVMTAETGTYRWMAPEVIEHKPYNHKADVFSYAIVLWELLTG 477
Query: 259 LTPFDNMTPEQAAYAVSYK 277
P+ +TP QAA V K
Sbjct: 478 DIPYAFLTPLQAAVGVVQK 496
>At5g50180 protein kinase ATN1-like protein
Length = 346
Score = 203 bits (516), Expect = 1e-52
Identities = 110/235 (46%), Positives = 148/235 (62%), Gaps = 11/235 (4%)
Query: 52 EEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSE 111
E +W D L +G K G H+++Y+G YK + VAIK+V + E E++A + +F E
Sbjct: 10 EPKWQIDPQLLFVGPKIGEGAHAKVYEGKYKNQTVAIKIVHRGETPEEIAK-RDSRFLRE 68
Query: 112 VSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALD 171
V +L ++H N++ F+ ACK+P V I+TE L GG+LRKYL + P + + + ALD
Sbjct: 69 VEMLSRVQHKNLVKFIGACKEP-VMVIVTELLQGGTLRKYLLNLRPACLETRVAIGFALD 127
Query: 172 IARGMQYLHSQGIIHRDLKSENLLLGED-MCVKVVDFGISCLESQCGSAKGFTGTYRWMA 230
IARGM+ LHS GIIHRDLK ENLLL D VK+ DFG++ ES TGTYRWMA
Sbjct: 128 IARGMECLHSHGIIHRDLKPENLLLTADHKTVKLADFGLAREESLTEMMTAETGTYRWMA 187
Query: 231 PEMI--------KEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
PE+ ++KH+ KVD YSF IVLWELL PF+ M+ QAAYA ++K
Sbjct: 188 PELYSTVTLRLGEKKHYNHKVDAYSFAIVLWELLHNKLPFEGMSNLQAAYAAAFK 242
>At5g40540 protein kinase - like protein
Length = 353
Score = 202 bits (514), Expect = 2e-52
Identities = 108/237 (45%), Positives = 150/237 (62%), Gaps = 11/237 (4%)
Query: 50 EGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFT 109
E + +W D L +G K G H++IY+G YK + VAIK+V + E E++A E +F
Sbjct: 14 ELDPKWVVDPQHLFVGPKIGEGAHAKIYEGKYKNKTVAIKIVKRGESPEEIAK-RESRFA 72
Query: 110 SEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLA 169
EVS+L ++H N++ F+ ACK+P + I+TE L GG+LRKYL P S+ + + + A
Sbjct: 73 REVSMLSRVQHKNLVKFIGACKEP-IMVIVTELLLGGTLRKYLVSLRPGSLDIRVAVGYA 131
Query: 170 LDIARGMQYLHSQGIIHRDLKSENLLLGED-MCVKVVDFGISCLESQCGSAKGFTGTYRW 228
LDIAR M+ LHS G+IHRDLK E+L+L D VK+ DFG++ ES TGTYRW
Sbjct: 132 LDIARAMECLHSHGVIHRDLKPESLILTADYKTVKLADFGLAREESLTEMMTAETGTYRW 191
Query: 229 MAPEMI--------KEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
MAPE+ ++KH+ KVD YSF IVLWEL+ PF+ M+ QAAYA ++K
Sbjct: 192 MAPELYSTVTLRHGEKKHYNHKVDAYSFAIVLWELIHNKLPFEGMSNLQAAYAAAFK 248
>At3g27560 putative protein kinase, ATN1
Length = 356
Score = 200 bits (508), Expect = 8e-52
Identities = 104/237 (43%), Positives = 149/237 (61%), Gaps = 11/237 (4%)
Query: 50 EGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFT 109
E + +W D L +G K G H+++Y+G Y+ + VAIK++ + E E++A + +F
Sbjct: 14 ELDPKWLVDPRHLFVGPKIGEGAHAKVYEGKYRNQTVAIKIIKRGESPEEIAK-RDNRFA 72
Query: 110 SEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLA 169
E+++L ++H N++ F+ ACK+P + I+TE L GG+LRKYL P + + L + A
Sbjct: 73 REIAMLSKVQHKNLVKFIGACKEP-MMVIVTELLLGGTLRKYLVSLRPKRLDIRLAVGFA 131
Query: 170 LDIARGMQYLHSQGIIHRDLKSENLLLGED-MCVKVVDFGISCLESQCGSAKGFTGTYRW 228
LDIAR M+ LHS GIIHRDLK ENL+L D VK+ DFG++ ES TGTYRW
Sbjct: 132 LDIARAMECLHSHGIIHRDLKPENLILSADHKTVKLADFGLAREESLTEMMTAETGTYRW 191
Query: 229 MAPEMI--------KEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
MAPE+ ++KH+ KVD YSF IVLWEL+ PF+ M+ QAAYA ++K
Sbjct: 192 MAPELYSTVTLRQGEKKHYNHKVDAYSFAIVLWELILNKLPFEGMSNLQAAYAAAFK 248
>At4g35780 unknown protein
Length = 570
Score = 199 bits (506), Expect = 1e-51
Identities = 96/231 (41%), Positives = 147/231 (63%), Gaps = 5/231 (2%)
Query: 47 IKGEGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEK 106
I +G +EW DM QL I K A G + +++G Y ++VAIK++ + ++ +
Sbjct: 277 IPTDGTDEWEIDMKQLKIEKKVACGSYGELFRGTYCSQEVAIKILKPERVNAEML----R 332
Query: 107 QFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVL 166
+F+ EV ++ +RH N++ F+ AC + P CI+TE++ GS+ +LH + + +L
Sbjct: 333 EFSQEVYIMRKVRHKNVVQFIGACTRSPNLCIVTEFMTRGSIYDFLHKHKG-VFKIQSLL 391
Query: 167 KLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTY 226
K+ALD+++GM YLH IIHRDLK+ NLL+ E VKV DFG++ ++++ G TGTY
Sbjct: 392 KVALDVSKGMNYLHQNNIIHRDLKTANLLMDEHEVVKVADFGVARVQTESGVMTAETGTY 451
Query: 227 RWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
RWMAPE+I+ K + + DV+S+ IVLWELLTG P+ +TP QAA V K
Sbjct: 452 RWMAPEVIEHKPYDHRADVFSYAIVLWELLTGELPYSYLTPLQAAVGVVQK 502
>At5g01850 protein kinase ATN1-like protein
Length = 333
Score = 194 bits (493), Expect = 5e-50
Identities = 109/235 (46%), Positives = 146/235 (61%), Gaps = 11/235 (4%)
Query: 52 EEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSE 111
EE D L IG K G H ++Y+G Y + VAIK+V++ + + + LE +F E
Sbjct: 8 EESLLVDPKLLFIGSKIGEGAHGKVYQGRYGRQIVAIKVVNRGSKPDQQSS-LESRFVRE 66
Query: 112 VSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALD 171
V+++ ++H N++ F+ ACK P + I+TE L G SLRKYL P + L L L ALD
Sbjct: 67 VNMMSRVQHHNLVKFIGACKDP-LMVIVTELLPGMSLRKYLTSIRPQLLHLPLALSFALD 125
Query: 172 IARGMQYLHSQGIIHRDLKSENLLLGED-MCVKVVDFGISCLESQCGSAKGFTGTYRWMA 230
IAR + LH+ GIIHRDLK +NLLL E+ VK+ DFG++ ES TGTYRWMA
Sbjct: 126 IARALHCLHANGIIHRDLKPDNLLLTENHKSVKLADFGLAREESVTEMMTAETGTYRWMA 185
Query: 231 PEMI--------KEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
PE+ ++KH+ KVDVYSFGIVLWELLT PF+ M+ QAAYA ++K
Sbjct: 186 PELYSTVTLRQGEKKHYNNKVDVYSFGIVLWELLTNRMPFEGMSNLQAAYAAAFK 240
>At4g38470 protein kinase like protein
Length = 545
Score = 186 bits (472), Expect = 1e-47
Identities = 91/182 (50%), Positives = 124/182 (68%), Gaps = 5/182 (2%)
Query: 77 YKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVF 136
YKG Y ++VAIK++ D DL EK+F EV ++ +RH N++ F+ AC KPP
Sbjct: 293 YKGTYCSQEVAIKVLKPERLDSDL----EKEFAQEVFIMRKVRHKNVVQFIGACTKPPHL 348
Query: 137 CIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLL 196
CI+TE++ GGS+ YLH Q+ L + K+A+DI +GM YLH IIHRDLK+ NLL+
Sbjct: 349 CIVTEFMPGGSVYDYLHKQKG-VFKLPTLFKVAIDICKGMSYLHQNNIIHRDLKAANLLM 407
Query: 197 GEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELL 256
E+ VKV DFG++ +++Q G TGTYRWMAPE+I+ K + K DV+S+GIVLWELL
Sbjct: 408 DENEVVKVADFGVARVKAQTGVMTAETGTYRWMAPEVIEHKPYDHKADVFSYGIVLWELL 467
Query: 257 TG 258
TG
Sbjct: 468 TG 469
>At3g58640 unknown protein
Length = 809
Score = 171 bits (434), Expect = 3e-43
Identities = 94/247 (38%), Positives = 143/247 (57%), Gaps = 11/247 (4%)
Query: 36 WSKYLVSPGAAIKGE-GEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQP 94
W K L SP K EEW+ D S+L +G + G +++G++ DVAIK+ +
Sbjct: 526 WDKVLGSPMFQNKPLLPYEEWNIDFSELTVGTRVGIGFFGEVFRGIWNGTDVAIKVFLE- 584
Query: 95 EEDEDLACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHH 154
+DL + F +E+S+L LRHPN+I F+ AC KPP +ITEY+ GSL LH
Sbjct: 585 ---QDLTAENMEDFCNEISILSRLRHPNVILFLGACTKPPRLSLITEYMEMGSLYYLLHL 641
Query: 155 Q-EPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLE 213
+ + LK+ DI RG+ +H GI+HRD+KS N LL VK+ DFG+S +
Sbjct: 642 SGQKKRLSWRRKLKMLRDICRGLMCIHRMGIVHRDIKSANCLLSNKWTVKICDFGLSRIM 701
Query: 214 SQCGSAKGFT---GTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQA 270
+ G+ T GT WMAPE+I+ + ++K D++S G+++WEL T P++ + PE+
Sbjct: 702 T--GTTMRDTVSAGTPEWMAPELIRNEPFSEKCDIFSLGVIMWELCTLTRPWEGVPPERV 759
Query: 271 AYAVSYK 277
YA++Y+
Sbjct: 760 VYAIAYE 766
>At5g11850 MAP3K like protein kinase
Length = 880
Score = 166 bits (419), Expect = 2e-41
Identities = 87/227 (38%), Positives = 134/227 (58%), Gaps = 8/227 (3%)
Query: 54 EWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVS 113
+W L IG + G + +Y+ + +VA+K D+D + QF SE+
Sbjct: 601 KWEIMWEDLQIGERIGIGSYGEVYRAEWNGTEVAVKKFL----DQDFSGDALTQFKSEIE 656
Query: 114 LLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIA 173
++L LRHPN++ F+ A +PP F I+TE+L GSL + LH H + +++ALD+A
Sbjct: 657 IMLRLRHPNVVLFMGAVTRPPNFSILTEFLPRGSLYRLLHRPN-HQLDEKRRMRMALDVA 715
Query: 174 RGMQYLHSQG--IIHRDLKSENLLLGEDMCVKVVDFGISCLESQCG-SAKGFTGTYRWMA 230
+GM YLH+ ++HRDLKS NLL+ ++ VKV DFG+S ++ S+K GT WMA
Sbjct: 716 KGMNYLHTSHPTVVHRDLKSPNLLVDKNWVVKVCDFGLSRMKHHTYLSSKSTAGTPEWMA 775
Query: 231 PEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
PE+++ + +K DVYSFG++LWEL T P+ + P Q AV ++
Sbjct: 776 PEVLRNEPANEKCDVYSFGVILWELATSRVPWKGLNPMQVVGAVGFQ 822
>At2g31010 putative protein kinase
Length = 375
Score = 165 bits (418), Expect = 2e-41
Identities = 83/227 (36%), Positives = 134/227 (58%), Gaps = 6/227 (2%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
+EW D S+L +G + G +++GV+ DVAIKL + +DL + F +E+
Sbjct: 47 QEWDIDFSELTVGTRVGIGFFGEVFRGVWNGTDVAIKLFLE----QDLTAENMEDFCNEI 102
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQ-EPHSVPLHLVLKLALD 171
S+L +RHPN++ F+ AC KPP +ITEY+ GSL +H + + H L++ D
Sbjct: 103 SILSRVRHPNVVLFLGACTKPPRLSMITEYMELGSLYYLIHMSGQKKKLSWHRRLRMLRD 162
Query: 172 IARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGIS-CLESQCGSAKGFTGTYRWMA 230
I RG+ +H I+HRDLKS N L+ + VK+ DFG+S + + GT WMA
Sbjct: 163 ICRGLMCIHRMKIVHRDLKSANCLVDKHWTVKICDFGLSRIMTDENMKDTSSAGTPEWMA 222
Query: 231 PEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
PE+I+ + T+K D++S G+++WEL T P++ + PE+ +AV+++
Sbjct: 223 PELIRNRPFTEKCDIFSLGVIMWELSTLRKPWEGVPPEKVVFAVAHE 269
>At5g66710 protein kinase ATN1-like protein
Length = 405
Score = 163 bits (412), Expect = 1e-40
Identities = 97/235 (41%), Positives = 133/235 (56%), Gaps = 19/235 (8%)
Query: 54 EWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVS 113
E D+ + IG G S +Y+G+++ + QP+ L+ K+F EV
Sbjct: 63 ELLVDVKDISIGDFIGEGSSSTVYRGLFRRVVPVSVKIFQPKRTSALSIEQRKKFQREVL 122
Query: 114 LLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIA 173
LL RH NI+ F+ AC +P + IITE + G +L+K++ P + L L + ALDIA
Sbjct: 123 LLSKFRHENIVRFIGACIEPKLM-IITELMEGNTLQKFMLSVRPKPLDLKLSISFALDIA 181
Query: 174 RGMQYLHSQGIIHRDLKSENLLL-GEDMCVKVVDFGISCLESQCGSAKGF----TGTYRW 228
RGM++L++ GIIHRDLK N+LL G+ VK+ DFG++ E+ KGF GTYRW
Sbjct: 182 RGMEFLNANGIIHRDLKPSNMLLTGDQKHVKLADFGLAREET-----KGFMTFEAGTYRW 236
Query: 229 MAPEMI--------KEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVS 275
MAPE+ ++KH+ KVDVYSF IV WELLT TPF AYA S
Sbjct: 237 MAPELFSYDTLEIGEKKHYDHKVDVYSFAIVFWELLTNKTPFKGKNNIFVAYAAS 291
>At1g67890 putative protein kinase (At1g67890)
Length = 765
Score = 160 bits (405), Expect = 7e-40
Identities = 90/217 (41%), Positives = 128/217 (58%), Gaps = 7/217 (3%)
Query: 62 LLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVSLLLPLRHP 121
L IG + G +Y G++ DVA+K+ S+ E E++ F EVSL+ LRHP
Sbjct: 487 LTIGEQIGQGSCGTVYHGLWFGSDVAVKVFSKQEYSEEIIT----SFKQEVSLMKRLRHP 542
Query: 122 NIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLH- 180
N++ F+ A P CI+TE+L GSL + L + + L + +A DIARGM YLH
Sbjct: 543 NVLLFMGAVASPQRLCIVTEFLPRGSLFRLLQRNKS-KLDLRRRIHMASDIARGMNYLHH 601
Query: 181 -SQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPEMIKEKHH 239
S IIHRDLKS NLL+ + VKV DFG+S ++ + GT +WMAPE+++ +
Sbjct: 602 CSPPIIHRDLKSSNLLVDRNWTVKVADFGLSRIKHETYLTTNGRGTPQWMAPEVLRNEAA 661
Query: 240 TKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSY 276
+K DVYSFG+VLWEL+T P++N+ Q AV +
Sbjct: 662 DEKSDVYSFGVVLWELVTEKIPWENLNAMQVIGAVGF 698
>At5g50000 protein kinase
Length = 385
Score = 160 bits (404), Expect = 1e-39
Identities = 89/241 (36%), Positives = 132/241 (53%), Gaps = 20/241 (8%)
Query: 54 EWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEE---DEDLACFLEKQFTS 110
EW D S+L+I A G +++G+Y +DVA+KL+ EE E L F
Sbjct: 74 EWEIDPSKLIIKTVLARGTFGTVHRGIYDGQDVAVKLLDWGEEGHRSEAEIVSLRADFAQ 133
Query: 111 EVSLLLPLRHPNIITFVAACKKPP----------------VFCIITEYLAGGSLRKYLHH 154
EV++ L HPN+ F+ A + C++ EYL GG+L+ YL
Sbjct: 134 EVAVWHKLDHPNVTKFIGATMGASGLQLQTESGPLAMPNNICCVVVEYLPGGALKSYLIK 193
Query: 155 QEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLE- 213
+ +V++LALD+ARG+ YLHSQ I+HRD+K+EN+LL + VK+ DFG++ +E
Sbjct: 194 NRRRKLTFKIVVQLALDLARGLSYLHSQKIVHRDVKTENMLLDKTRTVKIADFGVARVEA 253
Query: 214 SQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYA 273
S G TGT +MAPE++ + +K DVYSFGI LWE+ P+ ++T + A
Sbjct: 254 SNPNDMTGETGTLGYMAPEVLNGNPYNRKCDVYSFGICLWEIYCCDMPYPDLTFSEVTSA 313
Query: 274 V 274
V
Sbjct: 314 V 314
>At1g73660 putative protein kinase
Length = 1030
Score = 159 bits (403), Expect = 1e-39
Identities = 88/220 (40%), Positives = 133/220 (60%), Gaps = 8/220 (3%)
Query: 61 QLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVSLLLPLRH 120
++ +G + G + +Y+G + +VA+K D+DL ++F SEV ++ LRH
Sbjct: 747 EITVGERIGLGSYGEVYRGDWHGTEVAVKKFL----DQDLTGEALEEFRSEVRIMKKLRH 802
Query: 121 PNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLH 180
PNI+ F+ A +PP I+TE+L GSL + +H + + L++ALD ARGM YLH
Sbjct: 803 PNIVLFMGAVTRPPNLSIVTEFLPRGSLYRLIHRPN-NQLDERRRLRMALDAARGMNYLH 861
Query: 181 SQG--IIHRDLKSENLLLGEDMCVKVVDFGISCLE-SQCGSAKGFTGTYRWMAPEMIKEK 237
S I+HRDLKS NLL+ ++ VKV DFG+S ++ S S+K GT WMAPE+++ +
Sbjct: 862 SCNPMIVHRDLKSPNLLVDKNWVVKVCDFGLSRMKHSTYLSSKSTAGTAEWMAPEVLRNE 921
Query: 238 HHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
+K DVYS+G++LWEL T P+ M P Q AV ++
Sbjct: 922 PADEKCDVYSYGVILWELFTLQQPWGKMNPMQVVGAVGFQ 961
>At3g06630 protein kinase, putative
Length = 671
Score = 158 bits (400), Expect = 3e-39
Identities = 90/218 (41%), Positives = 130/218 (59%), Gaps = 8/218 (3%)
Query: 62 LLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVSLLLPLRHP 121
L IG + G +Y G++ DVA+K+ S+ E E + K F EVSL+ LRHP
Sbjct: 434 LTIGEQIGRGSCGTVYHGIWFGSDVAVKVFSKQEYSESVI----KSFEKEVSLMKRLRHP 489
Query: 122 NIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLH- 180
N++ F+ A P CI++E++ GSL + L + + + +ALDIARGM YLH
Sbjct: 490 NVLLFMGAVTSPQRLCIVSEFVPRGSLFRLLQ-RSMSKLDWRRRINMALDIARGMNYLHC 548
Query: 181 -SQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCG-SAKGFTGTYRWMAPEMIKEKH 238
S IIHRDLKS NLL+ + VKV DFG+S ++ Q ++K GT +WMAPE+++ +
Sbjct: 549 CSPPIIHRDLKSSNLLVDRNWTVKVADFGLSRIKHQTYLTSKSGKGTPQWMAPEVLRNES 608
Query: 239 HTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSY 276
+K D+YSFG+VLWEL T P++N+ Q AV +
Sbjct: 609 ADEKSDIYSFGVVLWELATEKIPWENLNSMQVIGAVGF 646
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,199,057
Number of Sequences: 26719
Number of extensions: 321336
Number of successful extensions: 4183
Number of sequences better than 10.0: 1019
Number of HSP's better than 10.0 without gapping: 972
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 1152
Number of HSP's gapped (non-prelim): 1119
length of query: 286
length of database: 11,318,596
effective HSP length: 98
effective length of query: 188
effective length of database: 8,700,134
effective search space: 1635625192
effective search space used: 1635625192
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0166.14


